Abstract
Autophagy is a bulk degradation process that promotes survival under metabolic stress, but it can also be a means of cell death if executed to completion. Monoallelic loss of the essential autophagy gene beclin1 causes susceptibility to metabolic stress, but also promotes tumorigenesis. This raises the paradox that the loss of a survival pathway enhances tumor growth, where the exact mechanism is not known. Here, we show that compromised autophagy promoted chromosome instability. Failure to sustain metabolism through autophagy was associated with increased DNA damage, gene amplification, and aneuploidy, and this genomic instability may promote tumorigenesis. Thus, autophagy maintains metabolism and survival during metabolic stress that serves to protect the genome, providing an explanation for how the loss of a survival pathway leads to tumor progression. Identification of this novel role of autophagy may be important for rational chemotherapy and therapeutic exploitation of autophagy inducers as potential chemopreventive agents.
Keywords: Autophagy, beclin1, genomic instability, apoptosis, cancer
Autophagy is an evolutionarily conserved catabolic process involving regulated turnover and elimination of proteins and cellular organelles, such as peroxisomes, mitochondria, and endoplasmic reticulum, through the lysosomal degradation pathway (Mizushima 2005). The process of autophagy is characterized by the formation of double-membrane cytosolic vesicles, known as autophagosomes, that are essential for the lysosomal targeting of these organelles. In yeast, a number of autophagy-related genes (referred to as atg) have been identified that regulate the formation of autophagosomes and the autophagy process (Klionsky et al. 2003). Several mammalian homologs of these yeast genes have been identified (Levine and Klionsky 2004), among which the essential autophagy genes atg5 and atg7 have been most informative in demonstrating a role for autophagy in maintaining metabolism and homeostasis in mammalian development.
Autophagy, constitutively active at low levels, is robustly activated under metabolic stress. Autophagy plays an important role in development, as mice deficient in autophagy due to complete deficiency of beclin1 (atg6/vps30), another essential autophagy gene, die early in embryogenesis (Yue et al. 2003). Mice lacking atg5 fail to survive the neonatal starvation period and die perinatally, suggesting that autophagy plays an important role in the maintenance of energy homeostasis (Kuma et al. 2004). Thus, autophagy functions as an alternative cellular energy source to maintain normal metabolism during development and starvation by recycling cytoplasm and macromolecules (Jin and White 2007). Furthermore, targeted deletion of atg5 and atg7 in the central nervous system results in accumulation of polyubiquitinated proteins leading to neurodegeneration, revealing a housekeeping role for autophagy in the regulation of long-lived or damaged proteins to prevent neurodegeneration (Komatsu et al. 2005; Hara et al. 2006). The protective effects of autophagy also include immune-related functions such as defense against pathogens (Kirkegaard et al. 2004) and T lymphocyte development (Pua et al. 2006). Thus, autophagy plays a critical role in development, survival, and maintenance of homeostasis.
Despite its predominant role as a survival pathway, progressive autophagy can result in cell death if allowed to proceed to completion under persistent stress and during development. In Drosophila, autophagic cell death plays an important role during salivary gland development (Baehrecke 2003). In mammals, autophagy proteins Beclin1 and Atg7 are necessary for the autophagic cell death induced by the caspase-8 inhibitor zVAD-fmk in L929 cells as well as in human U937 cells (Yu et al. 2004). Furthermore, apoptosis-resistant bax−/−/bak−/− mouse embryonic fibroblasts (MEFs) undergo autophagic cell death in response to apoptosis inducers in a Beclin1- and Atg5-dependent manner (Shimizu et al. 2004). Autophagy-like cell death is visualized in Alzheimer’s (Cataldo et al. 1994) and Parkinson’s (Anglade et al. 1997) and other neurodegenerative diseases (Klionsky and Emr 2000; Gomez-Santos et al. 2003). However, evidence in support of autophagy as a cell death mechanism is limited mostly to pharmacological induction of autophagy in cell lines in vitro, and direct evidence in a physiological context is frequently lacking. Moreover, it is not clear whether the induction of autophagy under the above circumstances represents the activation of a specific pathway for cell death or merely a failed attempt to promote cell survival under stress.
Quite contradictory to its survival-promoting function, defective autophagy is implicated in tumorigenesis, as haploinsuffieciency in the essential autophagy gene beclin1 is commonly observed in sporadic human ovarian, breast, and prostate cancers (Aita et al. 1999), and allelic loss of beclin1 promotes the incidence of sporadic malignancies in mice (Qu et al. 2003; Yue et al. 2003). In addition, oncogenic signals such as that of the Class I phosphatidylinositol-3-kinase (PI3K) pathway suppress autophagy, but if or how this contributes to tumorigenesis is not known.
We have shown that defects in apoptosis due to either Bax/Bak deficiency or Bcl-2 expression reveal autophagy in response to metabolic stress, which is required for cell survival (Degenhardt et al. 2006). Allelic loss of beclin1 impairs both constitutive and stress-induced autophagy, causing susceptibility to metabolic stress in immortalized baby mouse kidney epithelial (iBMK) cells (Degenhardt et al. 2006). Despite impairment in autophagy-mediated survival, the autophagy-defective beclin1+/− iBMK cells are more tumorigenic than the wild-type cells, and the tumorigenicity is further accentuated by defects in apoptosis conferred by Bcl-2 expression (Degenhardt et al. 2006). In tumors, autophagy localizes to metabolically stressed regions, suggesting that tumor cells exploit autophagy to survive metabolic stress (Degenhardt et al. 2006). Thus, defects in apoptosis manifest an autophagy-mediated survival pathway, and concomitant defects in apoptosis and autophagy render cells susceptible to metabolic stress while promoting tumorigenesis (Nelson et al. 2004; Degenhardt et al. 2006). However, these two apparently contradictory roles of autophagy lead to the paradox that the loss of a survival pathway promotes tumorigenesis.
Despite several studies linking autophagy to development, protein turnover, maintenance of cell organelles, and energy homeostasis, there is no satisfactory explanation for the intuitively contradictory roles of autophagy as a survival pathway and a tumor suppressor mechanism. We report here for the first time that autophagy can function to protect the genome. Defects in autophagy, conferred by either allelic loss of beclin1 or deficiency in atg5, lead to impaired survival in metabolic stress and an enhanced DNA damage response. Furthermore, loss of Beclin1 function promotes gene amplification and chromosomal instability, resulting in aneuploidy, all of which are hallmarks of cancer, suggesting that autophagy plays a major role in the maintenance of genomic integrity.
