Abstract
Natriuretic peptides (NPs) are major cardiovascular and osmoregulatory hormones in vertebrates. Although tetrapods generally have three subtypes, atrial NP (ANP), B-type NP (BNP), and C-type NP (CNP), some teleosts lack BNP, and sharks and hagfish have only one NP. Thus, NPs have diverged during fish evolution, possibly reflecting changes in osmoregulatory systems. In this study, we found, by cDNA cloning, four distinct CNPs (1 through 4) in the medaka (Oryzias latipes) and puffer fish (Takifugu rubripes), although to our knowledge no more than two CNPs have been isolated from a vertebrate species. Predicted mature CNP-1 was most similar, and CNP-4 was most dissimilar, to mammalian CNPs. However, synthetic CNP-4 most potently activated OlGC1, a medaka CNP-specific receptor (NPR-B) expressed in cultured cells, whereas CNP-1 and CNP-3 most activated OlGC7 and OlGC2, two medaka homologues of the ANP/BNP receptor (NPR-A), respectively. Linkage mapping in medaka followed by comparative genomic analyses among fishes and humans located four CNP genes in separate medaka chromosomes corresponding to human chromosomes 1, 2, 12, and 17. From conserved synteny, the following evolutionary history of NPs was evoked: (i) four CNP genes were generated by chromosomal duplications before the divergence of elasmobranchs; (ii) the CNP-3 gene generated ANP and BNP genes through tandem duplication before the divergence of tetrapods and teleosts; (iii) CNP-1 and -2 genes were retained in the teleost lineage but not in the tetrapod lineage; (iv) the CNP-3 gene disappeared from the tetrapod lineage after divergence of amphibians; and (v) the CNP-4 gene is retained in humans as CNP.
The natriuretic peptide (NP) family is a group of hormones that regulate circulatory and fluid homeostasis in vertebrates (1, 2). Tetrapods, including mammals, typically have three subtypes: atrial NP (ANP), B-type NP (BNP), and C-type NP (CNP). In teleosts, however, BNP has not been found, though another subtype, ventricular NP, has been isolated from the eel and rainbow trout (3, 4). Sharks appear to have only CNP (5), and the more primitive hagfish has only one NP, named EbuNP (6). Clearly the NP family has differentiated during fish evolution, probably reflecting the changes in osmoregulatory systems promoting adaptation to various osmotic environments. All NPs share a conserved ring structure of 17 aa, which is generally accompanied by short “head” and “tail” segments, i.e., extensions from amino and carboxyl termini of the ring, respectively (1, 2). Among them, CNP is characterized by the absence of the tail segment (1). Studies in mammals and eels have suggested that CNP is functionally distinguished from other NPs, in that CNP is expressed abundantly in the brain and acts as an autocrine/paracrine factor, whereas other NPs are expressed mainly in the heart and circulate in the blood (1). In addition, CNP has a high affinity to NP receptor (NPR)-B, whereas other NPs prefer NPR-A (7, 8).
We isolated NPs from two teleosts, the medaka Oryzias latipes and the puffer fish Takifugu rubripes, because characterization of NPs in genetically well characterized species is important to understanding molecular and functional evolution of the NP family. Medaka is a widely used teleost model species (9, 10) that is also suitable for studies of osmoregulatory systems (11), and the puffer fish offers an advantage by virtue of its small genome size (12). In addition, it has been suggested that the moderate phylogenetic distance between these two species is suitable for comparative genomics (9, 10). In this paper, we show the existence in these two teleosts of four cDNAs, each of which encodes a distinct CNP isoform, whereas no more than one type of CNP has been, to our knowledge, isolated from any vertebrate species, except for the bullfrog Rana catesbeiana, which has an additional CNP (CNP-II) (13). Functional differentiation of four predicted CNPs was suggested on the basis of synthetic medaka peptides and three medaka NPRs expressed in COS cells. Subsequently, we demonstrated the relationship among the four CNPs and previously known NPs by phylogenetic analyses. Localization of four genes on medaka chromosomes and their corresponding positions in the human genome were found by linkage mapping in medaka, followed by comparative genomic analyses among the medaka, puffer fish, and human. Finally, based on the gene synteny conserved among fishes and humans in combination with the comparison of exon/intron arrangements, we propose an evolutionary history of the NP family from ancestral to extant vertebrates, through chromosomal and tandem gene duplications, followed by losses of specific genes in each class of vertebrates.
Materials and Methods
cDNA Cloning. Orange-red medaka were obtained from Sato Fish Farm (Yatomi, Japan). The puffer fish was obtained from Oita Biological Technology Center (Nippon Suisan Kaisha, Tsurumi, Japan). The heart and brain were isolated from 200 adult medaka of both sexes and from 1 immature puffer fish (sex unknown) after anesthesia with 2-phenoxyethanol. cDNA clones encoding four CNPs were isolated by RACE using the specific primers shown in Table 1, according to Inoue et al. (14).
