Abstract
Prokaryotes rely on a distant tubulin homolog, FtsZ, for assembling the cytokinetic ring essential for cell division, but are otherwise generally thought to lack tubulin-like polymers that participate in processes such as DNA segregation. Here we characterize a protein (TubZ) from the Bacillus thuringiensis virulence plasmid pBtoxis, which is a member of the tubulin/FtsZ GTPase superfamily but is only distantly related to both FtsZ and tubulin. TubZ assembles dynamic, linear polymers that exhibit directional polymerization with plus and minus ends, movement by treadmilling, and a critical concentration for assembly. A point mutation (D269A) that alters a highly conserved catalytic residue within the T7 loop completely eliminates treadmilling and allows the formation of stable polymers at a much lower protein concentration than the wild-type protein. When expressed in trans, TubZ(D269A) coassembles with wild-type TubZ and significantly reduces the stability of pBtoxis, demonstrating a direct correlation between TubZ dynamics and plasmid maintenance. The tubZ gene is in an operon with tubR, which encodes a putative DNA-binding protein that regulates TubZ levels. Our results suggest that TubZ is representative of a novel class of prokaryotic cytoskeletal proteins important for plasmid stability that diverged long ago from the ancient tubulin/FtsZ ancestor.
Keywords: FtsZ, tubulin, cytoskeleton
Actin and tubulin are central components of a complex cytoskeleton essential for growth and division of eukaryotic cells. Bacteria also contain cytoskeletal proteins related to actin and tubulin, but in many cases their functions within cells are not well understood (van den Ent et al. 2001a; Carballido-Lopez and Errington 2003b; Moller-Jensen and Lowe 2005; Michie and Lowe 2006; Shih and Rothfield 2006). The bacterial tubulin homolog FtsZ is highly conserved in prokaryotes where it assembles a cytokinetic ring at midcell required for cell division (Romberg and Levin 2003; Margolin 2005; Dajkovic and Lutkenhaus 2006). The FtsZ ring recruits other cell division proteins to the site of septum formation and constricts as the invaginating septum grows inward (Goehring and Beckwith 2005; Margolin 2005; Dajkovic and Lutkenhaus 2006). Biochemical and structural similarities suggest that FtsZ and tubulin evolved from a common ancestor (Nogales et al. 1998a; van den Ent et al. 2001a; Errington 2003; Vaughan et al. 2004), but phylogenetic analysis demonstrates that they diverged very early in the evolution of the three domains of life, likely more than a billion years ago (Nogales et al. 1998a; van den Ent et al. 2001a; Errington 2003; Vaughan et al. 2004). Aside from FtsZ, most bacteria are assumed to lack dynamic tubulin-related polymers that might play a role in intracellular transport. However, a few bacteria have multiple FtsZ-related proteins, and one (Prosthecobacter) encodes α- and β-tubulin homologs (BtubA/BtubB) (Jenkins et al. 2002; Sontag et al. 2005) although the function and localization of these proteins remains unclear.
At least five different families of bacterial actins have been described that are distantly related (<20% identity) to each other and to eukaryotic actin (van den Ent et al. 2001b; Carballido-Lopez and Errington 2003b; Amos et al. 2004; Carballido-Lopez 2006; Michie and Lowe 2006; Shih and Rothfield 2006). MreB and its close relative Mbl form dynamic helical filaments adjacent to the cell membrane and move rapidly through the cell (Carballido-Lopez and Errington 2003a, b; Defeu Soufo and Graumann 2004). MreB is required for maintaining the shape of the cell and has also been implicated in chromosome segregation (Jones et al. 2001; Defeu Soufo and Graumann 2005; Gitai et al. 2005; Carballido-Lopez et al. 2006). Other families of bacterial actins, such as ParM and AlfA, catalyze the segregation of plasmid DNA (Jensen and Gerdes 1997; Gerdes et al. 2004; Becker et al. 2006). ParM assembles two-stranded, actin-like filaments that display dynamic instability in vitro and is thought to function in vivo by polymerizing between newly replicated plasmids, pushing them apart (Moller-Jensen et al. 2002, 2003; van den Ent et al. 2002; Garner et al. 2004, 2007). Similarly, AlfA forms dynamic polymers required for plasmid segregation in Bacillus subtilis (Becker et al. 2006). A fifth family of actin, MamK from Magnetospirillum, forms filaments required for maintaining magnetosome organization and function (Komeili et al. 2006; Pradel et al. 2006). Thus, bacterial actin homologs participate in a variety of cellular functions, including the separation of DNA molecules, a function performed in eukaryotic cells by tubulin.
Bacterial cells also contain the ParA family of ATPases that comprise a cytoskeletal element (Gerdes et al. 2004; Leonard et al. 2005; Hayes and Barilla 2006). ParA proteins are required for plasmid and chromosomal DNA segregation in many bacteria and have recently been shown to assemble polymers both in vivo and in vitro (Barilla et al. 2005; Lim et al. 2005; Adachi et al. 2006; Ebersbach et al. 2006; Fogel and Waldor 2006; Bouet et al. 2007). Although the mechanisms of ParA-mediated segregation are not fully understood, it is thought that the ability of ParA proteins to polymerize and depolymerize underlies their dynamic localization. Rapid movement of ParA is most likely coupled to DNA movement via a DNA-binding protein that allows ParA polymers to either push or pull DNA to its proper location.
Molecular transport mechanisms likely arose long ago, and are mediated in bacteria by both distant actin relatives and the unrelated ParA proteins. Curiously, no transport process in microbial cells has been ascribed to any tubulin-related protein, leaving it unclear as to when tubulin-based molecular transport mechanisms evolved. Here we investigate the segregation of plasmid pBtoxis in Bacillus thuringiensis subsp. israeliensis. This 126-kb plasmid encodes crystalline insecticidal toxins required for B. thuringiensis virulence (Berry et al. 2002). In our preliminary studies, we found that this plasmid is very stably inherited, but a survey of the complete plasmid DNA sequence did not reveal any obvious homologs of either the actin or ParA families. We therefore searched for genes that might play a role in pBtoxis stability and identified one, gene 156, that encodes a protein (TubZ) distantly related to both tubulin and FtsZ. Previous work suggested that a mini-plasmid derived from pBtoxis containing the tubZ region was capable of plasmid replication in B. thuringiensis (Tang et al. 2006). TubZ is also distantly related to RepX, encoded by pXO1 of Bacillus anthracis, which was recently reported to be required for replication of a mini-pXO1 plasmid (Tinsley and Khan 2006). Here we characterize the pBtoxis tubRZ region. We show that TubZ assembles dynamic polymers required for pBtoxis stability and propose that TubZ represents a novel type of tubulin-based cytoskeletal element in bacteria.
Results
Members of the tubulin superfamily distinct from both tubulin and FtsZ
Several protein sequences from Archaea and Bacteria are annotated in the GenBank database as FtsZ or “FtsZ-like” but phylogenetic analysis demonstrates that they are only distantly related to tubulin and FtsZ and, in fact, fall into separate, loosely related groups (Fig. 1; Vaughan et al. 2004). Many of the archaeal proteins (Fig. 1A, pink) cluster together on the phylogenetic tree. The bacterial proteins identified thus far are encoded by plasmids of the genus Bacillus and each is highly divergent from the others (Fig. 1A, red; Vaughan et al. 2004). These divergent FtsZ relatives are clearly on a different branch of the phylogenetic tree from tubulin and FtsZ, as supported by robust bootstrap values (99% and 93%) but how they are related to each other is less clear. Inspection of the genome sequences of the four Bacilli included in our analysis revealed that each also has a chromosomal ftsZ gene encoding a homolog closely related to the well-characterized Escherichia coli cell division protein FtsZ, making it unlikely that the more distantly related “FtsZ-like” proteins are essential for cell division. The Archaea included in our analysis also have more than one tubulin/FtsZ-related gene and some genera (Halobacterium and Haloarcula) have five such genes: two closely related to FtsZ, and three that belong to the distantly related cluster.
Figure 1.