Results
Allelic loss of beclin1 renders iBMK cells susceptible to metabolic stress in vitro and impairs autophagy in vivo
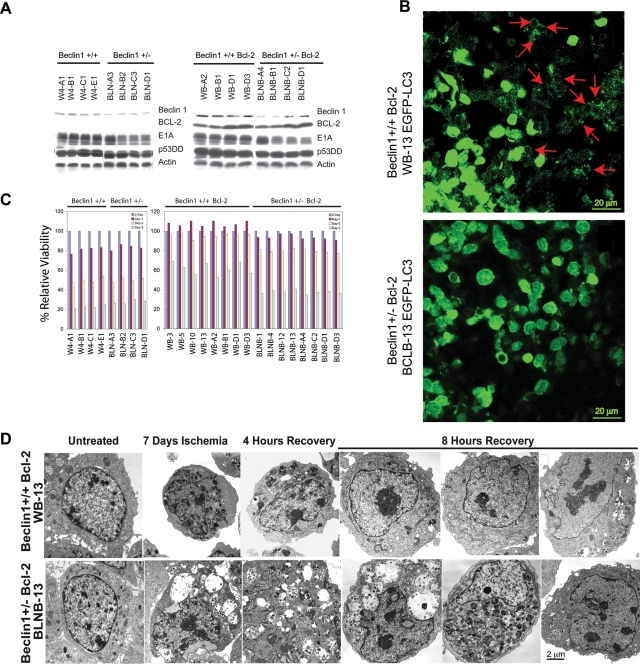
In order to understand the role of impaired autophagy conferred by allelic loss of beclin1 in tumorigenesis, primary baby mouse kidney epithelial cells were immortalized from mutant mice, expanding the panel of independent cell lines originally isolated (Degenhardt et al. 2006). Several independent clones of these iBMK cells were then also engineered to express the anti-apoptotic protein Bcl-2. All expressed the immortalization genes (E1A and p53DD), and beclin1+/− cells showed reduced levels of Beclin1 compared with beclin1+/+ iBMK cells (Fig. 1A).
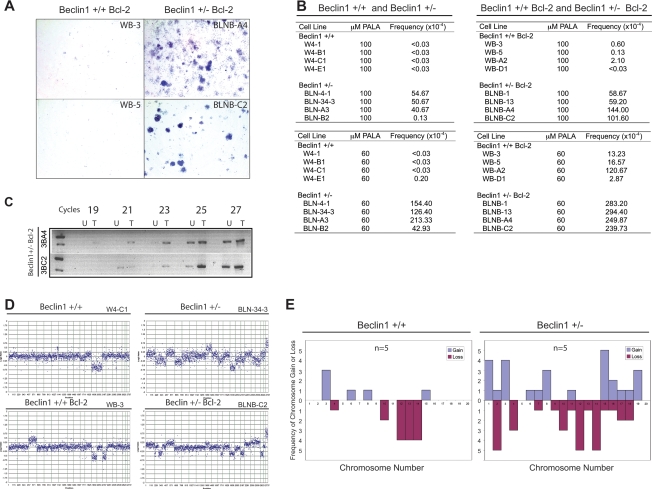
Figure 1.
Allelic loss of beclin1 impairs cell survival and recovery in vitro and results in defective autophagy in vivo. A panel of beclin1+/+ and beclin1+/− iBMK cells with and without Bcl-2 were subjected to ischemia viability assay and recovery was analyzed for ultrastructure morphology by EM. (A) Western blots showing protein expression in beclin1+/+ and beclin1+/− iBMK cells with and without Bcl-2. (B) Allelic loss of beclin1 impairs autophagy in tumors in vivo. beclin1+/+ and beclin1+/− iBMK cells were engineered to stably express EGFP-LC3 and were injected subcutaneously into nude mice for tumor formation. Photomicrograph of beclin1+/+ and beclin1+/− tumor sections 1 d post-implantation showing robust activation of autophagy in the wild-type beclin1+/+ iBMK cells and impaired autophagy in the autophagy-defective beclin1+/− iBMK cells. (C) Allelic loss of beclin1 results in susceptibility to metabolic stress. Viability of beclin1+/+ and beclin1+/− iBMK cells with and with out Bcl-2 showing diminished survival of the beclin1+/− iBMK cells in response to ischemia. (D) Electron micrograph showing impaired recovery of beclin1+/− iBMK cells expressing Bcl-2 following ischemic stress.
Previous work demonstrated that allelic loss of beclin1 in iBMK cells results in a profound defect in autophagy that confers increased susceptibility to metabolic stress in apoptosis-defective cells (Degenhardt et al. 2006), which was reproduced in all the cell lines in the panel (Fig. 1C; see below). This enhanced sensitivity to metabolic stress was also associated with enhanced tumorigenicity, as previously reported (Degenhardt et al. 2006). To measure autophagy, beclin1+/+and beclin1+/− cells stably expressing Bcl-2 were engineered to express the autophagy marker EGFP-LC3 and were monitored in vivo for cytoplasmic-to-membrane translocation, indicative of autophagy. Autophagy was robustly activated in beclin1+/+ iBMK cells overexpressing Bcl-2, which was profoundly diminished due to allelic loss of beclin1 (Degenhardt et al. 2006). Wild-type iBMK cells expressing Bcl-2 demonstrated massive induction of autophagy in the tumor tissue, indicated by punctate cytoplasmic LC3 translocation (Fig. 1B, red arrows). In stark contrast, tumors from the autophagy-defective beclin1+/− iBMK cells expressing Bcl-2 displayed predominantly diffuse localization, indicative of an autophagy defect (Fig. 1B). As tumor allografts suffer from a lack of oxygen and nutrition at early stages of implantation before adequate vascularization is established (Nelson et al. 2004), this observation suggests that tumor cells exploit the autophagy pathway to survive intermittent metabolic stress in vivo. This is consistent with our earlier observation that defects in apoptosis reveal the autophagy survival pathway, and autophagy localizes to metabolically stressed regions in tumors (Degenhardt et al. 2006).
In order to confirm that beclin1+/− iBMK cells have impaired autophagic survival, viability under ischemia was determined as described previously (Degenhardt et al. 2006) using a 1% oxygen gas mixture and medium without glucose that simulates metabolic stress conditions in the tumor microenvironment in vitro (Nelson et al. 2004). There was no significant difference in sensitivity observed in the apoptosis-competent beclin1+/+ and beclin1+/− cells without Bcl-2. In contrast, beclin1+/− cells expressing Bcl-2 showed substantial sensitivity to ischemia when compared with wild-type cells (Fig. 1C), suggesting that metabolic stress is a potent trigger for apoptosis, which readily eliminated apoptosis-competent cells before autophagy could be manifested.
Allelic loss of beclin1 impairs recovery from metabolic stress
To further understand how this impaired autophagy response to metabolic stress may impact homeostasis, beclin1+/+ and beclin1+/− iBMK cells expressing Bcl-2 were subjected to ischemia for 7 d and allowed to recover, and ultrastructural morphology was monitored by electron microscopy. Wild-type beclin1+/+ iBMK cells expressing Bcl-2 transitioned from normal to a condensed morphology by the end of 7 d in ischemia (Fig. 1D) and retained a high level of viability under these conditions (Degenhardt et al. 2006). Most wild-type cells regained their normal morphology by 4 h of recovery, and almost all did so by 8 h. In stark contrast, mutant beclin1+/− iBMK cells expressing Bcl-2 typically showed enhanced necrotic morphology and cell death with highly vacuolated cytoplasm, and there were fewer signs of reversion to normal morphology even after 8 h of recovery (Fig. 1D). These observations suggested that autophagy not only promotes survival but also facilitates recovery from metabolic stress, suggesting that autophagy is a specific process of adaptation to intermittent nutrient depletion to enable high-fidelity restoration of normal cellular functions.