Table 1. Primers used for cloning and RT-PCR analyses.
| Name | Sequence | Purposes |
|---|---|---|
| CNP-A9 | TCCTCGGTTCCATCCCCTCCGT | 5′-RACE of medaka CNP-1 |
| RT-PCR of medaka CNP-1 | ||
| CNP-S17 | GAGCAGAACCGCCTCGACCAA | 3′-RACE of medaka CNP-1 |
| RT-PCR of medaka CNP-1 | ||
| CNP-A11 | TTGGTAGTTTAATGCTGGCAGGT | Amplification of medaka CNP-2 |
| RT-PCR of medaka CNP-2 | ||
| CNP-S14 | GACGGCTTGGTGACCTGAGAC | Amplification of medaka CNP-2 |
| RT-PCR of medaka CNP-1 | ||
| CNP-A16 | GAATCAAAGTTTTACTGCAACATG | 5′-RACE of medaka CNP-3 |
| CNP-S21 | GACAACAGACCGGAACCAGAA | 3′-RACE of medaka CNP-3 |
| RT-PCR of medaka CNP-3 | ||
| CNP-A20 | ACTCACACATGCACTCACACGT | 5′-RACE of medaka CNP-4 |
| CNP-S22 | GCTTTGGAGTTAAGTGCGCCTT | 3′-RACE of medaka CNP-4 |
| RT-PCR of medaka CNP-4 | ||
| CNP-A17 | CAGCCCAGTCCGCTAAAGGA | RT-PCR of medaka CNP-3 |
| CNP-A21 | GCAGCCCATTCCACTGATGGT | RT-PCR of medaka CNP-4 |
| FuguC1-A1 | CCGAAGCATCCTCGGTTCCA | 5′-RACE of puffer fish CNP-1 |
| FuguC2-A1 | CCAAAGCATCCTCTTCCTCC | 5′-RACE of puffer fish CNP-2 |
| FuguC3-A1 | CCGAAGCAGCTCCTCAGACC | 5′-RACE of puffer fish CNP-3 |
| FuguC4-A1 | CCGAAGCAGCCACTCCGTGA | 5′-RACE of puffer fish CNP-4 |
| FuguC1-S1 | ACAGCTCCGAGCTCCGACAG | 3′-RACE of puffer fish CNP-1 |
| FuguC1-S2 | AAGACCAACAACAGTCCTGGT | 3′-RACE of puffer fish CNP-1 |
| FuguC2-S1 | TCTGGACCCGTCCCGACAG | 3′-RACE of puffer fish CNP-2 |
| FuguC2-S3 | AGAATCTTCCGGAGCGTGTT | 3′-RACE of puffer fish CNP-2 |
| FuguC3-S1 | TGAGGGAGCAGACGCTGCTT | 3′-RACE of puffer fish CNP-3 |
| FuguC3-S2 | AGACGCTGCTTCGTCTCTCT | 3′-RACE of puffer fish CNP-3 |
| FuguC4-S1 | GAGAGGGGCACTTGAGGAG | 3′-RACE of puffer fish CNP-4 |
| FuguC4-S2 | CTTTGGAGTAGAGGACGGAC | 3′-RACE of puffer fish CNP-4 |
RT-PCR. Tissues for RT-PCR analyses were isolated from eight adult males and four adult females after anesthesia as above. Separation of total RNA, reverse transcription, and PCR were performed as described (15) by using the specific primers shown in Table 1. Numbers of amplification cycles were 27, 30, 30, 30, and 24 for CNP-1, -2, -3, -4, and GAPDH transcripts, respectively.
Receptor Activation Assays. Activities of four predicted CNPs were compared by using synthetic medaka CNPs and three endogenous NP receptors, OlGC1 (16), OlGC2, and OlGC7 (17) expressed in COS-7 cells. Synthetic CNPs were obtained from the Peptide Institute (Minoo, Japan). Expression of receptors in COS-7 cells was performed as described (18). Forty-eight hours after transfection, 10–6 to 10–11 M synthetic CNPs were added and incubated for 10 min. cGMP concentrations were determined according to Koller et al. (7). Experiments were quadruplicated and data were indicated as means ± SEM. Repeated-measures ANOVA with Fisher's probable least-squares difference was used for statistic analyses. P < 0.05 was recognized to be significant.
Phylogenetic Analysis. Amino acid sequences of four CNP precursors of medaka and puffer fish, CNP precursors of vertebrates, and piscine cardiac NP precursors were aligned by using the clustal w alignment program. Phylogenetic trees were depicted by using the neighbor-joining method. The phylogenetic tree of enolase isozymes was constructed by using the same method.