Phylogenetic analysis of the tubulin superfamily. (A) Phylogenetic tree showing the relationship between tubulins (blue), FtsZ (green), and TubZ (Archaea, pink; Bacillus, red). The tree was calculated using the neighbor-joining method, and bootstrap values representing confidence levels are indicated for select branches. Sequence abbreviations are listed below. Accession numbers are provided in the Materials and Methods and Supplemental Material. For each of the proteins from Bacillus plasmids, the name of the plasmid as well as the ORF number is noted on parentheses. (B) Alignment of conserved motifs for selected sequences. Each motif (loops T1, T4, T6, T7) as previously annotated (Nogales et al. 1998a) is shown with universally conserved residues indicated in red, residues conserved in FtsZ indicated in green, and residues conserved in tubulin indicated in blue. Sequence abbreviations are as follows: (Ec) E. coli FtsZ; (Bs) B. subtilis FtsZ; (Hm1) Haloarcula marismortui FtsZ1; (Hm4) H. marismortui FtsZ4; (Hh4) Halobacterium NRC-1 FtsZ4; (Hm5) H. marismortui FtsZ5; (Hh5) Halobacterium NRC-1 FtsZ5; (Bm) B. megaterium pBM400 orf44; (Bc) B. cereus pBc218 orf139; (Ba) B. anthracis pXO1 orf45; (Bt) B. thuringiensis pBtoxis orf156; (SsA) Sus scrofa α tubulin; (SsB) S. scrofa β tubulin; (HsG) Homo sapiens γ tubulin; (HsE) H. sapiens ε tubulin; (HsD) H. sapiens δ tubulin.
We refer to the pBtoxis (B. thuringiensis) protein as TubZ since it is different from but related to both tubulin and FtsZ. A comparison of TubZ and other “TubZ”-like sequences from both the Bacilli and the Archaea (Fig. 1A [red and pink], B) with those of FtsZ and tubulin (Fig. 1A [green and blue], B) confirm that all have amino acid motifs that define the tubulin superfamily (Nogales et al. 1998a, b; Lowe et al. 2001; Nogales 2001). Notably, these include the tubulin signature motif in loop T4 [GGGTG(S/T)G] which is involved in GTP binding, specifically through interaction with the α and β phosphates of GTP. Within each of the motifs aligned in Figure 1B are key amino acids that are typically conserved only in tubulins (blue), only in FtsZ (green) or are conserved between both (red). Surprisingly, many of these distant prokaryotic tubulin homologs contain a combination of tubulin-specific and FtsZ-specific residues. For example, TubZ from B. thuringiensis (Bt) and Bacillus cereus (Bc) are similar to FtsZ in loop T4 (Fig. 1B, green) but similar to tubulin within loop T6 (Fig. 1B, blue). These results, together with previously published phylogenetic analyses, suggest that both the Bacillus plasmid-encoded proteins and the archaeal proteins form distinct groups that share an ancient, common ancestor with tubulin and FtsZ.
Another notable difference between tubulin and FtsZ, as compared with TubZ, is the sequence within the T7 loop, a region that plays an important role in GTP catalysis. The sequence GxxNxDxxD/E is conserved in both tubulins and FtsZ, and these residues contribute to the hydrolysis of GTP bound to an adjacent subunit in the polymer (Nogales et al. 1998a, b; Lowe et al. 2001; Nogales 2001). The Bacillus and archaeal TubZ-like sequences contain the DXXD/E portion of this motif; however, all lack either the essential asparagine, the conserved glycine, or both (Fig. 1B). The asparagine (Ec_FtsZ 207) plays a role in catalysis and mutations that alter this residue render FtsZ unable to hydrolyze GTP or to polymerize (Wang et al. 1997; Lu et al. 2001). Divergence in the T7 loop of TubZ might indicate that these proteins do not polymerize in a GTP-dependent manner (Vaughan et al. 2004).
Many bacterial FtsZ sequences contain a conserved C-terminal motif (LxIPxFL; EcFtsZ: 372–378) that mediates interactions with other cell division proteins, such as ZipA and FtsA (Addinall and Holland 2002; Lutkenhaus 2002; Romberg and Levin 2003; Vaughan et al. 2004; Goehring and Beckwith 2005; Margolin 2005). These residues are essential for cell division, and are conserved in FtsZ from organisms as divergent as E. coli and B. subtilis. The Bacillus TubZ proteins lack this motif, suggesting that they are not part of the cell division machinery. Consistent with this observation, B. thuringiensis and Bacillus megaterium can grow, divide, and sporulate without these plasmids, demonstrating that TubZ is not required for cell division during growth or sporulation.
A TubZ-GFP (green fluorescent protein) fusion protein forms dynamic polymers in B. thuringiensis
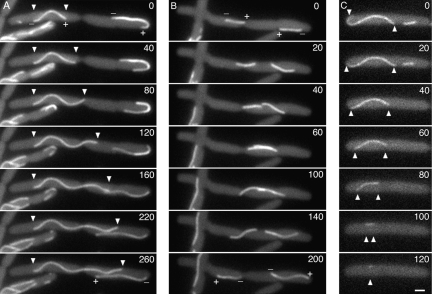
The ability to assemble into dynamic polymers is central to the function of both tubulin and FtsZ, but the significant divergence of TubZ sequences, particularly within loop T7, raised doubts as to whether these proteins share this or any other properties with their distant relatives. To determine if TubZ polymerizes, we constructed a GFP fusion to B. thuringiensis TubZ and recombined it into pBtoxis resulting in a strain (JP2172 = pBtoxis∷tubZ-gfp) in which tubZ-gfp is expressed under control of the native promoter and is the only full-length copy of the gene within the cell. When this strain was grown on an agarose pad at 30°C, TubZ-GFP assembled long polymers that often extended from one cell pole to the other (Fig. 2A). In time-lapse microscopy, TubZ-GFP polymers were very dynamic, translocating throughout the cell (Fig. 2B,C; Supplementary Movie S1), curving when necessary to travel around the pole of the cell, and continuing movement in the opposite direction (Fig. 2C). During translocation, filaments appeared to be polarized with a leading end (plus end) and a trailing end (minus end) that never reversed direction relative to each other. The rate of movement of the plus end through the cell (∼1.8 μm/min) (n = 12; three representative plots are shown in Fig. 2D) was consistent from cell to cell. The average rate of movement of the minus ends was similar to that of the plus ends (average = 1.5 μm/min, ±0.72; n = 7), but exhibited slightly more variability, with movement occasionally punctuated by brief pauses (Fig. 2E, arrow).
Figure 2.
TubZ-GFP assembles dynamic polymers in B. thuringiensis. (A) Fluorescence microscopy of B. thuringiensis expressing TubZ-GFP. Bar, 1 μm. (B,C) Time-lapse microscopy of TubZ-GFP movement. The plus (+) and minus (−) ends of each filament are noted to indicate the direction of movement. Images at 30-sec intervals are presented. (D) Rate of plus end movement versus time. The different points (squares, triangles, circles) represent measurements for three individual filaments. (E) Rate of minus end movement versus time. The arrow indicates a pause in this rate, likely due to a pause in depolymerization. The different points (squares, triangles, circles) represent measurements for three individual filaments.
TubZ-GFP forms dynamic polymers in E. coli
To determine if TubZ-GFP assembly and dynamics are dependent on proteins or factors found only in B. thuringiensis, we expressed TubZ-GFP from its native promoter (along with the upstream tubR gene) in E. coli. Surprisingly, TubZ-GFP assembled dynamic polymers in E. coli with rates of directional movement similar to those observed in B. thuringiensis (Fig. 3A–D; Supplementary Movies S2, S3). Plus end rates averaged 1.48 ± 0.63 μm/min (n = 24), with a maximal rate of 3.5 μm/min (Fig. 3F). Polymers ranged in length from a single focus to >2 μm in length and also were relatively flexible, existing in either straight or curved conformations adjacent to the cell membrane (Fig. 3A–C). While most cells (85%) contained only a single filament, when two filaments were present (15%), they were just as likely to travel in the same direction as in opposite directions from each other, suggesting that directional movement of these filaments was not obligatorily coupled (Fig. 3D; Supplementary Movie S3). The fact that B. thuringiensis TubZ-GFP displayed the same general dynamic properties when expressed in E. coli (which has no native TubZ) indicates that both polymerization and movement may be intrinsic properties of the protein. This, along with the observation that filaments did not appear to follow a prescribed path, suggests that TubZ-GFP does not rely on a defined cytoskeletal element such as MreB for movement.
Figure 3.