Autophagy suppresses the DNA damage in response to metabolic stress
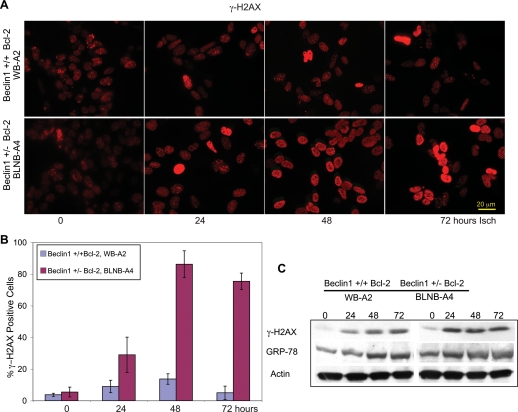
It was unclear how defects in autophagy could diminish survival while enhancing tumorigenesis. One possible explanation is that by impairing survival to metabolic stress, defective autophagy may increase DNA damage, and thereby the mutation rate in the remaining viable cells, facilitating tumor progression. Analogously, mutations in genes critical for DNA repair create sensitivity to DNA damage but enhance tumorigenesis by increasing the mutation rate (Symington 2002). In order to test this hypothesis, we evaluated wild-type and autophagy-deficient iBMK cell lines expressing Bcl-2 for Ser 139 phosphorylation of histone H2AX (γ-H2AX), signifying DNA double-strand breaks (DSBs) and activation of the DNA damage response in response to metabolic stress (ischemia). Both beclin1+/+ and beclin1+/− iBMK cells expressing Bcl-2 showed a low basal level of nuclear γ-H2AX staining (Fig. 2A,B). Wild-type beclin1+/+ iBMK cells expressing Bcl-2 showed a modest induction of γ-H2AX levels in response to metabolic stress gradually over the course of 3 d (Fig. 2A,B). In contrast, autophagy-defective beclin1+/− iBMK cells expressing Bcl-2 showed a remarkable increase in levels of γ-H2AX staining in response to metabolic stress (Fig. 2B). Analysis of γ-H2AX levels by Western blotting provided similar results, showing elevated γ-H2AX levels in the beclin1+/− cells expressing Bcl-2 (Fig. 2C). As a defect in autophagy may also be associated with other forms of stress such as induction of the unfolded protein response (UPR), levels of GRP-78, an early marker for UPR, were also examined. However, no difference in GRP-78 induction in ischemia was evident in wild-type and beclin1 mutant cells at these time points (Fig. 2C). Thus, the increased levels of γ-H2AX in the beclin1 mutant cells suggested that defects in autophagy may promote DNA damage induced by metabolic stress.
Figure 2.
Allelic loss of beclin1 promotes DNA damage response under metabolic stress. Representative beclin1+/+ (WB-A2) and beclin1+/− (BLNB-A4) iBMK cell lines expressing Bcl-2 were subjected to ischemia stress and the DNA damage response was measured by the induction of γ-H2AX by immunofluorescence and Western blotting. (A) Representative photomicrographs showing accumulation of γ-H2AX in the beclin1+/− iBMK cell line expressing Bcl-2 (BLNB-A4) as a marker of DNA damage. (B) Quantitation of γ-H2AX-positive cells in beclin1+/+ (WB-A2) and beclin1+/− (BLNB-A4) iBMK cell lines expressing Bcl-2 in A. The percentage of nuclei with positive γ-H2AX foci was tabulated. Data represent mean ± SD. Note enhanced accumulation of γ-H2AX in the beclin1+/− iBMK cell line expressing Bcl-2 (BLNB-A4) compared with the wild type. (C) Western blot time course showing γ-H2AX (marker of DNA damage) and GRP-78 (marker of UPR) in beclin1+/+ (WB-A2) and beclin1+/− (BLNB-A4) iBMK cell lines, both expressing Bcl-2.
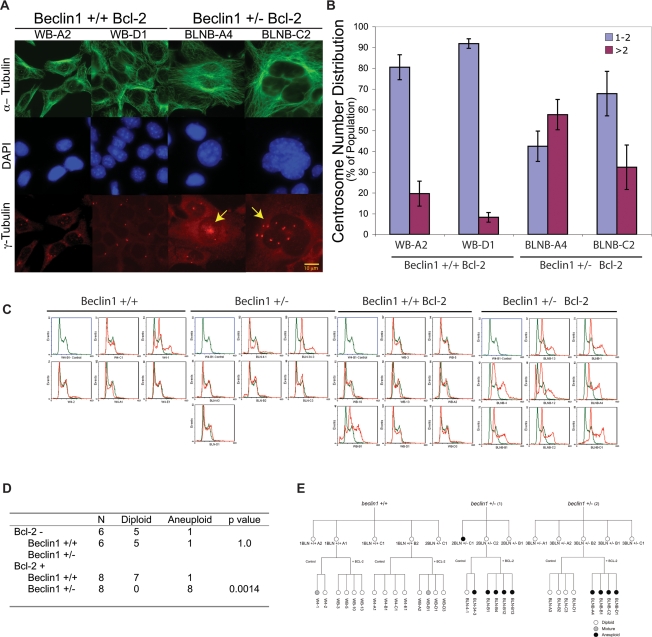
Allelic loss of beclin1 increases the frequency of centrosome abnormalities
How deficiency in autophagy-mediated survival may promote tumorigenesis was further suggested by the accumulation of large cells with abnormally shaped nuclei under standard tissue culture conditions specifically in the beclin1+/− iBMK cells expressing Bcl-2. To characterize the morphological differences between beclin1+/+ and beclin1+/− iBMK cells expressing Bcl-2, the microtubule network, DNA, and centrosomes were examined. beclin1+/+ iBMK cells expressing Bcl-2, with an intact autophagy pathway, maintained a normal cell size with a uniform microtubule framework, nuclear shape, and predominantly normal centrosome numbers (one or two centrosomes per cell) (Fig. 3A). In contrast, autophagy-defective beclin1+/− iBMK cells expressing Bcl-2 showed profound microtubule and centrosome abnormalities, including heterogeneity in cell and nuclear size and shape (Fig. 3A) and an increase in the percentage of cells with centrosome abnormalities including increased centrosome number (Fig. 3B). Abnormalities in the microtubule framework can arise due to abnormally large cell size, and, moreover, supernumerary centrosomes and large nuclei are defining traits of excess DNA content as well as genomic instability (Nigg 2002). These abnormalities in beclin1+/− iBMK cells suggested that autophagy and maintenance of metabolism might be crucial for limiting DNA damage and maintaining genome integrity.
Figure 3.