Linkage Mapping and Comparative Genomic Analyses. Linkage mapping was performed as described (19). Typing data and primer sequences for mapping are available at http://mbase.bioweb.ne.jp/~dclust/medaka_top.html. Puffer fish sequences and human chromosome data were obtained from http://fugu.hgmp.mrc.ac.uk and www.ncbi.nlm.nih.gov/genome/guide/human, respectively. Positions of introns in the four CNP genes of the puffer fish were identified by aligning cDNA sequences with genomic sequences.
Results
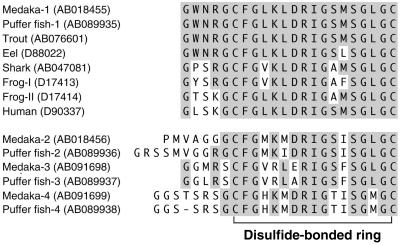
Structure of Four CNPs. Four cDNAs, each of which encodes a distinct CNP, were isolated from both medaka and puffer fish. We designated four CNPs, CNP-1, -2, -3, and -4, in the order of isolation of cDNA. CNP-1, -2, and -4 were isolated from the brain, and CNP-3 was isolated from the heart in both species. Comparison of predicted mature CNPs showed that they are structurally distinct (Fig. 1). CNP-1 was most similar to CNPs from other species, including mammals; the sequence of the ring was identical among medaka CNP-1, puffer fish CNP-1, and human CNP. In three other CNPs, up to five amino acid residues were substituted, even in the highly conserved ring domain. The sequences of CNP-4 were the most different from CNP-1 and CNPs of other species (Fig. 1).
Fig. 1.
Comparison of amino acid sequences of CNPs predicted from four distinct cDNAs isolated from medaka and puffer fish. Amino acid residues identical to medaka CNP-1 are shaded. GenBank accession numbers are in parentheses.
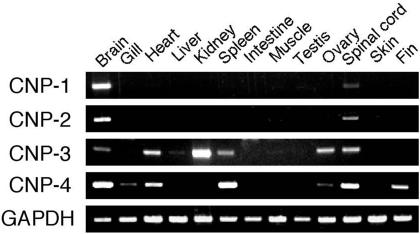
Tissue Distribution of Transcripts in Medaka. RT-PCR revealed that CNP-1 and -2 genes were specifically expressed in the CNS (Fig. 2). In contrast, expression of CNP-3 and -4 genes was detected in various peripheral tissues in addition to the CNS. For example, the CNP-4 gene was expressed in the spleen, heart, and fin, in addition to the brain and spinal cord, and was weakly expressed in the gill and ovary (Fig. 2). The expression pattern of the CNP-3 gene is more distinct; expression levels in the kidney, ovary, heart, and spleen were higher than that in the brain (Fig. 2).
Fig. 2.
Tissue distribution of transcripts of the four genes analyzed by RT-PCR. One microgram of total RNA was reverse transcribed and subjected to PCR. PCR products were electrophoresed on 1.2% agarose gels and stained by using ethidium bromide.
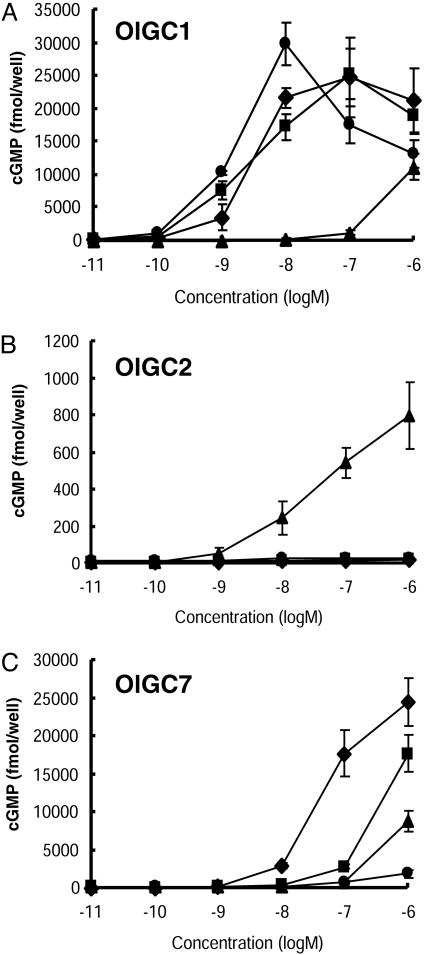
Activities of Four Medaka CNPs on Homologous Receptors. All four CNPs activated OlGC1, the medaka homologue of NPR-B (16); CNP-4 exhibited the greatest activity in the range from 10–10 to 10–8 M, followed by CNP-1 and -2, and CNP-3 showed the least activity (Fig. 3A). OlGC2 (17), a medaka homologue of NPR-A that is specific to ANP and BNP (7, 8), was activated by CNP-3 but not by the other three CNPs (Fig. 3B). On OlGC7, another homologue of NPR-A (17), CNP-1 revealed the highest activity, followed by CNP-2, whereas the other two CNPs had low activities (Fig. 3C).