TubZ-GFP assembles dynamic polymers in E. coli. Time-lapse microscopy of E. coli strain JP2167 expressing TubZ-GFP with time points indicated in seconds. The plus (+) end indicates the forward direction of movement. Bar, 1 μm. (A–C) Single filaments translocate within the cell. (D) Two filaments move simultaneously in opposite directions. (E) FRAP analysis demonstrates that translocation occurs by treadmilling. A prebleach image and post-bleach images collected at 12-sec intervals are shown. For clarity, the example shown is a cell treated with cephalexin. (F) Rate of plus end movement versus time for three individual filaments (squares, triangles, circles). (G) Rate of minus end movement versus time for two individual filaments (triangles, circles). (H) Model for the treadmilling mechanism of TubZ movement in vivo. New subunits assemble at the plus end (●) while subunits are removed from the minus end (○).
Movement of TubZ-GFP occurs by a treadmilling mechanism
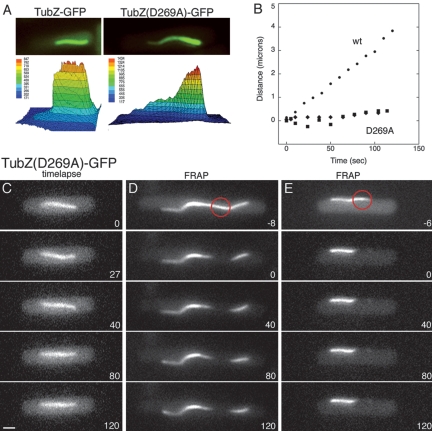
In eukaryotic cells, filament translocation can occur by at least two different mechanisms, including (1) sliding of a static polymer by means of a motor protein, or (2) treadmilling of a dynamic polymer in which there is simultaneous plus end polymerization and minus end depolymerization. To distinguish between these two possibilities, we performed FRAP (fluorescence recovery after photobleaching) experiments in which a small region of a filament was bleached with a laser (488 nm) and the fate of the bleached region followed using time-lapse microscopy. Before photobleaching, three images were collected to establish the directionality of filament movement (indicated by + and − in Fig. 3E). After photobleaching, filaments continued moving at the same rates as before; however, the bleached portion (Fig. 3E, white arrow) did not move and fluorescence did not recover (n = 20 cells; average filament length = 2.65 ± 1.08) (Fig. 3E; Supplementary Movie S4). Rather, TubZ-GFP filaments continued to grow at the plus ends and shrink at the minus ends, consistent with a treadmilling mechanism as depicted in Figure 3H. The lack of recovery in the bleached region is also consistent with an end assembly mechanism, suggesting that neither lateral association of new subunits nor significant sliding of protofilaments occurs. Similar results were obtained with FRAP analysis of TubZ-GFP filaments in B. thuringiensis (data not shown). These results are the first example of treadmilling for a prokaryotic tubulin.
To further understand TubZ polymer dynamics, we examined TubZ-GFP in E. coli cells treated with cephalexin, an inhibitor of cell division. Cephalexin-treated cells continue to grow but fail to divide and form long, multinucleate cells in which TubZ-GFP movement (polymerization) can be followed over greater distances. In these very long cells, TubZ-GFP filaments grew to >10 μm in length and moved at rates comparable with those in shorter cells (Fig. 4A,B). The filament on the left in Figure 4A (Supplementary Movie S5) elongated at the plus end at a rate of 0.98 μm/min with little disassembly from the minus end, validating our conclusion from photobleaching experiments that filaments are capable of polymerizing unidirectionally and demonstrating that minus end depolymerization is not required for plus end growth.
Figure 4.
Assembly dynamics of TubZ-GFP filaments in cephalexin-treated cells. Time-lapse microscopy of E. coli strain JP2167 expressing TubZ-GFP after incubation with 10 μg/mL cephalexin for 1 h with time points indicated in seconds. (A) Elongation and movement of TubZ-GFP. The plus (+) and minus (−) ends are marked by white triangles. The plus (+) end of the filament grew at a rate of 0.98 μm/min while the minus (−) end remained stationary. (Right) A second polymer in the cell translocated around the cell pole and continued movement in the opposite direction. (B) Movement of two filaments in opposite directions. Plus (+) and minus (−) ends of each are noted. (C) Rapid disassembly of TubZ-GFP filaments. A filament, with the ends noted by white triangles, rapidly shrank apparently from both ends after cells were incubated on a glass slide without nutrients. Bar, 1 μm.
Microtubules display dynamic instability in which pausing of elongation is followed by a switch to either rapid depolymerization (catastrophe) or to repolymerization (rescue) (Mitchison and Kirschner 1984; Desai and Mitchison 1997; Gadde and Heald 2004). We did not observe dynamic instability for TubZ; however, when cells were incubated on a glass slide (instead of on an agarose pad with nutrients) for an extended period of time, conditions that could deplete cellular energy reserves, complete depolymerization occurred as depicted in Figure 4C and Supplementary Movie S6. The overall rate of filament shrinkage was 2.25 ± 0.65 μm/min, with disassembly appearing to occur from both ends.
TubZ polymerizes without TubR and has a critical concentration for assembly
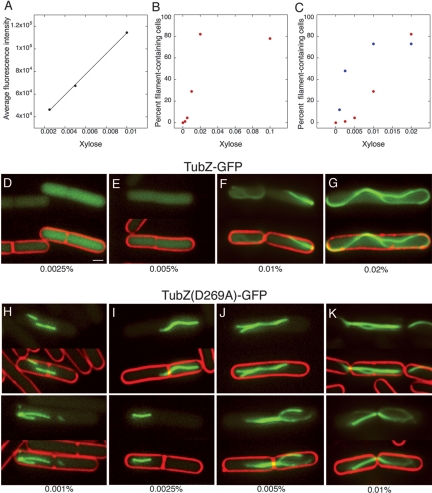
For all of the experiments described thus far in both E. coli and B. thuringiensis, the assembly of TubZ-GFP was examined in the presence of the upstream gene tubR, which is likely coexpressed with tubZ. To determine if TubZ is able to assemble polymers without any possible assistance from TubR, we constructed a plasmid (pRL218) expressing TubZ-GFP from a xylose-inducible promoter and transformed it into a strain of B. thuringiensis lacking pBtoxis (therefore lacking native tubRZ). The strain was grown with varying concentrations of xylose and TubZ-GFP expression was monitored by quantitating GFP fluorescence and determining the percentage of polymer-containing cells in the population (Fig. 5). As shown in Figure 5A, GFP fluorescence increased linearly with increasing xylose concentration. In contrast, the number of polymer-containing cells increased dramatically over a very small range of xylose concentrations (Fig. 5B). At low expression levels (0.0025% or 0.005% xylose), most cells contained only diffuse, cytoplasmic fluorescence (<5% polymer-containing cells) (Fig. 5D,E). At higher xylose concentrations, the percentage of polymer-containing cells increased rapidly (the lower range is replotted in Fig. 5C, red circles). A twofold increase in xylose concentration (0.005%–0.01%) generated a nearly sixfold increase (5%–29%) in the percentage of cells containing polymers, and with another twofold increase in xylose (to 0.02%) the percentage reached 80%, where it plateaued. The plateau can be explained by the ∼20% of cells that never produce GFP due to loss of the plasmid during cell growth. Similar results were obtained when TubZ-GFP was induced in strains containing pBtoxis (data not shown). These results not only demonstrate that TubR is not required for TubZ-GFP assembly but also provide evidence of a critical concentration for assembly.
Figure 5.
TubZ-GFP has a critical concentration for assembly in B. thuringiensis. (A) Average fluorescence intensity versus xylose concentration for expression of wild-type TubZ-GFP. Protein expression was quantified by measuring fluorescence intensity of individual cells expressed per unit area. Averages (n = 100) for each xylose concentration are plotted. (B) Percentage of filament-containing cells plotted against xylose concentration for JP2209 expressing wild-type TubZ-GFP. Values from the lower range (0%–0.02%) are replotted in C for clarity. (C) Percentage of filament-containing cells plotted against xylose concentration for JP2209 expressing wild-type TubZ-GFP (red) and for JP2217 expressing TubZ(D269A)-GFP (blue). A narrow range of xylose concentrations (0%–0.02%) is plotted to compare the difference between the wild type and the mutant. (D–G) Images of JP2209 expressing wild-type TubZ-GFP after growth in 0.0025% (D), 0.005% (E), 0.01% (F), and 0.02% (G) xylose. Bar, 1 μm. (H–K) Images of JP2217 expressing TubZ(D269A)-GFP after growth in 0.001% (D), 0.0025% (E), 0.005% (F), and 0.01% (G) xylose.