Allelic loss of beclin1 promotes nuclear, centrosome, and ploidy abnormalities. beclin1+/+ and beclin1+/− iBMK cells with and without Bcl-2 were analyzed by fluorescence microscopy and also for DNA content by flow cytometry. (A) Representative photomicrographs of beclin1+/+ and beclin1+/− iBMK cells expressing Bcl-2 (WB-A2 and WB-D1, BLNB-A4, and BLNB-C2) stained by indirect immunofluorescence for microtubules (anti-α-tubulin), DNA (DAPI), and centrosomes (anti-γ-tubulin). Note the extensive microtubule network, the heterogeneous nuclear size, and the accumulation of structural and numerical abnormalities in centrosomes in beclin1+/− mutants (indicated by arrows). (B) Quantitation of centrosome numbers in A. Percentage of cells with normal centrosome numbers (one or two; blue bars) and with supernumerary centrosomes (more than two; purple bars). Data represent the mean ± SD. (C) Flow cytometry analysis of a panel of beclin1+/+ and beclin1+/− iBMK cell lines showing their DNA content. A beclin1+/+ iBMK cell line (W4-B1) with a diploid DNA content was used as normal control for the analysis (shown in green in each panel). Note the high prevalence of an aneuploid DNA content in the beclin1+/− mutants expressing Bcl-2. (D) Summary of the DNA content analyses in C showing the P value by two-tailed Fisher’s Exact test. (E) Lineage of the cell lines used in the study with their ploidy status, showing independent, genotype-specific emergence of ploidy abnormalities.
Allelic loss of beclin1 manifests ploidy abnormalities in an apoptosis-deficient background
To investigate the potential for genomic instability in beclin1 mutant cells, the DNA content of multiple, independent beclin1+/+ and beclin1+/− iBMK cell lines along with their Bcl-2-expressing counterparts was determined by flow cytometry. Apoptosis-competent beclin1+/+ and beclin1+/− iBMK cell lines were predominantly diploid, with only one cell line out of six from each displaying a mixed population of diploid and aneuploid cells (Fig. 3C). Therefore, allelic loss of beclin1 did not grossly alter ploidy, presumably due to the negative selection for aneuploidy imposed by a functional apoptotic pathway. Flow cytometry analysis of Bcl-2-expressing beclin1+/+ and beclin1+/− iBMK cell lines, however, revealed a significant increase in the frequency of aneuploidy associated with beclin1 deficiency (Fig. 3C). All eight beclin1+/− iBMK cell lines expressing Bcl-2 were aneuploid, while only one out of eight Bcl-2-expressing beclin1+/+ iBMK cell lines showed aneuploidy (Fig. 3C,D). Ploidy abnormalities arose independently and in a genotype-specific manner (Fig. 3E). Overall, these observations suggested that impairment of autophagy promoted abnormalities in centrosome and DNA content that were preserved in an apoptosis-defective background.
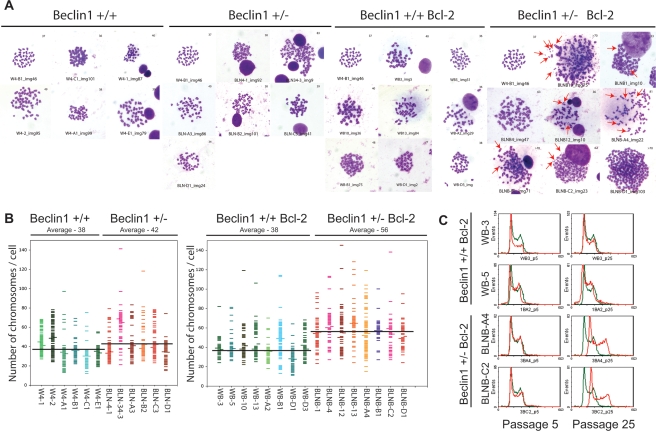
Allelic loss of beclin1 and defective autophagy promote numerical and structural chromosome abnormalities
Increases in centrosome numbers and cellular DNA content are often caused by a numerical increase in chromosome numbers. To further investigate the role of allelic loss of beclin1 and impaired autophagy in the accumulation of DNA content abnormalities, we determined the average metaphase chromosome number of beclin1+/+ and beclin1+/− iBMK cells with and without Bcl-2. Autophagy-competent beclin1+/+ iBMK cells showed a near-normal diploid chromosome number with a group average of 38 (the normal mouse chromosome number is 40) irrespective of Bcl-2 expression (Fig. 4A,B), and allelic loss of beclin1 had a marginal impact on the average chromosome number when apoptosis was functional (group average, 42) (Fig. 4A,B). The wild-type beclin1+/+ iBMK cells expressing Bcl-2 also showed a near-normal diploid chromosome number (group average, 38). In contrast, beclin1+/− iBMK cells expressing Bcl-2 displayed a profound increase in metaphase chromosomes with a near-triploid average chromosome number (group average, 56) (Fig. 4A,B), suggesting that autophagy suppresses aneuploidy. Along with numerical chromosomal abnormalities, beclin1+/− iBMK cells expressing Bcl-2 also displayed significant structural abnormalities, including the presence of double-minute chromosomes (Fig. 4A, red arrows), suggesting the possibility of increased gene amplification (Gebhart 2005).
Figure 4.
beclin1+/− iBMK cells demonstrate chromosomal instability. The panel of beclin1+/+ and beclin1+/− iBMK cells with and without Bcl-2 was analyzed for chromosome number in metaphase spreads and a subset of these cell lines were analyzed for ploidy changes in tissue culture by flow cytometry. (A) Cells were treated with nocodazole (0.5 μg/mL) and metaphase chromosomes were prepared (Brown and Baltimore 2000). The panel shows representative photomicrographs from at least 100 Geimsa-stained mitotic figures from each genotype. The diploid beclin1+/+ iBMK cell line, W4-B1, is shown as a control for each genotype. Note the enhanced numerical and structural abnormalities in the beclin1+/− iBMK cells expressing Bcl-2, with arrows indicating double-minute chromosomes. (B) Scatter plot of the chromosome numbers showing the mean (data point to the left) and the median (data point to the right) values in each cell line. One-hundred metaphases were scored for each cell line. Average of the mean chromosome numbers in each group is shown as a horizontal black line for each genotype. (C) Flow cytometry analysis of passages 5 and 25 of Bcl-2-expressing beclin1+/+ and beclin1+/− iBMK cell lines (red) showing transition from the diploid to aneuploid state. The cell lines were maintained in parallel in the same culture conditions. The diploid beclin1+/+ iBMK cell line, W4-B1 (shown in green), was used as a control.
As these iBMK cell lines were derived from normal primary epithelial cells, defective autophagy may facilitate gradual accumulation of chromosome aberrations permitted by inactivation of checkpoints (p53 and pRb pathways) and are further manifested by impaired apoptosis. In order to determine when these ploidy abnormalities arose, we examined the early passages (Passage 5) of both beclin1+/+ and beclin1+/− Bcl-2-expressing iBMK cell lines shown in Figure 3C for their DNA content. Indeed, seven of the eight early passage beclin1+/+ iBMK cell lines expressing Bcl-2 showed a diploid DNA content, and aneuploidy was observed in only one cell line (WB-B1), as was observed before. In contrast, aneuploidy was evident in six of the eight beclin1+/− iBMK cell lines expressing Bcl-2 as early as passage 5. Interestingly, two of the eight early passage beclin1+/− iBMK cell lines expressing Bcl-2 (BLNB-A4 and BLNB-C2) that showed aneuploidy at a later passage number (Fig. 3C) were diploid at passage 5, suggesting that aneuploidy in these two cell lines arose during passage in tissue culture. Continued propagation in vitro, however, resulted in conversion of the diploid beclin1+/−, but not the beclin1+/+, iBMK cells expressing Bcl-2 to aneuploid cells (Fig. 4C). This suggested that normal tissue culture constituted a level of metabolic stress that may be sufficient to manifest as genomic instability in autophagy-deficient cells. Thus, constitutive autophagy functions to suppress chromosomal instability that occurs gradually over time in culture, perhaps due to low levels of constitutive metabolic stress, and that apoptosis plays a role in the elimination of these cells from the population. Indeed, a constitutive level of autophagy in the range of 1%–10% of the cell population is readily apparent in Bcl-2-expressing or bax/bak-deficient iBMK cells, which is reduced by allelic loss of or RNA interference for beclin1 (Degenhardt et al. 2006).