Fig. 3.
Dose-dependent activation of NP receptors OlGC1 (a NPR-B homologue), OlGC2, and OlGC7 (NPR-A homologues) by synthetic CNP ligands. Receptors were expressed in COS-7 cells, and cGMP concentrations were measured by ELISA. CNP-1, -2, -3, and -4 are indicated by ♦, ▪, ▴, and •, respectively. (A) Differences between CNP-4, CNP-1 and -2, and CNP-3 were significant in the range from 10–10 to 10–8 M. (B) The activity of CNP-3 was higher than other CNPs in the range from 10–9 to 10–6 M. (C) Activities of four CNPs were significantly different in the range from 10–9 to 10–6 M, except between CNP-3 and -4.
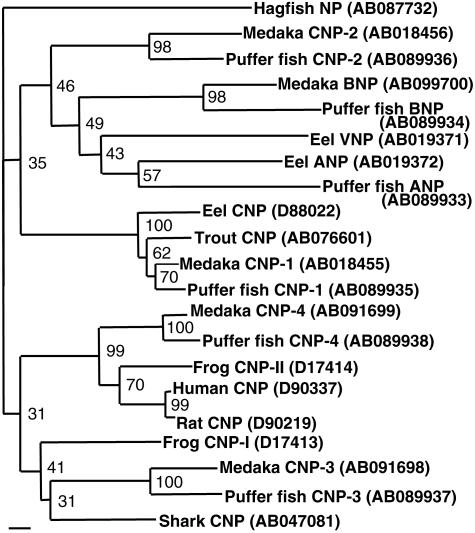
Phylogenetic Analysis. In phylogenetic analyses of the whole predicted precursor sequences, CNP-1 formed a clade with known teleost CNPs, CNP-3 with shark CNPs and frog CNP-I, and CNP-4 with the frog CNP-II and mammalian CNPs, whereas CNP-2 was at the independent position (Fig. 4). Thus, the structurally most dissimilar (CNP-4), not the most similar (CNP-1), is suggested to be the orthologue of mammalian CNPs. However, bootstrap values at nodes connecting different clades were low because of highly variable prosegment sequences. Thus, the relationship among four CNPs was not clear in this phylogenetic tree.
Fig. 4.
Phylogenetic tree of NP precursor sequences constructed by using the neighbor-joining method. Piscine ANP, BNP, and ventricular NP (VNP) sequences are included. The number at each node represents bootstrap value in percent. Hagfish NP was used as an outgroup. Bar = 0.1 genetic distance. GenBank accession numbers are in parentheses.
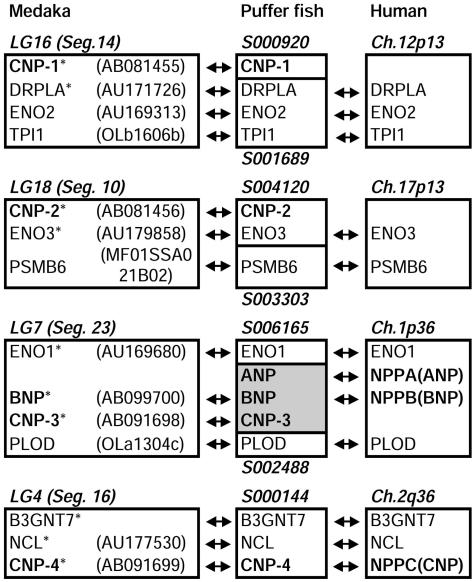
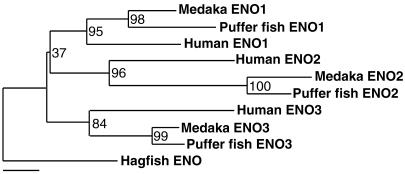
Linkage and Comparative Genomic Analyses. Linkage mapping in medaka showed that four CNP genes were located on separate linkage groups (LGs), i.e., on separate chromosomes (Fig. 5). We attempted to identify the relationship between medaka LGs containing CNP genes and human chromosomes. First, the puffer fish genome and medaka EST databases were screened for homologues of human genes around the CNP locus (NPPC, 2q24–qter) (Fig. 5). As a result, linkage of CNP (NPPC)-nucleolin (NCL)-β-1,3-N-acetylglucosamyltransferase 7 (B3GNT7) genes on human chromosome 2 (around 2q36) was found to be conserved in the puffer fish scaffold S000144 and medaka LG4, male segment 16 (LG4, Seg.16) containing the CNP-4 gene. The “male segment” in medaka LG is the regional unit, in which included genes exhibit no recombination among 39 panel individuals (19). Second, we found the linkages of three other CNP genes to three enolase gene paralogues, respectively, because the CNP-1 gene was found to be linked to enolase 2 (ENO2) gene at LG16, Seg.14 of medaka, both CNP-2 gene and enolase 3 (ENO3) gene were on S004120 of puffer fish, and enolase 1 (ENO1) gene is located near the loci of ANP (NPPA) and BNP (NPPB) on human chromosome 1 (1p36). Although the information about enolase genes in fish is limited at present, especially for enolase 2 (20), identities of enolase paralogues in medaka and puffer fish were easily determined by a phylogenetic analysis (Fig. 6). By mapping the medaka loci corresponding to human genes around three enolase loci, conserved linkages between medaka LG16, Seg.14 containing CNP-1 gene and human 12p13, between medaka LG18, Seg.10 containing CNP-2 gene and human 17p13, and between medaka LG7, Seg.23 containing CNP-3 gene and human chromosomes 1p36 were identified (Fig. 5). blast searches revealed no human counterparts of the three CNP genes, which are supposed to be lost or deformed during the course of evolution. In a genomic DNA contig, 5216.2 of another puffer fish, Tetraodon nigroviridis, it was found that the CNP-3 gene is tandemly located with ANP and BNP genes in the same orientation (Fig. 5).