A mutation in the putative GTPase domain of TubZ affects polymer formation
A mutation in the putative T7 loop of TubZ, changing aspartate to alanine at position 269, was constructed and characterized in vivo. In tubulins and FtsZ (Ec_FtsZ 212) this residue is required for GTPase activity and similar mutations eliminate hydrolysis but not GTP binding (Dai et al. 1994; Lu et al. 2001; Scheffers et al. 2001; Stricker and Erickson 2003). When TubZ(D269A)-GFP was expressed at various concentrations from the xylose promoter in B. thuringiensis (Fig. 5C,H–K), its assembly properties were dramatically different from those of wild-type TubZ-GFP. In contrast to the wild-type protein, which first accumulated as a soluble pool at low expression levels and then assembled into polymers with increasing frequency at higher expression levels, we detected very little diffuse, cytoplasmic fluorescence within the cell at any expression level. Instead, even when expressed at very low levels (0.001% xylose), a significant percentage of cells (15%) contained polymers (Fig. 5C, blue circles). These polymers were typically short (<0.5 μm in length) and occasionally branched (Fig. 5H). At a slightly higher level of expression (0.005% xylose), where wild-type still assembled very few (<5%) polymers, 50% of the cells expressing TubZ(D269A)-GFP contained filaments (Fig. 5C) that were longer (>1 μm), more branched, and sometimes trapped within the septum (Fig. 5J). The percentage of cells containing polymers reached a plateau near 80% (similar to wild type), but at a lower expression level (0.01% xylose) (Fig. 5C, blue circles), and many contained filaments trapped across the cell septum, inhibiting cell separation (Fig. 5K). Quantitation of GFP fluorescence confirms that expression levels are similar to those of wild type (data not shown), suggesting that the mutant protein accumulates polymers at a much lower protein concentration.
Wild-type TubZ-GFP filaments, whether expressed in E. coli or B. thuringiensis, are uniformly fluorescent. In the example shown in Figure 6A, left panel, quantitation of fluorescence intensity versus length indicates a <5% deviation over the length of a polymer and minimal variation between polymers in different cells (data not shown), suggesting that the filament itself is composed of a consistent number of protofilaments. In contrast, TubZ(D269A)-GFP filaments were nonuniform (Fig. 6A, right panel) and frequently branched at the ends (Fig. 5H–K) suggesting irregular lateral association of polymers.
Figure 6.
TubZ(D269A)-GFP forms irregular, static filaments. Polymer structure and dynamics were examined for TubZ(D269A)-GFP induced with 0.0025% xylose in strain JP2217. At this xylose concentration, filaments of the mutant are similar in length to wild-type TubZ-GFP. (A) A comparison of the uniformity of filaments in wild-type (left panel) versus TubZ(D269A) (right panel). Both filaments are within the focal plane. A three-dimensional graph showing the pixel intensity for each image is shown to highlight the differences between the two. (B) Rates of movement of TubZ(D269A)-GFP of two filaments (squares and diamonds) are presented as distance traveled versus time. Rate of movement of a wild-type TubZ-GFP filament (circles) is included for comparison. (C) Time-lapse microscopy of a TubZ(D269A)-GFP filament with time points indicated in seconds. Bar 1 μm. (D,E) FRAP analysis of TubZ(D269A)-GFP filaments. A small region (indicated by the red circle) was bleached with an argon laser and images captured at 40-sec intervals post-bleach are presented.
TubZ(D269A)-GFP filaments were also less dynamic than wild type (Fig. 6B). In time-lapse microscopy experiments, the filaments appeared to be completely static, with little to no movement of either end (Fig. 6B,C). However, it remained possible that the apparently static filaments were actually exchanging subunits from a cytoplasmic pool without obvious movement of the polymer itself, as has been reported for FtsZ-GFP rings (Stricker et al. 2002) and for AlfA-GFP filaments in B. subtilis (Becker et al. 2006). To test for this possibility, we performed FRAP analysis on TubZ(D269A)-GFP filaments (Fig. 6D,E). When either the center or the end of a polymer was bleached, there was no evidence of fluorescence recovery, indicating that neither subunit exchange within the filaments nor treadmilling in place occurred. Taken together, these observations suggest that the mutant protein retains the ability to assemble into polymers but is deficient in disassembly, and as a result, produces static filaments and exhibits a lower apparent critical concentration for assembly.
Dynamic TubZ is required for stability of pBtoxis
If TubZ participates in either replication or segregation of pBtoxis, then perturbing its expression level or polymerization dynamics might adversely affect the stability of pBtoxis. We first measured the stability of pBtoxis and pBtoxis∷tubZ-gfp after growth and sporulation in LB liquid cultures. Colonies resulting from germinated spores were tested for the presence of the plasmid using PCR and the results are shown in Table 1. In a wild-type strain, 100% of the spores contained pBtoxis demonstrating that this plasmid is very stably inherited. In contrast, for strains containing tubZ-gfp in place of tubZ, pBtoxis stability was temperature sensitive. JP2172 grew at a rate comparable with wild type at all temperatures tested (data not shown), but pBtoxis∷tubZ-gfp was lost at higher rates at 37°C (69% retention) compared with 25°C, where it was mostly stable (97%). This suggests that TubZ is important for maintenance of the native pBtoxis plasmid and is reminiscent of FtsZ-GFP fusions, which are also temperature sensitive in certain strains (Levin et al. 1999). All of our experiments were performed at 25°C or 30°C where the fusion protein appeared to be nearly as functional as wild type and we obtained similar results at both temperatures.
Table 1.
Stability of pBtoxis
aPercentage of colonies from germinated spores containing pBtoxis.
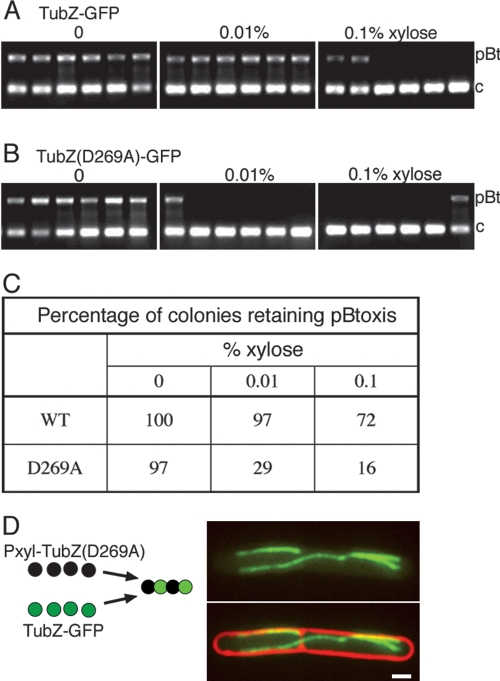
We attempted to integrate the TubZ(D269A)-GFP fusion into pBtoxis, but were unable to obtain the correct integrants, perhaps due to the deleterious effects of the mutant protein on the plasmid. We therefore expressed either TubZ-GFP or TubZ(D269A)-GFP in trans from a xylose controlled promoter in a strain containing pBtoxis. Based on the properties of TubZ(D269A)-GFP we anticipated that this mutant might have a dominant-negative effect on the function of the wild-type protein within the cell. These strains were grown with varying concentrations of xylose and the presence of pBtoxis was detected using PCR (Fig. 7). When wild-type TubZ-GFP was expressed in trans, pBtoxis was maintained efficiently both without xylose (100% retention) and with 0.01% xylose (97% retention) (Fig. 7A,C). However, overexpression of wild-type TubZ-GFP (0.1% xylose) resulted in the production of long twisted polymers (Fig. 5G) and significant plasmid loss, with only 72% of cells retaining pBtoxis (Fig. 7A,C). This suggests that proper expression levels of TubZ may be important for both normal polymerization dynamics and plasmid stability. Strikingly, when the TubZ(D269A)-GFP mutant protein was expressed in trans, pBtoxis was very unstable. In the presence of 0.01% xylose, only 29% of cells retained pBtoxis and this number decreased to 16% at the highest xylose concentration (0.1%) (Fig. 7B,C). Thus, TubZ(D269A)-GFP is a dominant-negative mutant protein that disrupts plasmid stability.