Allelic loss of beclin1 is associated with gene amplification and chromosome gains and losses
To arrive at the mechanism of enhanced chromosome instability induced by the allelic loss of beclin1, particularly the formation of double-minute chromosomes, we explored the possibility that autophagy suppressed DNA damage and thereby gene amplification. Gene amplification, a major mechanism of oncogene activation and resistance to chemotherapy, results from DNA DSBs (Little and Chartrand 2004). It is facilitated by the inactivation of the p53 DNA damage checkpoint (Livingstone et al. 1992; White et al. 1994), increased oxidative stress (in particular by H2O2) (Mondello et al. 2002), and defects in DNA repair (Lin et al. 2001). Moreover, gene amplification is the known mechanism of resistance to N-phosphonacetyl-l-aspartate (PALA) that prevents de novo pyrimidine biosynthesis through the inhibition of carbamoyl-P synthetase/aspartate transcarbamylase/dihydroorotase (CAD) enzyme complex and selects for CAD gene amplification (Livingstone et al. 1992). To assay for the propensity for gene amplification, we tested for the frequency of PALA resistance in beclin1+/+ and beclin1+/− iBMK cell lines.
Autophagy-defective, beclin1+/− iBMK cells displayed ∼50-fold to >1000-fold increase in the rate of colony formation in PALA in comparison with the beclin1+/+ iBMK cells at five times the PALA LD50, independent of apoptosis deficiency (Fig. 5A,B). Inhibition of apoptosis further enhanced the frequency of PALA resistance in beclin1+/− cell lines, but only by several-fold (Fig. 5B). It should be noted that beclin1+/− colonies grew in the presence of PALA (Fig. 5A), indicating that this was true drug resistance and not merely failure of autophagic cell death. beclin1+/+ iBMK cells without Bcl-2 expression showed substantially lower frequencies for PALA resistance, which was marginally increased by an apoptotic block by Bcl-2 (Fig. 5B). Indeed, PALA-resistant clones demonstrated amplification of the CAD gene (Fig. 5C). These data suggested that defective autophagy promotes drug resistance by gene amplification, which is further enhanced by defective apoptosis.
Figure 5.
Allelic loss of beclin1 promotes PALA resistance and chromosome gain and losses. Multiple independent clones of beclin1+/+ and beclin1+/− iBMK cell lines with and without Bcl-2 were subjected to PALA selection (Livingstone et al. 1992) and aCGH (Kallioniemi et al. 1992). (A) Representative photomicrographs showing PALA-resistant colonies (100 μM). PALA selection was performed using three and five times the LD50 concentration (60 μM and 100 μM, respectively). (B) Quantitation of the frequency of PALA resistance in beclin1+/+ and beclin1+/− iBMK cells with or without Bcl-2. The frequency of PALA resistance was determined as the number of resistance colonies per 3 × 105 cells. (C) PALA-resistant clones display CAD gene amplification in CAD-specific PCR. Panel shows PCR reaction product (for indicated number of cycles) (756 bp) from the genomic DNA isolated from untreated (U) or PALA-treated (T) beclin1+/− iBMK cells expressing Bcl-2. (D) Representative whole-genome DNA copy number profile by aCGH for beclin1+/+ (W4-B1, W4-C1, WB-3, WB-A2, WB-D1) and beclin1+/− (BLN-4-1, BLN-34-3, BLNB-13, BLNB-A4, BLNB-C2) iBMK cell lines with and without Bcl-2 showing enhanced gains and losses of chromosomes in the beclin1+/− mutants. Plotted are log2 transformed hybridization ratios of the genomic DNA isolated from the cell line to genomic DNA from the normal isogenic mouse kidney. (E) Summary of specific chromosome gains (blue bars) and losses (purple bars) in beclin1+/+ and beclin1+/− iBMK cell lines (n = 5 for each). Complete or partial chromosomal dose variations are represented as frequencies. Note the significant and widespread chromosomal dosage variations in the beclin1+/− iBMK cells (two-tailed Fisher’s Exact test; P = 0.003).
The increased frequency of gene amplification observed in the beclin1+/− iBMK cells prompted us to analyze the chromosomes of beclin1+/+ and beclin1+/− iBMK cell lines for random gains and losses by microarray-based comparative genome hybridization (aCGH) (Kallioniemi et al. 1992). Random chromosomal gains or losses occurred with low frequency in the autophagy-competent beclin1+/+ iBMK cell lines and were limited to fewer chromosomes in comparison with beclin1+/− iBMK cell lines, which showed widespread chromosomal gains and losses in nearly all of the chromosomes (Fig. 5D,E). As aCGH detects only relative copy number variations and not ploidy, the increased chromosomal gains and losses observed in the aCGH data do not address absolute chromosome variations that were apparent in the mitotic spreads and the DNA content analysis (Figs. 3, 4). Nevertheless, these data suggested that beclin1 heterozygocity enhanced gene dosage variations, which may accompany the gross ploidy abnormalities observed by flow cytometry analysis.
Inhibition of pyrimidine biosynthesis triggers autophagy that suppresses DNA damage
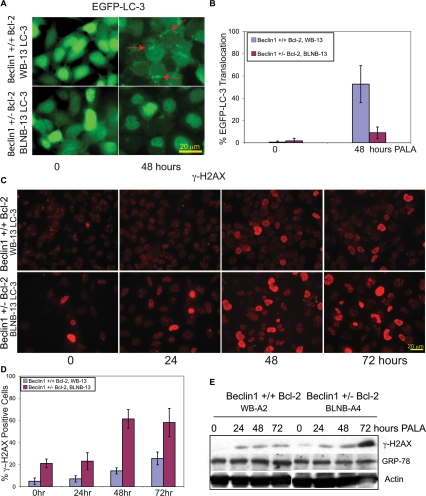
The induction of gene amplification in the beclin1 mutant cells raised the intriguing possibility that PALA and the resulting pyrimidine starvation, which itself may be a form of metabolic stress, promotes autophagy induction. Therefore, we tested the relative induction of autophagy in beclin1+/+ and beclin1+/− iBMK cells in response to PALA by cytoplasmic-to-membrane translocation of EGFP-LC3 in stably expressing cells. Constitutive autophagy levels were low in both beclin1+/+ and beclin1+/− iBMK cells expressing Bcl-2. Within 48 h of PALA exposure (60 μM), wild-type beclin1+/+ iBMK cells expressing Bcl-2 showed significant induction of autophagy (50%), and this induction of autophagy was significantly diminished (9%) in the mutant beclin1+/− iBMK cells expressing Bcl-2 (Fig. 6A,B). This demonstrated that pharmacologic depletion of pyrimidines triggers autophagy.
Figure 6.