Fig. 5.
Comparison of positions of NP gene loci in medaka, puffer fish, and human genomes. Arrows indicate corresponding loci. Each locus is named after those in the human genome except for fish NP loci, which are indicated by using the name of subtypes. NP loci are boldfaced. Position of each box is indicated in italic, as the male segments (Segs.) in linkage groups (LGs) of medaka, scaffold number of Takifugu rubripes, and positions in the human chromosomes (Ch.). The shaded box is contig 5216.2 of Tetraodon nigroviridis. Asterisks indicate medaka loci mapped in the present study.
Fig. 6.
Phylogenetic tree of enolase genes, which are linked to three CNP genes in medaka, puffer fish, and humans, by using the neighbor-joining method. The number at each node represents bootstrap value in percent. Hagfish enolase (AB025326) was used as an outgroup. Bar = 0.02 genetic distance.
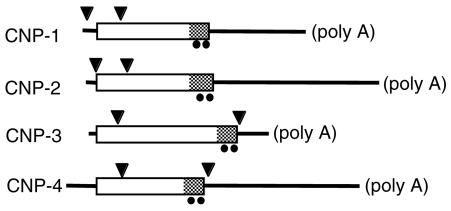
Exon/Intron Arrangements. All four CNP genes of the puffer fish contained two introns (Fig. 7). In the CNP-3 and -4 genes, one is nearly 100 bases downstream from the start codon and the other is around the last cysteine codon. On the other hand, positions of introns were found to shift in CNP-1 and CNP-2 genes, in which the second intron of CNP-3 and -4 is missing and an additional intron is observed around the start codon.
Fig. 7.
Positions of introns (arrowheads) in four CNP genes of the puffer fish T. rubripes. Boxes and lines indicate coding and noncoding regions of cDNA sequences, respectively. The mature CNP domain is shaded. Filled circles indicate the positions of two cysteine residues that form the ring structure.
Discussion
Distinct Activities of Four CNPs. In this study, we isolated four cDNAs, each of which encodes a structurally distinct CNP. It was suggested that the four predicted CNPs are functionally differentiated because each of the four medaka CNPs exhibited different activities on three medaka receptors, OlGC1, OlGC2, and OlGC7. Because OlGC1 is a homologue of the mammalian NPR-B (16), activation of OlGC1 by all four CNPs is as predicted. However, an unexpected finding is that CNP-4, which has the structure most differentiated from mammalian CNPs, exhibited higher activity on OlGC1 than did CNP-1, which is structurally most similar to CNPs of other vertebrates (Fig. 1). It seems that amino acid substitutions in CNP-4 increased the activity on OlGC1. In NPs, amino acid residues important for ligand–receptor interaction are under study using crystal structure analyses (21). The information obtained in this study will contribute to such structure–activity studies. Because OlGC1 is expressed in the spleen, heart, gill, and ovary (16), CNP-4 is thought to function as a paracrine/autocrine factor in these and other peripheral organs. It is also intriguing that OlGC7 and OlGC2, medaka homologues of NPR-A, were also activated by some of four CNPs. OlGC7 was most activated by CNP-1 and CNP-2. Unlike CNP-4, the expression of CNP-1 and CNP-2 genes was CNS-specific. These two CNPs may function separately from CNP-4 through OlGC7. OlGC2 was activated only by CNP-3. Of the two medaka homologues of NPR-A, OlGC2 is functionally more similar to mammalian NPR-A (18). In addition, CNP-3 exhibited the lowest activity on OlGC1, and the CNP-3 gene exhibited higher expression in the heart than in the brain. Thus, it seems that CNP-3 is functionally more similar to ANP and BNP than to CNP, which supports the idea that ANP and BNP genes are derived from the CNP-3 gene (see below).