Figure 7.
Disruption of TubZ dynamics by overexpression or by mutation affects the stability of pBtoxis. (A–C) Colony PCR (n = 100) of strains expressing either wild-type TubZ-GFP or TubZ(D269A)-GFP after sporulation in the presence of the indicated concentrations of xylose was performed to detect the presence of pBtoxis. The larger PCR product (labeled “pBt”) was amplified from pBtoxis and the smaller PCR product (labeled “c”) was amplified from the B. thuringiensis chromosome as a control. (A) Representative agarose gels from PCR analysis of JP2219 (expressing wild-type TubZ-GFP). (B) Representative agarose gels from PCR analysis of JP2227 [expressing TubZ(D269A)-GFP]. (C) Summary of results from the PCR analysis. (D) TubZ(D269A) coassembles with wild-type TubZ-GFP. The left panel depicts the predicted behavior of the wild-type and mutant subunits when expressed in the same cell. The right panel is a representative image from fluorescent microscopy of JP2241 showing GFP alone (top) and GFP with FM4-64-stained membranes (bottom). Bar, 1 μm.
TubZ(D269A) coassembles with wild-type TubZ
One possible explanation for the strong dominant-negative effect of TubZ(D269A)-GFP on the stability of pBtoxis is that the mutant protein coassembles with the wild-type protein to form a mixed nonfunctional polymer. To test this possibility, we expressed TubZ(D269A) without a GFP fusion from the xylose promoter in trans (from pRL228) in a strain that also contained pBtoxis∷tubZ-GFP. Wild-type TubZ-GFP expressed from pBtoxis is normally very dynamic (Fig. 2); however, in this strain (JP2241), induction of TubZ(D269A) with 0.01% xylose caused the wild-type protein to form abnormal filaments that were static, branched, and trapped within septa (Fig. 7D). This behavior is similar to that seen for polymers of the mutant alone, demonstrating that the mutant and wild-type TubZ-GFP coassembled within the cell.
An upstream ORF, TubR, regulates TubZ levels
Well-studied plasmid segregation systems typically require not only a cytoskeletal protein but also a DNA-binding protein that tethers the plasmid to the cytoskeleton to mediate separation. Such a DNA-binding protein also frequently serves as a negative regulator of the operon encoding both proteins (Gerdes et al. 2004; Hayes and Barilla 2006). Immediately upstream of tubZ is a gene (tubR) encoding a protein with weak sequence similarity to helix–turn–helix DNA-binding proteins that could play this role. To test whether this protein might serve a regulatory role, we created a plasmid containing an in-frame deletion of tubR in the tubRZ operon and measured TubZ-GFP levels by Western blot analysis. These experiments were carried out in E. coli, since we were not able to move this mutation onto the native plasmid. In the absence of the tubR gene, TubZ-GFP levels were 4.5-fold higher than when tubR was present (Fig. 8). This mutation causing increased expression was fully complemented in trans by expressing tubR from an arabinose-inducible promoter on a compatible plasmid (Fig. 8). These results suggest that TubR is a negative regulator of TubZ levels.
Figure 8.
Regulation of TubZ by TubR. TubR was expressed in trans from an arabinose-inducible promoter in E. coli, and the effect on TubZ-GFP expression was determined by Western blotting using anti-GFP antibody. (Top) A diagram of the tubRZ operon on plasmids pRL165 (wild type) and pCUS12 (ΔtubR). (Bottom) Western blotting showing the relative amounts of TubZ-GFP expressed from either pRL165 or pCUS12 in the presence or absence of TubR expressed in trans from pRL186 (PBAD-tubR). TubZ-GFP expression in the presence of plasmid pBAD33 (PBAD) is included as a control. TubZ-GFP bands were quantified and values were adjusted for the amount of cells loaded on the gel. (Lane 5) Wild-type levels were arbitrarily set to 1.0 and the relative levels of other samples are noted below each band.
Discussion
We have characterized one member (TubZ) of a diverse protein family that is distantly related to both tubulin and FtsZ. Our studies demonstrate that TubZ-GFP from B. thuringiensis pBtoxis assembles dynamic polymers in vivo that share many characteristics with eukaryotic cytoskeletal proteins. TubZ-GFP polymers appear to be polarized, with distinct plus and minus ends. FRAP studies indicate that the polymers translocate through the cytoplasm by a treadmilling mechanism. Formation of TubZ-GFP polymers was highly dependent on protein concentration, suggesting the existence of a critical concentration for assembly, similar to polymer-forming proteins that assemble cooperatively such as tubulin and actin. A TubZ mutant predicted to be defective in GTP hydrolysis assembled polymers at expression levels significantly (five to 10 times) lower than the wild-type protein. Similar mutations in other cytoskeletal proteins also affect polymerization. For example, a mutation in ParM that reduces ATP hydrolysis simultaneously lowers the critical concentration for assembly and dramatically alters its assembly dynamics in vivo (Moller-Jensen et al. 2002; Garner et al. 2004). The mutant TubZ polymers also were static in both time-lapse microscopy and FRAP experiments. Thus, the TubZ(D269A) mutation appears to alter a key amino acid required for polymerization dynamics, as predicted from its position in the putative T7 loop that is critical for tubulin polymerization dynamics.
Several lines of evidence suggest that dynamic TubZ polymers are required for stability of plasmid pBtoxis in B. thuringiensis. First, expression of a dominant-negative TubZ(D269A) mutant significantly destabilized pBtoxis, even when the mutant protein was expressed at very low levels. We have shown that the mutant protein coassembles with wild type (Fig. 7), thereby trapping wild type in static, nonfunctional polymers. Second, overexpression of wild-type TubZ-GFP resulted in the formation of abnormally long and twisted polymers, and this also destabilized the plasmid. These two findings, suggesting that normal TubZ polymerization dynamics are required for plasmid stability, are somewhat parallel to results from plasmid partitioning systems in other bacteria. Mutations predicted to block ATP hydrolysis in the partitioning ATPases ParM and AlfA also trap the proteins in nonfunctional polymers and simultaneously disrupt plasmid segregation (Moller-Jensen et al. 2002; Becker et al. 2006). A third line of evidence that TubZ affects plasmid stability is that a plasmid (pBtoxis∷tubZ-GFP) expressing TubZ-GFP from its native promoter was temperature sensitive for stability. Thermal sensitivity is not surprising, since many FtsZ-GFP fusions are also temperature sensitive for function in vivo (Levin et al. 1999). Finally, a recent report suggests that the tubZ region is necessary for the formation of a mini-pBtoxis replicon, perhaps by playing a role in plasmid replication or segregation (Tang et al. 2006). Taken together, these results implicate TubZ as a novel type of bacterial tubulin required for maintenance of plasmid pBtoxis.
What specific role might TubZ play in plasmid stability? TubZ could play a role in plasmid replication, as suggested for RepX of plasmid pXO1 in B. anthracis (Tinsley and Khan 2006). However, given its dynamic properties, we favor a model in which TubZ is at least partly required for plasmid segregation. In other plasmid partitioning systems, polymerization dynamics are central to accurate DNA segregation. For example, ParM assembles dynamically unstable polymers that can be captured by ParR bound to parC DNA, resulting in subsequent polymerization of ParM bidirectionally between two plasmids pushing them apart (Moller-Jensen et al. 2003; Garner et al. 2004, 2007). By analogy, directional polymerization of TubZ might also be capable of driving plasmids apart, if the polymers were coupled to plasmid DNA by a DNA-binding protein. In keeping with the proposed role of TubZ in plasmid segregation, TubR, encoded immediately upstream of TubZ, is a trans-acting regulatory protein that also has DNA-binding activity as expected (data not shown). TubZ and TubR therefore have many of the properties of other plasmid segregation systems, including a cytoskeletal protein and a DNA-binding protein that governs expression of the operon, and could possibly tether the plasmid to the cytoskeletal element. We therefore suggest that they act together to form the central components of the TubZ stability region.
At least four distinct types of TubZ-like sequences are found on different Bacillus plasmids. These distant tubulin relatives might all play an important role in plasmid segregation, replication, or possibly both in a coupled manner. However, as shown in Figure 1, RepX, TubZ, and the other plasmid-encoded tubulin relatives are highly divergent from each other (<25% identity), leaving open the possibility that these proteins have different functions within the cell. The functions of the archaeal TubZ-like proteins, which are also distantly related and form their own separate, loosely related family, are unknown, but the fact that several of these are encoded on archaeal plasmids suggests that a function in plasmid stability might be conserved between this family and the Bacillus TubZ proteins.