PALA-induced autophagy is impaired leading to the accumulation of DNA damage response in beclin1+/− cells and not in the beclin1+/+ cells. Alleleic loss of beclin1 results in impaired autophagy and sustains enhanced DNA damage response in response to PALA treatment when compared with the corresponding wild-type cell line. (A) Representative photomicrographs of beclin1+/+ (WB-13 LC3) and beclin1+/− (BLNB-13 LC3) iBMK cell lines stably expressing EGFP-LC3, showing the lack of EGFP-LC3 localization in the beclin1+/− (BLNB-13 LC3) iBMK cell lines indicating impaired autophagy in response to PALA treatment (60 μM). (B) Quantitation of the percentage of EGFP-LC3 translocation indicative of autophagy in A as described previously (Degenhardt et al. 2006). Data represent mean ± SD. Note that autophagy induction in the beclin1+/− was approximately fivefold less than the wild-type cells even after 48 h of PALA treatment. (C) Representative photomicrographs of γ-H2AX immunostaining in beclin1+/+ iBMK cells expressing Bcl-2 (WB-13 LC3) and beclin1+/− iBMK cells expressing Bcl-2 (BLNB-13 LC3) following PALA treatment showing enhanced DNA damage response in the beclin1+/− iBMK cells expressing Bcl-2 compared with the wild-type cells. (D) Quantitation of percentage of γ-H2AX-positive cells is shown in C. Data represent mean ± SD. (E) Western blots showing γ-H2AX and GRP-78 levels in beclin1+/+ (WB-A2) and beclin1+/− (BLNB-A4) iBMK cells, both expressing Bcl-2.
PALA depletes pyrimidine nucleotide (dNTP) pools, and in checkpoint-deficient cells this can induce replication stress and DNA damage. Autophagy-deficient cells may be particularly prone to this highly specific form of metabolic stress, which may enhance replication stress and DNA damage. Therefore, we tested the DNA damage response following PALA exposure in both beclin1+/+ and beclin1+/− iBMK cells by assessing the accumulation of γ-H2AX, an early marker for DSBs. Low levels of basal DNA damage were observed in both beclin1+/+ and beclin1+/− iBMK cells expressing Bcl-2 (Figs. 2B,C, 6C,D). γ-H2AX staining was dramatically increased following PALA exposure in the mutant beclin1+/− iBMK cells expressing Bcl-2, while in the wild-type cells this increase was significantly less (Fig. 6C,D). Indeed, Western blot analysis confirmed accumulation of γ-H2AX in beclin1+/− iBMK cells (Fig. 6E). Furthermore, no induction of GRP-78 indicative of UPR was observed (Fig. 6E).
Deficiency in atg5 confers susceptibility to metabolic stress
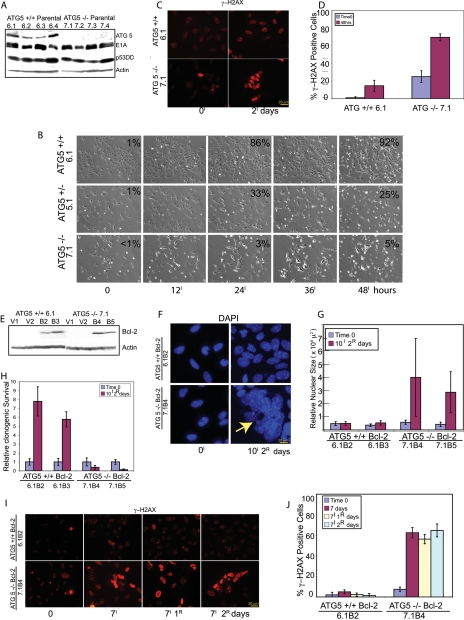
In order to establish if the susceptibility to metabolic stress and genomic instability were due to defective autophagy, and not limited to allelic loss of beclin1, immortalized iBMK cell lines were derived from mice deficient for the essential autophagy gene atg5 (Kuma et al. 2004). Multiple independent iBMK cell lines were derived from atg5+/+, atg5+/−, and atg5−/− mice, and Western blot analysis confirmed the absence of ATG5 protein expression in atg5−/− (Fig. 7A) and reduced expression in the atg5+/− (data not shown) compared with atg5+/+ iBMK cell lines (Fig. 7A). All iBMK cell lines expressed E1A and p53DD as expected (Fig. 7A). To confirm that loss of ATG5 impaired autophagy under metabolic stress, the wild-type, heterozygous, and deficient ATG5 iBMK cells (6.1, 5.1, and 7.1) transiently expressing LC3-EGFP were subjected to metabolic stress, and the percentage of membrane translocation indicative of autophagy was determined. All cell lines had low levels of EGFP-LC3 translocation under normal growth conditions, whereas metabolic stress induced a dramatic up-regulation of autophagy in the atg5+/+ that was reduced in the atg5+/− and negligible in the atg5−/− iBMK cell lines (Fig. 7B, percentage autophagy indicated in the top right corner of the panels). In order to test if defective autophagy due to loss of ATG5 also impaired survival under metabolic stress, these cell lines were monitored by long-term computerized video time-lapse microscopy (CVTL). atg5+/+ iBMK cell lines were minimally affected at 24 h with morphological signs of apoptosis becoming apparent at 48 h of metabolic stress, as expected (Fig. 7B). In comparison with the atg5+/+, at the same time points, apoptosis was accelerated in atg5+/− and was markedly pronounced in the atg5−/− iBMK cell lines (Fig. 7B). Thus, deficiency in ATG5 compromises both autophagy and survival in metabolic stress, as does allelic loss of beclin1 (Fig. 1C; Degenhardt et al. 2006), but does so to a greater extent. This suggests that impaired survival to metabolic stress in cancer cells is independent of the means of autophagy inactivation as reported previously (Degenhardt et al. 2006).
Figure 7.
ATG5 deficiency impairs survival and promotes DNA damage response under metabolic stress. (A) Western blot showing protein expression levels in parental atg5+/+ and atg5−/− iBMK cell lines. (B) Frames from 2-d time-lapse videos of atg5+/+ (6.1), atg5+/− (5.1), and atg5−/− (7.1) cells performed as described previously (Degenhardt et al. 2006), showing differential survival under ischemia for the indicated time points. Numbers represent percentage of cells with LC3-EGFP translocation from a total of 300 cells, indicative of autophagy at the indicated time points. (C) Representative photomicrographs showing induction of γ-H2AX in atg5+/+ and atg5−/− parental iBMK cell lines (6.1 and 7.1) in response to ischemia treatment for 48 h. (D) Quantitation of γ-H2AX-positive cells in atg5+/+ (6.1) and atg5−/− (7.1) iBMK cell lines in C. Data represent mean ± SD. (E) Western blot showing protein expression levels for two independent atg5+/+ and atg5−/− iBMK cell lines expressing Bcl-2 (6.1B2, 6.1B3, 7.1B4, and 7.1B5 respectively) and corresponding vector controls (6.1V1, 6.1V2, 7.1V1, and 7.1V2). (F) Representative photomicrographs of atg5+/+ and atg5−/− iBMK cells expressing Bcl-2 showing DAPI-stained nuclei untreated (left) or treated with metabolic stress (right) for 10 d followed by 2 d of recovery. Note the gross abnormality in nuclear size and shape in the atg5−/− cells (yellow arrow). (G) Quantitation of nuclear size in atg5+/+ and atg5−/− iBMK cell lines expressing Bcl-2, shown in F. Data represent mean ± SD. (H) Relative clonogenic survival in atg5+/+ and atg5−/− iBMK cell lines expressing Bcl-2 (6.1B2 and 7.1B4) following 10 d ischemia and 2 d of recovery. Data represent mean ± SD. (I) Representative photomicrographs showing induction of γ-H2AX in atg5+/+ and atg5−/− iBMK cell lines expressing Bcl-2 (6.1B2 and 7.1B4). (J) Quantitation of percentage of γ-H2AX-positive nuclei in Bcl-2-expressing atg5+/+ and atg5−/− iBMK cells shown in I. Data represent mean ± SD.