Paralogous Relationship Among Previously Known CNPs. Because only one CNP to our knowledge had to date been isolated from any vertebrate species except for the bullfrog (13), and because these CNPs have structures that are conserved among different species (Fig. 1), it was thought that previously known CNPs are orthologous with one another. However, in the phylogenetic tree based on CNP precursors (Fig. 4), previously known vertebrate CNPs are clustered with different isoforms of the medaka and puffer fish: eel and trout CNPs with CNP-1, frog CNP-I and shark CNPs with CNP-3, and mammalian CNPs and frog CNP-I with CNP-4. Thus, it is clear from this analysis that CNPs previously known in different classes of vertebrates were derived from different paralogues, which well explains the paradox that sequences of prosegments of CNP precursors are quite dissimilar among different vertebrate classes despite striking similarity within the same vertebrate class (1). However, it was difficult to estimate either the relationship among clusters or the time of divergence from this tree because of variable prosegment sequences.
Generation of Four CNP Genes by Chromosomal Duplication. Linkage mapping in medaka located four CNP genes on separate chromosomes (Fig. 5). We found conserved syntenies around the four CNP genes among medaka, puffer fish, and humans. Conserved linkages of CNP-1, -2, and -3 with enolase paralogs suggest that they were generated by the block duplication of the chromosome (22, 23). Because extensive similarity among other regions of human chromosomes 2, 12, and 17 has been pointed out by others (24–26), it is possible that large-scale duplication occurred. Although the timing of duplication of CNP genes was unpredictable from phylogenetic analyses of CNP genes as mentioned above, it is predictable from enolase genes linked to CNP genes. Duplications of enolase paralogues have been estimated to occur between cyclostome/gnathostome and chondrichthyan/osteichthyan divergences, because separation of vertebrate-type enolases is evident in a shark Chiloscyllium punctatum (20) but not in cyclostomes (27). Thus, duplications of CNP genes linked to enolase genes are estimated to have occurred between 564 and 528 million years ago according to Kumar and Hedges (28). Although we cannot conclude at present whether these duplications correspond to “two rounds of duplications” of whole chromosome proposed in vertebrates (29), it is evident that duplications of CNP genes are not specific to the teleost lineage.
The Order of CNP Gene Duplications. Changes in exon/intron arrangement offered keys to estimate the order of duplication. The positions of introns in puffer fish CNP-3 and CNP-4 genes were different from those of CNP-1 and CNP-2 genes (Fig. 7). Because hagfish NP also has an intron at two bases after the stop codon (unpublished data), the original exon/intron arrangement is of the CNP-3/4 type. Thus, it is evident that the common ancestor of CNP-1 and -2 genes branched from the CNP-3 gene after the establishment of the linkage with the enolase gene, and then CNP-1 and -2 separated after rearrangement of exon/intron structure. The earliest divergence of the CNP-4 gene, before the establishment of the linkage with the ancestral enolase gene, is highly probable considering the independent gene linkage (Fig. 5), although generation by later translocation from CNP-3 cannot be excluded.
Generation of ANP and BNP Genes from the CNP-3 Gene. In the genome of another puffer fish, Tetraodon nigroviridis, the CNP-3 gene is tandemly localized with ANP and BNP genes in the same orientation (Fig. 5), which suggests that the ANP and BNP genes were generated by tandem duplication of the CNP-3 gene. Functional similarities of CNP-3 to ANP and BNP mentioned above also support this idea. Although it is clear that these tandem duplications occurred before the divergence of teleosts and tetrapods and after the separation of CNP-1 and -2 from CNP-3, we are not able to determine at present whether these duplications occurred before or after the radiation of elasmobranchs because genomic information on elasmobranchs is limited.