The sequence similarities within the GTP-binding and hydrolysis domains between TubZ, tubulin, and FtsZ suggest that they share a common ancestor, yet the differences indicate that they diverged very early in the evolution of this superfamily, perhaps predating the split between Bacteria, Archaea, and Eukarya >2 billion years ago. TubZ defines a new prokaryotic cytoskeletal protein that may be an evolutionary link between the tubulin-based cytoskeletal proteins of eukaryotes and the FtsZ-based cytokinetic ring of prokaryotes.
Materials and methods
Phylogenetic analysis
Sequences were aligned using TCoffee and ClustalW and phylogenetic trees were constructed using Phylip version 3.6 (Felsenstein 1989). A bootstrap consensus tree of 100 trees is shown. The 100 trees were generated by resampling the data set and creating a distance matrix using a PAM matrix to assign various weights to amino acid substitutions. Neighbor joining was used to assemble the 100 distance matrices into 100 trees. Similar trees were obtained regardless of the methods used.
Figure 1B included the following sequences (accession numbers): E. coli FtsZ, (Ec) NP_308126; B. subtilis FtsZ (Bs), NP_389412; Haloarcula marismortui FtsZ1 (Hm1), YP_135405; H. marismortui FtsZ4 (Hm4), AAV48016; Halobacterium NRC-1 FtsZ4 (Hh4), NP_279378; H. marismortui FtsZ5 (Hm5), YP_137684; Halobacterium NRC-1 FtsZ5 (Hh5), NP_395771; B. megaterium pBM400 orf44 (Bm), NP_799504; B. cereus pBc218 orf139 (Bc), ZP_00236418; B. anthracis pXO1 orf45 (Ba), NP_052741; B. thuringiensis pBtoxis orf156 (Bt), CAD30186; Sus scrofa α tubulin (sA), P02550; S. scrofa β tubulin (SsB), P02554; Homo sapiens γ tubulin (HsG), NP_001061; H. sapiens ε tubulin (HsE), Q9UJT0; H. sapiens δ tubulin (HsD), Q9UJT1.
Accession numbers for additional sequences included in Figure 1A are provided in the Supplemental Material.
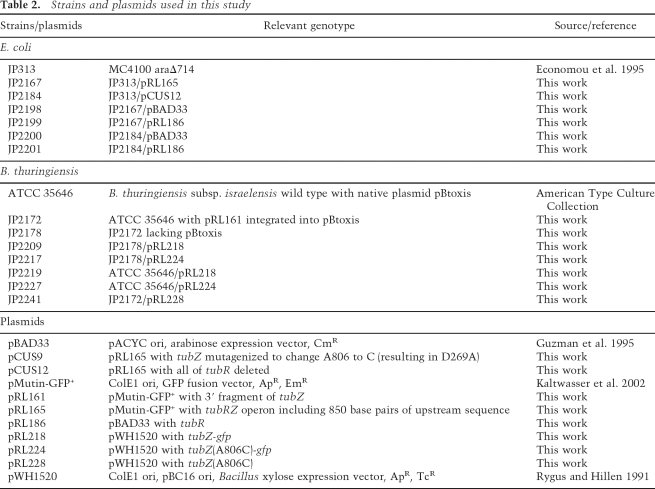
Bacterial strains, plasmids, and growth conditions
Strains and plasmids used in this study are listed in Table 2. See the Supplemental Material for details of plasmid constructions. B. thuringiensis strains were grown at either 25°C or 30°C in LB media supplemented when necessary with 0.5 μg/mL erythromycin or 10 μg/mL tetracycline. JP2178, a B. thuringiensis strain isogenic to JP2172 but lacking pBtoxis, was isolated during a screen for loss of pBtoxis from JP2172 grown at 37°C. The absence of pBtoxis was confirmed by PCR analysis and by the observation that sporulating cultures do not produce crystal toxin.
Table 2.
Strains and plasmids used in this study
The strains (JP2209 and JP2217) used for experiments involving modulated expression of TubZ were grown overnight at 30°C in LB 10 μg/mL tetracycline with (for microscopy) or without (for plasmid loss screen) 0.1% glucose. These cultures were then used to inoculate LB 10 μg/mL tetracycline containing varying concentrations of xylose (0%–0.2%) as indicated in the results. For microscopy, cultures were allowed to grow for ∼3–5 h at 30°C before imaging.
E. coli strains were typically grown at 37°C in LB media supplemented with 100 μg/mL ampicillin. When cephalexin was used, it was added to the media at a final concentration of 10 μg/mL and cells were allowed to continue growing for 1 h before imaging (Spratt 1977; Pogliano et al. 1997). For TubR repression studies, strains (JP2198, JP2199, JP2200, JP2201) were grown at 30°C in LB media supplemented with 100 μg/mL ampicillin and 30 μg/mL chloramphenicol, with or without 0.1% arabinose as indicated.
DNA manipulations
All DNA manipulations were done using standard protocols (Sambrook et al. 1989). Materials utilized included restriction enzymes and T4 ligase from New England Biolabs, dNTPs from Applied Biosystems, shrimp alkaline phosphatase from Promega, and DNA purification kits from Qiagen. Site-directed mutagenesis was performed as described (Sawano and Miyawaki 2000) using polynucleotide kinase and Taq ligase from New England Biolabs and Pfu turbo from Stratagene. PCR for cloning was done using the Triplemaster kit and PCR for screening was done using the Master Taq kit (Eppendorf). All PCRs were carried out in an Eppendorf Mastercycler EP. Plasmids were transformed into both E. coli and B. thuringiensis by electroporation using 0.2-cm cuvettes (Genessee) and a Bio-Rad Gene-pulser Xcell.
Plasmid stability assays
Cultures were grown for ∼15 generations in LB and then allowed to sporulate, after which nonsporulating cells were killed by heating for 20 min at 75°C. Dilutions of the spore suspension were plated onto LB and resulting colonies were used for PCR reactions directly. A fraction of a colony was added to 50 μL of ddH2O and heated in a microwave for 1 min at high power, after which 5 μL of this suspension was added to a 50-μL PCR reaction. PCR was performed as described above using the following primer pairs: Bt036-1 and Bt036-2, which amplify a region of gene 36 from pBtoxis, and BtiTrxBfwd and BtiTrxBrev, which amplify a region of the trxB gene from the chromosome. Between 100 and 400 colonies were measured for each condition.
Microscopy
Cells were affixed to a slide either with (E. coli) or without (B. thuringiensis) poly-L-lysine and visualized with or without 0.5 μg/mL membrane stain FM 4-64 (Pogliano et al. 1999). Images were collected using a DeltaVision optical sectioning microscope (Applied Precision) equipped with a Quantifiable Laser Module (QLM) for photobleaching experiments. Imaging of B. thuringiensis was performed in an environmental chamber at 30°C. Cells were taken directly from a fresh overnight colony and placed on an agarose pad containing 50% CH media (Pogliano et al. 1999). Optical sections were collected with a spacing of 0.15 μm. E.coli cells expressing TubZ-GFP (JP2167) were imaged at 37°C. For photobleaching experiments, three prebleach images were collected, after which cells were exposed for 0.5 sec to a laser (488 nm) at 50% power and post-bleach images were collected at the indicated time points. The images of TubZ are presented without deconvolution processing; however, images of stained membranes were deconvolved for clarity.
Western blotting
Proteins were separated on 10% polyacrylamide gels, blotted to PVDF membranes, and probed with anti-GFP antibody (Invitrogen) and anti-mouse IgG Cy5 conjugate (Jackson ImmunoResearch). The Cy5 conjugate was then detected using a Typhoon imager (GE Healthcare).