ATG5 deficiency promotes activation of the DNA damage response
To determine if the enhanced susceptibility to metabolic stress conferred by ATG5 deficiency also resulted in an enhanced DNA damage response, as observed with allelic loss of beclin1, the appearance of γ-H2AX nuclear staining was monitored under normal and stress conditions. Autophagy-defective, atg5−/− cells had a slightly higher level of γ-H2AX staining under normal growth conditions that was substantially increased by metabolic stress compared with wild-type atg5+/+ cells (Fig. 7C,D).
To test if autophagy dependent on ATG5 further enabled viability and suppressed DNA damage under metabolic stress in cells with a defect in apoptosis, as does Beclin1, atg5+/+ and atg5−/− iBMK cell lines were engineered to express Bcl-2 along with vector controls (Fig. 7E). When subjected to metabolic stress for 10 d followed by recovery for 2 d, Bcl-2-expressing atg5−/− iBMK cell lines displayed gross abnormalities in nuclear morphology (increased size and irregular shape indicative of chromosome instability) (Fig. 7F,G), associated with dramatic impairment in clonogenic survival compared with Bcl-2-expressing atg5+/+ iBMK cells (Fig. 7H). Although atg5 deficiency reduced survival to metabolic stress even in the presence of an apoptotic block (data not shown), as observed for allelic loss of beclin1 (Fig. 1C; Degenhardt et al. 2006), this survival deficit was greatly accentuated upon replenishment of nutrients and oxygen (Fig. 7H). Impaired recovery following metabolic stress was also observed in Bcl-2-expressing beclin1+/− iBMK cells (Fig. 1D). This suggested that defects in autophagy resulted in increased genome damage in stress conditions that were manifested more prominently when cells were returned to normal growth conditions. To test this, Bcl-2-expressing atg5+/+ and atg5−/− iBMK cell lines were monitored for γ-H2AX-positive nuclei under stress conditions and recovery. Bcl-2-expressing atg5+/+ iBMK cells showed few γ-H2AX-positive nuclei, the occurrence of which was modestly increased by stress, as expected (Fig. 7I,J). In contrast, Bcl-2-expressing atg5−/− iBMK cells displayed a marked increase in γ-H2AX-positive nuclei during both stress and recovery (Fig. 7I,J). Thus, inactivation of autophagy by either allelic loss of beclin1 or deficiency in atg5 amplified the cellular response to DNA damage and increased the susceptibility to metabolic stress. This supports a role for the autophagy pathway in suppressing genome damage and enabling survival of cancer cells under conditions of starvation. Inactivation of cell cycle checkpoints and apoptosis in cancer cells may allow further manifestation of genome damage resulting from defective autophagy, contributing to tumor progression.
Discussion
The results described here demonstrate that autophagy functions to limit DNA damage and chromosomal instability, which may explain its tumor suppressor activity. Thus, our study provides the first mechanistic link between the regulation of metabolism and genome integrity. We report here that impaired autophagy rendered beclin1+/− and atg5−/− iBMK cells susceptible to metabolic stress in vitro. This impaired survival in autophagy-deficient cells, however, was overcome by an increased mutation rate and chromosomal instability resulting in accelerated progression to aneuploidy, the hallmark of cancer, providing an explanation as to why the loss of this survival pathway can function to suppress tumorigenesis. We found that these abnormalities in autophagy-deficient cells were also accompanied by an increased DNA damage response. Interestingly, pyrimidine starvation triggered autophagy, the defect in which was sufficient to promote a DNA damage response and gene amplification in autophagy-deficient cells. Hence, our data suggest that constitutive and stress-induced autophagy function to limit DNA damage and maintain genomic integrity, thereby identifying a novel, but not necessarily the only, role for defective autophagy in promoting oncogenesis.
Defective autophagy, metabolism, and chromosomal instability
We have shown earlier that a defect in apoptosis enables survival to metabolic stress through autophagy, and that concomitant impairment of apoptosis and autophagy pathways results in necrotic cell death in response to metabolic stress, functioning as a non-cell-autonomous mechanism to promote tumorigenesis (Degenhardt et al. 2006). Our current findings suggest that autophagy also has a more direct role in suppressing tumorigenesis through the maintenance of genome integrity. Metabolic stress is a common occurrence in human solid tumors (Folkman 2003) that functions as a potent trigger for apoptosis (Nelson et al. 2004). Therefore, autophagy as an alternate means for energy production may be beneficial for tumor cells to mitigate metabolic stress. In normal development, the lack of autophagy in atg5−/− mice impairs survival in heart tissue during neonatal starvation, leading to early lethality (Kuma et al. 2004). Nevertheless, defects in autophagy promote tumorigenesis despite conferring an impediment to survival, as autophagy-defective beclin1+/− iBMK cells are more tumorigenic than the autophagy-competent beclin1+/+ iBMK cells (Degenhardt et al. 2006). Therefore, the enhanced gene amplification and chromosomal instability observed in the mutant beclin1+/− cells expressing Bcl-2 may enable cells to overcome the survival deficit by promoting tumorigenesis through an increased mutation rate. This observation is analogous to the increased tumorigenicity of cells with defects in DNA repair that also creates sensitivity to DNA-damaging agents and ionizing radiation while facilitating the accumulation of oncogenic mutations (Symington 2002).
Metabolic stress, gene amplification, and tumorigenesis
Tumorigenesis results from the accumulation of mutations that, our evidence suggests, is facilitated by deficient metabolism due to defective autophagy. Autophagy deficiency in atg5−/− neonates results in lethal levels of energy depletion and low amino acid concentrations in plasma and tissues that impair normal heart function (Kuma et al. 2004). Insufficient generation of ATP for vital cellular processes such as DNA replication or repair in autophagy-deficient cells may lead to stalled replication forks and breakage–fusion–bridge cycles leading to gene amplification and tumorigenesis (Hellman et al. 2002; Narayanan et al. 2006; Jin and White 2007). Moreover, the basal housekeeping function of autophagy is required for protein quality control and homeostasis, as neuronal cells impaired for autophagy accumulate polyubiquitinated proteins (Hara et al. 2006; Komatsu et al. 2006). In keeping with this finding, we observed that autophagy-defective beclin1+/− and atg5−/− cells with Bcl-2 also showed poor recovery from ischemic stress, with accumulation of vacuolated cytoplasm and other cellular aggregates. Damage to critical cellular proteins that control mitosis or centrosome function in autophagy-deficient cells can be a cause of chromosomal instability. In addition, excessive generation of reactive oxygen species due to inefficient removal of damaged mitochondria is another possible mechanism by which defective autophagy can promote DNA damage and tumorigenesis (Jin 2006). Whether any or all of these mechanisms are responsible for genome alterations in autophagy-defective cells remains to be determined.