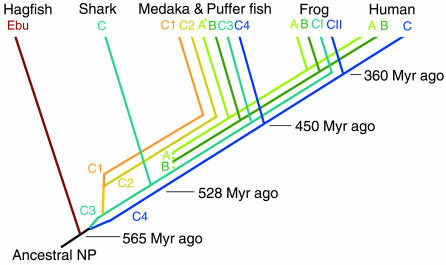
The Evolutionary History of the NP Family. The lineage of the NP family proposed by the present study is summarized in Fig. 8. First, four CNP genes were generated by chromosomal duplications before the divergence of elasmobranchs in the order noted above. Subsequently, the CNP-3 gene generated ANP and BNP genes through tandem duplication before the divergence of tetrapods and teleosts. After such duplications, specific lineages may have been lost in each vertebrate class during the course of evolution. For example, it seems that frogs have lost CNP-1 and CNP-2 lineages, although they still retain CNP-3 and CNP-4 lineages as CNP-I and CNP-II, respectively. After branching of amphibians, tetrapods may have lost the CNP-3 gene, although mammals maintain ANP and BNP genes derived from the CNP-3 lineage and a CNP gene derived from the CNP-4 lineage. As for sharks, results obtained in the present study suggest that the present constitution of a single CNP is also a result of losses of three CNPs, because duplications of CNPs had already occurred before the divergence of elasmobranchs. In addition, if tandem duplications of ANP and BNP genes from CNP-3 had occurred before the divergence of elasmobranchs as discussed above, they may have also lost ANP and BNP genes.
Fig. 8.
Lineage of NP genes. Different colors indicate different lineages. A, ANP; B, BNP; C, CNP; C1-C4, CNP-1–CNP-4 of teleosts; CI and CII, CNP-I and CNP-II of the frog, respectively; Ebu, hagfish NP. Time of divergence is indicated in million years (Myr) ago according to Kumar and Hedges (28). The dotted line indicates unspecified timing of divergence. *, Medaka ANP has not been found.
In contrast, it seems that teleosts still maintain the most CNP lineages. Because the phylogenetic distance between the medaka and puffer fishes is not very short (9), it is probable that many other teleost species also have at least some of the four CNP genes. In addition, at least some species have one more subtype, ventricular NP (1, 30), although we cannot presume the origin of this subtype in this study. Moreover, not only additional ligands, but also additional NP receptors, are found in teleosts (17, 31, 32). We showed in the present study that four CNPs have distinct activities, not only on a NPR-B-like receptor, but also on two NPR-A-like receptors. This pattern is different from the simple relationship in mammals, where ANP and BNP use NPR-A while CNP uses NPR-B. Fishes may have retained four CNP genes by modifying functions of each duplicated gene (33), and may have developed complicated NP systems to achieve body fluid homeostasis against a variety of aquatic environments, including hyperosmotic seawater, causing water loss and salt gain, and hypoosmotic fresh water, causing the opposite movements (30, 34). In the tetrapod lineage, unnecessary molecules may have been lost with the transition from water to land, where the retention of water is of primary importance in body fluid regulation.
Acknowledgments
We thank Prof. Christopher A. Loretz (State University of New York, Buffalo) for critical reading of the manuscript, Oita Biological Technology Center, Nippon Suisan Kaisha, for the kind gift of the puffer fish, and Dr. Susumu Hyodo and Ms. Sanae Hasegawa for valuable discussion. This work was supported by Grant-in-Aid for Creative Basic Research 12NP0201 from the Ministry of Education, Science, Sports, and Culture of Japan, and by Scientific Research (A) Award 13304063 from the Japan Society for the Promotion of Science.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NP, natriuretic peptide; ANP, atrial NP; BNP, B-type NP; CNP, C-type NP; LG, linkage group; NPR, NP receptor; Seg.n, male segment n.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB081455, AB081456, AB091698, and AB091699 for CNP-1 to -4 of medaka, and AB089935, AB089936, AB089937, and AB089938 for the puffer fish CNP-1 to -4, respectively).