Acknowledgments
We are grateful to Dan Broder, Amber Dance, and Kit Pogliano for critical reading of the manuscript and Arshad Desai for helpful discussions. This work was supported by a grant from the NIH (R01GM073898) to J.P.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1546107
References
- Adachi S., Hori K., Hiraga S., Hori K., Hiraga S., Hiraga S. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J. Mol. Biol. 2006;356:850–863. doi: 10.1016/j.jmb.2005.11.088. [DOI] [PubMed] [Google Scholar]
- Addinall S.G., Holland B., Holland B. The tubulin ancestor, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J. Mol. Biol. 2002;318:219–236. doi: 10.1016/S0022-2836(02)00024-4. [DOI] [PubMed] [Google Scholar]
- Amos L.A., van den Ent F., Lowe J., van den Ent F., Lowe J., Lowe J. Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr. Opin. Cell Biol. 2004;16:24–31. doi: 10.1016/j.ceb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Barilla D., Rosenberg M.F., Nobbmann U., Hayes F., Rosenberg M.F., Nobbmann U., Hayes F., Nobbmann U., Hayes F., Hayes F. Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. EMBO J. 2005;24:1453–1464. doi: 10.1038/sj.emboj.7600619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker E., Herrera N.C., Gunderson F.Q., Derman A.I., Dance A.L., Sims J., Larsen R.A., Pogliano J., Herrera N.C., Gunderson F.Q., Derman A.I., Dance A.L., Sims J., Larsen R.A., Pogliano J., Gunderson F.Q., Derman A.I., Dance A.L., Sims J., Larsen R.A., Pogliano J., Derman A.I., Dance A.L., Sims J., Larsen R.A., Pogliano J., Dance A.L., Sims J., Larsen R.A., Pogliano J., Sims J., Larsen R.A., Pogliano J., Larsen R.A., Pogliano J., Pogliano J. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 2006;25:5919–5931. doi: 10.1038/sj.emboj.7601443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C., O’Neil S., Ben-Dov E., Jones A.F., Murphy L., Quail M.A., Holden M.T., Harris D., Zaritsky A., Parkhill J., O’Neil S., Ben-Dov E., Jones A.F., Murphy L., Quail M.A., Holden M.T., Harris D., Zaritsky A., Parkhill J., Ben-Dov E., Jones A.F., Murphy L., Quail M.A., Holden M.T., Harris D., Zaritsky A., Parkhill J., Jones A.F., Murphy L., Quail M.A., Holden M.T., Harris D., Zaritsky A., Parkhill J., Murphy L., Quail M.A., Holden M.T., Harris D., Zaritsky A., Parkhill J., Quail M.A., Holden M.T., Harris D., Zaritsky A., Parkhill J., Holden M.T., Harris D., Zaritsky A., Parkhill J., Harris D., Zaritsky A., Parkhill J., Zaritsky A., Parkhill J., Parkhill J. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensissubsp. israelensis. Appl. Environ. Microbiol. 2002;68:5082–5095. doi: 10.1128/AEM.68.10.5082-5095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouet J.Y., Ah-Seng Y., Benmeradi N., Lane D., Ah-Seng Y., Benmeradi N., Lane D., Benmeradi N., Lane D., Lane D. Polymerization of SopA partition ATPase: Regulation by DNA binding and SopB. Mol. Microbiol. 2007;63:468–481. doi: 10.1111/j.1365-2958.2006.05537.x. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 2006;70:888–909. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-Lopez R., Errington J., Errington J. The bacterial cytoskeleton: In vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev. Cell. 2003a;4:19–28. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R., Errington J., Errington J. A dynamic bacterial cytoskeleton. Trends Cell Biol. 2003b;13:577–583. doi: 10.1016/j.tcb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R., Formstone A., Li Y., Ehrlich S.D., Noirot P., Errington J., Formstone A., Li Y., Ehrlich S.D., Noirot P., Errington J., Li Y., Ehrlich S.D., Noirot P., Errington J., Ehrlich S.D., Noirot P., Errington J., Noirot P., Errington J., Errington J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev. Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Dai K., Mukherjee A., Xu Y., Lutkenhaus J., Mukherjee A., Xu Y., Lutkenhaus J., Xu Y., Lutkenhaus J., Lutkenhaus J. Mutations in ftsZthat confer resistance to SulA affect the interaction of FtsZ with GTP. J. Bacteriol. 1994;176:130–136. doi: 10.1128/jb.176.1.130-136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajkovic A., Lutkenhaus J., Lutkenhaus J. Z ring as executor of bacterial cell division. J. Mol. Microbiol. Biotechnol. 2006;11:140–151. doi: 10.1159/000094050. [DOI] [PubMed] [Google Scholar]
- Defeu Soufo H.J., Graumann P.L., Graumann P.L. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 2004;5:789–794. doi: 10.1038/sj.embor.7400209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeu Soufo H.J., Graumann P.L., Graumann P.L. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 2005;6:10. doi: 10.1186/1471-2121-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J., Mitchison T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Ebersbach G., Ringgaard S., Moller-Jensen J., Wang Q., Sherratt D.J., Gerdes K., Ringgaard S., Moller-Jensen J., Wang Q., Sherratt D.J., Gerdes K., Moller-Jensen J., Wang Q., Sherratt D.J., Gerdes K., Wang Q., Sherratt D.J., Gerdes K., Sherratt D.J., Gerdes K., Gerdes K. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol. Microbiol. 2006;61:1428–1442. doi: 10.1111/j.1365-2958.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- Economou A., Pogliano J.A., Beckwith J., Oliver D.B., Wickner W., Pogliano J.A., Beckwith J., Oliver D.B., Wickner W., Beckwith J., Oliver D.B., Wickner W., Oliver D.B., Wickner W., Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- Errington J. Dynamic proteins and a cytoskeleton in bacteria. Nat. Cell Biol. 2003;5:175–178. doi: 10.1038/ncb0303-175. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—Phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Fogel M.A., Waldor M.K., Waldor M.K. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes & Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadde S., Heald R., Heald R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 2004;14:R797–R805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Garner E.C., Campbell C.S., Mullins R.D., Campbell C.S., Mullins R.D., Mullins R.D. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 2004;306:1021–1025. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- Garner E.C., Campbell C.S., Weibel D.B., Mullins R.D., Campbell C.S., Weibel D.B., Mullins R.D., Weibel D.B., Mullins R.D., Mullins R.D. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Moller-Jensen J., Ebersbach G., Kruse T., Nordstrom K., Moller-Jensen J., Ebersbach G., Kruse T., Nordstrom K., Ebersbach G., Kruse T., Nordstrom K., Kruse T., Nordstrom K., Nordstrom K. Bacterial mitotic machineries. Cell. 2004;116:359–366. doi: 10.1016/s0092-8674(04)00116-3. [DOI] [PubMed] [Google Scholar]
- Gitai Z., Dye N.A., Reisenauer A., Wachi M., Shapiro L., Dye N.A., Reisenauer A., Wachi M., Shapiro L., Reisenauer A., Wachi M., Shapiro L., Wachi M., Shapiro L., Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Goehring N.W., Beckwith J., Beckwith J. Diverse paths to midcell: Assembly of the bacterial cell division machinery. Curr. Biol. 2005;15:R514–R526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Guzman L.M., Belin D., Carson M.J., Beckwith J., Belin D., Carson M.J., Beckwith J., Carson M.J., Beckwith J., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F., Barilla D., Barilla D. The bacterial segrosome: A dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- Jenkins C., Samudrala R., Anderson I., Hedlund B.P., Petroni G., Michailova N., Pinel N., Overbeek R., Rosati G., Staley J.T., Samudrala R., Anderson I., Hedlund B.P., Petroni G., Michailova N., Pinel N., Overbeek R., Rosati G., Staley J.T., Anderson I., Hedlund B.P., Petroni G., Michailova N., Pinel N., Overbeek R., Rosati G., Staley J.T., Hedlund B.P., Petroni G., Michailova N., Pinel N., Overbeek R., Rosati G., Staley J.T., Petroni G., Michailova N., Pinel N., Overbeek R., Rosati G., Staley J.T., Michailova N., Pinel N., Overbeek R., Rosati G., Staley J.T., Pinel N., Overbeek R., Rosati G., Staley J.T., Overbeek R., Rosati G., Staley J.T., Rosati G., Staley J.T., Staley J.T. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc. Natl. Acad. Sci. 2002;99:17049–17054. doi: 10.1073/pnas.012516899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.B., Gerdes K., Gerdes K. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR–parCcomplex. J. Mol. Biol. 1997;269:505–513. doi: 10.1006/jmbi.1997.1061. [DOI] [PubMed] [Google Scholar]