References
- 1.Takei, Y. (2000) Int. Rev. Cytol. 194, 1–66. [DOI] [PubMed] [Google Scholar]
- 2.Kone, B. C. (2001) Cardiovasc. Res. 51, 429–441. [DOI] [PubMed] [Google Scholar]
- 3.Kawakoshi, A., Hyodo, S. & Takei, Y. (2001) Zool. Sci. 18, 861–868. [Google Scholar]
- 4.Kawakoshi, A., Hyodo, S., Yasuda, A. & Takei, Y. (2003) J. Mol. Endocrinol. 30, in press. [DOI] [PubMed]
- 5.Takei, Y., Takahashi, A., Watanabe, T. X., Nakajima, K. & Sakakibara, S. (1991) FEBS Lett. 282, 317–320. [DOI] [PubMed] [Google Scholar]
- 6.Takei, Y., Takano, M., Itahara, Y., Watanabe, T. X., Nakajima, K., Conklin, D. J., Duff, D. W. & Olson, K. R. (1994) Gen. Comp. Endocrinol. 96, 420–426. [DOI] [PubMed] [Google Scholar]
- 7.Koller, K. J., Lowe, D. G., Benette, G. L., Minamino, N., Kangawa, K., Matsuo, H. & Goeddel, D. V. (1994) Science 252, 120–123. [DOI] [PubMed] [Google Scholar]
- 8.Suga, S., Nakao, K., Hosoda, K., Mukoyama, M., Ogawa, Y., Shirakami, G., Arai, H., Saito, Y., Kambayashi, Y., Inouye, K. & Imura, H. (1992) Endocrinology 130, 229–239. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa, Y. (2000) BioEssays 22, 487–495. [DOI] [PubMed] [Google Scholar]
- 10.Wittbrodt, J., Shima, A. & Schartl, M. (2002) Nat. Rev. Genet. 3, 53–64. [DOI] [PubMed] [Google Scholar]
- 11.Inoue, K. & Takei, Y. (2002) Zool. Sci. 19, 727–734. [DOI] [PubMed] [Google Scholar]
- 12.Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. (2002) Science 297, 1301–1310. [DOI] [PubMed] [Google Scholar]
- 13.Kojima, M., Ohyama, Y., Miyamoto, K., Minamino, N., Kangawa, K. & Matsuo, H. (1994) J. Biol. Chem. 269, 13136–13140. [PubMed] [Google Scholar]
- 14.Inoue, K., Russel, M. J., Olson, K. R. & Takei, Y. (2003) Gen. Comp. Endocrinol. 130, 185–192. [DOI] [PubMed] [Google Scholar]
- 15.Inoue, K., Iwatani, H. & Takei, Y. (2003) Gen. Comp. Endocrinol. 131, 77–84. [DOI] [PubMed] [Google Scholar]
- 16.Takeda, K. & Suzuki, N. (1999) J. Biochem. 126, 104–114. [DOI] [PubMed] [Google Scholar]
- 17.Yamagami, S., Suzuki, K. & Suzuki, N. (2001) J. Biochem. 130, 39–50. [DOI] [PubMed] [Google Scholar]
- 18.Yamagami, S., Xu, S. H., Tsutsumi, M., Hori, H. & Suzuki, N. (2003) Zool. Sci. 20, 591–606. [DOI] [PubMed] [Google Scholar]
- 19.Naruse, K., Fukamachi, S., Mitani, H., Kondo, M., Matsuoka, T., Kondo, S., Hanamura, N., Morita, Y., Hasegawa, K., Nishigaki, R., et al. (2000) Genetics 154, 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tracy, M. J. & Hedges, S. B. (2000) Gene 259, 129–138. [DOI] [PubMed] [Google Scholar]
- 21.He, X.-L., Chow, D.-c., Martick, M. M. & Garcia, K. C. (2001) Science 293, 1657–1662. [DOI] [PubMed] [Google Scholar]
- 22.Hughes, A. L. (1998) Mol. Biol. Evol. 15, 854–870. [DOI] [PubMed] [Google Scholar]
- 23.Abi-Rached, L., Gilles, A., Shiina, T., Pontarotti, P. & Inoko, H. (2002) Nat. Genet. 31, 100–105. [DOI] [PubMed] [Google Scholar]
- 24.Ruddle, F. H., Bentley, K. L., Murtha, M. T. & Risch, N. (1994) Development (Cambridge, U.K.) Suppl. 120, 155–161. [PubMed] [Google Scholar]
- 25.Wolfe, K. H. (2001) Nat. Genet. 2, 333–341. [DOI] [PubMed] [Google Scholar]
- 26.McLysaght, A., Hokamp, K. & Wolfe, K. H. (2002) Nat. Genet. 31, 200–204. [DOI] [PubMed] [Google Scholar]
- 27.Kuraku, S., Hoshiyama, D., Katoh, K., Suga, H. & Miyata, T. (1999) J. Mol. Evol. 49, 729–735. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, S. & Hedges, S. B. (1998) Nature 392, 917–920. [DOI] [PubMed] [Google Scholar]
- 29.Meyer, A. & Schartl, M. (1999) Curr. Opin. Cell Biol. 11, 699–704. [DOI] [PubMed] [Google Scholar]
- 30.Loretz, C. A. & Pollina, C. (2000) Comp. Biochem. Physiol. A Physiol. 125, 169–187. [DOI] [PubMed] [Google Scholar]
- 31.Kashiwagi, M., Katafuchi, T., Kato, A., Inuyama, H., Ito, T., Hagiwara, H., Takei, Y. & Hirose, S. (1995) Eur. J. Biochem. 233, 102–109. [DOI] [PubMed] [Google Scholar]
- 32.Kusakabe, T. & Suzuki, N. (2000) Zool. Sci. 17, 131–140. [Google Scholar]
- 33.Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. & Postlethwait, J. (1999) Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takei, Y. & Hirose, S. (2002) Am. J. Physiol. 282, R940–R951. [DOI] [PubMed] [Google Scholar]