- Jones L., Carballido-Lopez R., Errington J., Carballido-Lopez R., Errington J., Errington J. Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Kaltwasser M., Wiegert T., Schumann W., Wiegert T., Schumann W., Schumann W. Construction and application of epitope- and green fluorescent protein-tagging integration vectors for Bacillus subtilis. Appl. Environ. Microbiol. 2002;68:2624–2628. doi: 10.1128/AEM.68.5.2624-2628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A., Li Z., Newman D.K., Jensen G.J., Li Z., Newman D.K., Jensen G.J., Newman D.K., Jensen G.J., Jensen G.J. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Leonard T.A., Moller-Jensen J., Lowe J., Moller-Jensen J., Lowe J., Lowe J. Towards understanding the molecular basis of bacterial DNA segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:523–535. doi: 10.1098/rstb.2004.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P.A., Kurtser I.G., Grossman A.D., Kurtser I.G., Grossman A.D., Grossman A.D. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim G.E., Derman A.I., Pogliano J., Derman A.I., Pogliano J., Pogliano J. Bacterial DNA segregation by dynamic SopA polymers. Proc. Natl. Acad. Sci. 2005;102:17658–17663. doi: 10.1073/pnas.0507222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J., Li H., Downing K.H., Nogales E., Li H., Downing K.H., Nogales E., Downing K.H., Nogales E., Nogales E. Refined structure of α β-tubulin at 3.5 A resolution. J. Mol. Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- Lu C., Stricker J., Erickson H.P., Stricker J., Erickson H.P., Erickson H.P. Site-specific mutations of FtsZ—Effects on GTPase and in vitro assembly. BMC Microbiol. 2001;1:7. doi: 10.1186/1471-2180-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. Dynamic proteins in bacteria. Curr. Opin. Microbiol. 2002;5:548–552. doi: 10.1016/s1369-5274(02)00376-4. [DOI] [PubMed] [Google Scholar]
- Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie K.A., Lowe J., Lowe J. Dynamic filaments of the bacterial cytoskeleton. Annu. Rev. Biochem. 2006;75:467–492. doi: 10.1146/annurev.biochem.75.103004.142452. [DOI] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M., Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J., Lowe J., Lowe J. Increasing complexity of the bacterial cytoskeleton. Curr. Opin. Cell Biol. 2005;17:75–81. doi: 10.1016/j.ceb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J., Jensen R., Lowe J., Gerdes K., Jensen R., Lowe J., Gerdes K., Lowe J., Gerdes K., Gerdes K. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 2002;21:3119–3127. doi: 10.1093/emboj/cdf320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Jensen J., Borch J., Dam M., Jensen R.B., Roepstorff P., Gerdes K., Borch J., Dam M., Jensen R.B., Roepstorff P., Gerdes K., Dam M., Jensen R.B., Roepstorff P., Gerdes K., Jensen R.B., Roepstorff P., Gerdes K., Roepstorff P., Gerdes K., Gerdes K. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol. Cell. 2003;12:1477–1487. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- Nogales E. Structural insight into microtubule function. Annu. Rev. Biophys. Biomol. Struct. 2001;30:397–420. doi: 10.1146/annurev.biophys.30.1.397. [DOI] [PubMed] [Google Scholar]
- Nogales E., Downing K.H., Amos L.A., Lowe J., Downing K.H., Amos L.A., Lowe J., Amos L.A., Lowe J., Lowe J. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 1998a;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- Nogales E., Wolf S.G., Downing K.H., Wolf S.G., Downing K.H., Downing K.H. Structure of the α β tubulin dimer by electron crystallography. Nature. 1998b;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Pogliano J., Pogliano K., Weiss D., Losick R., Beckwith J., Pogliano K., Weiss D., Losick R., Beckwith J., Weiss D., Losick R., Beckwith J., Losick R., Beckwith J., Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J., Osborne N., Sharp M.D., Abanes-DeMello A., Perez A., Sun Y.-L., Pogliano K., Osborne N., Sharp M.D., Abanes-DeMello A., Perez A., Sun Y.-L., Pogliano K., Sharp M.D., Abanes-DeMello A., Perez A., Sun Y.-L., Pogliano K., Abanes-DeMello A., Perez A., Sun Y.-L., Pogliano K., Perez A., Sun Y.-L., Pogliano K., Sun Y.-L., Pogliano K., Pogliano K. A vital stain for studying membrane dynamics in bacteria: A novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel N., Santini C.L., Bernadac A., Fukumori Y., Wu L.F., Santini C.L., Bernadac A., Fukumori Y., Wu L.F., Bernadac A., Fukumori Y., Wu L.F., Fukumori Y., Wu L.F., Wu L.F. Biogenesis of actin-like bacterial cytoskeletal filaments destined for positioning prokaryotic magnetic organelles. Proc. Natl. Acad. Sci. 2006;103:17485–17489. doi: 10.1073/pnas.0603760103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L., Levin P.A., Levin P.A. Assembly dynamics of the bacterial cell division protein FTSZ:Poised at the edge of stability. Annu. Rev. Microbiol. 2003;57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rygus T., Hillen W., Hillen W. Inducible high-level expression of heterologous genes in Bacillus megaterium using the regulatory elements of the xylose-utilization operon. Appl. Microbiol. Biotechnol. 1991;35:594–599. doi: 10.1007/BF00169622. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T., Fritsch E.F., Maniatis T., Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sawano A., Miyawaki A., Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers D.J., de Wit J.G., den Blaauwen T., Driessen A.J., de Wit J.G., den Blaauwen T., Driessen A.J., den Blaauwen T., Driessen A.J., Driessen A.J. Substitution of a conserved aspartate allows cation-induced polymerization of FtsZ. FEBS Lett. 2001;494:34–37. doi: 10.1016/s0014-5793(01)02310-9. [DOI] [PubMed] [Google Scholar]
- Shih Y.L., Rothfield L., Rothfield L. The bacterial cytoskeleton. Microbiol. Mol. Biol. Rev. 2006;70:729–754. doi: 10.1128/MMBR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag C.A., Staley J.T., Erickson H.P., Staley J.T., Erickson H.P., Erickson H.P. In vitro assembly and GTPase hydrolysis by bacterial tubulins BtubA and BtubB. J. Cell Biol. 2005;169:233–238. doi: 10.1083/jcb.200410027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B.G. Properties of the penicillin binding proteins of Escherichia coli K12. Eur. J. Biochem. 1977;72:341–342. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- Stricker J., Erickson H.P., Erickson H.P. In vivo characterization of Escherichia coli ftsZmutants: Effects on Z-ring structure and function. J. Bacteriol. 2003;185:4796–4805. doi: 10.1128/JB.185.16.4796-4805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J., Maddox P., Salmon E.D., Erickson H.P., Maddox P., Salmon E.D., Erickson H.P., Salmon E.D., Erickson H.P., Erickson H.P. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M., Bideshi D.K., Park H.W., Federici B.A., Bideshi D.K., Park H.W., Federici B.A., Park H.W., Federici B.A., Federici B.A. Minireplicon from pBtoxis of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 2006;72:6948–6954. doi: 10.1128/AEM.00976-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley E., Khan S.A., Khan S.A. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J. Bacteriol. 2006;188:2829–2835. doi: 10.1128/JB.188.8.2829-2835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F., Amos L., Lowe J., Amos L., Lowe J., Lowe J. Bacterial ancestry of actin and tubulin. Curr. Opin. Microbiol. 2001a;4:634–638. doi: 10.1016/s1369-5274(01)00262-4. [DOI] [PubMed] [Google Scholar]
- van den Ent F., Amos L., Lowe J., Amos L., Lowe J., Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001b;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- van den Ent F., Moller-Jensen J., Amos L.A., Gerdes K., Lowe J., Moller-Jensen J., Amos L.A., Gerdes K., Lowe J., Amos L.A., Gerdes K., Lowe J., Gerdes K., Lowe J., Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan S., Wickstead B., Gull K., Addinall S.G., Wickstead B., Gull K., Addinall S.G., Gull K., Addinall S.G., Addinall S.G. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 2004;58:19–29. doi: 10.1007/s00239-003-2523-5. [DOI] [PubMed] [Google Scholar]
- Wang X., Huang J., Mukherjee A., Cao C., Lutkenhaus J., Huang J., Mukherjee A., Cao C., Lutkenhaus J., Mukherjee A., Cao C., Lutkenhaus J., Cao C., Lutkenhaus J., Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]