Gene amplification, a consequence of permissive levels of DSBs, is the major mechanism of oncogene activation in solid tumors, as exemplified by myc, egfr, and erbB2 (Shen et al. 1986; Hennessy et al. 2005; Albertson 2006). Gene amplification also leads to acentric, extrachromosomal fragments called double minutes, a common chromosomal aberration found in human solid tumors (Albertson et al. 2003). Furthermore, most human tumors display aneuploidy in the form of a near triploid DNA content (Barlogie et al. 1982), but the mechanism by which this is achieved is not clearly understood. By suppressing DNA damage under stress, autophagy may suppress progression to aneuploidy as a tumor suppressor mechanism.
In summary, our data suggest that reduced autophagy promotes DNA damage, gene amplification, chromosome instability, and aneuploidy, all of which are clinically associated with tumor progression and poor prognosis. Similar findings with allelic loss of beclin1 in immortalized breast epithelial cells (V. Karantza-Wadsworth, pers. comm.) and immortalized, autophagy-defective atg5−/− mouse embryo fibroblasts (MEFs) (S. Jin, pers. comm.) suggest that these observations are independent of tissue type. Analogous to the mammalian central nervous system where targeted deletion of either atg5 or atg7 revealed the protective role of constitutive autophagy through prevention of the accumulation of polyubiquitinated proteins and neurodegeneration (Hara et al. 2006; Komatsu et al. 2006), in tumor cells, preventing the genome damage through autophagy may be an essential cell-autonomous mechanism for tumor suppression.
Autophagy defects, genomic instability, and novel therapeutic strategies
Identification of a novel role of autophagy in limiting chromosomal instability offers new insight into our understanding of how autophagy modulates genomic integrity and impacts tumorigenesis. Moreover, the finding that autophagy functions to limit chromosomal instability may have implications in rational chemotherapy and chemoprevention. As tumor cells frequently acquire defects in autophagy, pharmacological activation of autophagy may serve to limit genome damage and tumorigenesis. Alternatively, in established aggressive cancers with defects in apoptosis, inhibiting the autophagy survival pathway can be therapeutically exploited to promote other modes of cell death such as metabolic catastrophe, redirecting the cells to an acute necrotic cell death (Jin and White 2007).
Materials and methods
Generation of stable cell lines
Primary epithelial cells were isolated from beclin1+/+ and beclin1+/− mice (generously provided by Dr. Zhenyu Yue, Mount Sinai School of Medicine, New York, NY) and were immortalized by the inactivation of the retinoblastoma (Rb) and p53 pathways (Degenhardt et al. 2002, 2006), and four independent clones of each were transfected with the vector (W4-A1, W4-B1, W4-C1, W4-E1, and BLN-A3, BLN-B2, BLN-C3, BLN-D1) or were engineered to express Bcl-2 (WB-A2, WB-B1, WB-D1, WB-D3, and BLNB-A4, BLNB-B1, BLNB-C2, BLNB-D1), expanding the original pool of cell lines (Degenhardt et al. 2006) from 12 to 28. Primary epithelial cells were isolated from atg5+/+, atg5+/−, and atg5−/− newborn mice (kindly provided by Dr. Noboru Mizushima, Tokyo Metropolitan Institute of Medical Sciences, Tokyo, Japan) and immortalized as previously described (Degenhardt et al. 2002) to generate atg5+/+ (6.1), atg5+/− (5.1), and atg5−/− (7.1) iBMK cell lines. Wild-type atg5 (6.1) and atg5−/− (7.1) iBMK cells were then transfected with vector (6.1V2, 6.1V3) or were engineered to express Bcl-2 (6.1B4, 6.1B5).
Western blotting, immunofluorescence, and electron microscopy
Western blotting, immunofluorescence, and EM were performed as previously described (Nelson et al. 2004; Degenhardt et al. 2006). Western blotting and immunofluoresce were performed using the following antibodies: anti-Beclin1, anti-Bcl-2 (Santa Cruz Biotechnology); anti-p53 (Ab-1), anti-E1A, and anti-actin (Oncogene); anti-α-tubulin (Calbiochem); DNA (DAPI, Sigma); anti-γ-tubulin (Axel); anti-γH2AX (S139) (Upstate Biotechnology); anti-ATG5 (Mizushima et al. 2001).
DAPI staining and quantitation of nuclear size
DAPI staining of the nuclei was performed as described previously (Nelson et al. 2004). Relative nuclear sizes were calculated using Image-Pro Plus software. Briefly, representative photomicrographs (600×) were taken and nuclear boundaries were determined using histogram-based segmentation, and areas were calculated and relative nuclear sizes were expressed in μ2.
Measurement of the frequency of PALA-resistant clones
The LD50 of PALA in beclin1+/+ and beclin1+/− iBMK cells (105/10-cm dish) was determined by treating the cells with varying concentrations of PALA (0–100 μM) (Drug Synthesis and Chemistry Branch, National Cancer Institute) for 3 d and determining the cell viability by trypan blue exclusion. The frequency of PALA resistance was determined to be the number of PALA-resistant clones formed per 3 × 105 cells following treatment with 3× LD50 and 5× LD50 concentrations of PALA (60 μM and 100 μM, respectively) for 10–14 d. PALA-containing medium was replaced every 3–4 d. Colonies were fixed in methanol, stained with Geimsa, and photographed.
PCR for CAD gene amplification
Genomic DNA was isolated from multiple independent PALA-resistant (100 μM) colonies of beclin1+/− iBMK cells expressing Bcl-2. A 756-base-pair (bp) fragment flanking two exons that correspond to the Mus musculus CAD gene (accession no. NM_023525) from mouse chromosome 5 (accession no. AC_109608) was amplified from genomic DNA by PCR using the following set of primers: 4-fwd (5′-GGAGCTGGAGACTC CGACG-3′) and 4-rev (5′-CTAATGAACAGGAAGATCCGG TATC-3′).
The PCR reaction was performed for the indicated number of cycles (Fig. 6C) to compare relative amplification in the linear range.
aCGH
aCGH was performed as described previously (Kallioniemi et al. 1994). Briefly, genomic DNA from beclin1+/+ and beclin1+/− iBMK cells and the normal mouse tissue was isolated, fluorescent-labeled, and hybridized to a BAC array (Snijders et al. 2001) in triplicate. For each cell line, the data were plotted in genome order as the mean log2 ratio of the triplicate spots. An average log2 ratio of more than ±0.5 was considered as a loss or gain.
Acknowledgments
We thank Nathaniel Heintz and Zhenyu Yue for providing beclin1+/+ and beclin1+/− mice; Noboru Mizushima for providing atg5+/+, atg5+/−, and atg5−/− mice, the anti-mouse ATG5 antibody, and EGFP-LC3 plasmid; the Drug Synthesis and Chemistry Branch, National Cancer Institute, Bethesda, MD, for the generous gift of PALA; Haiyan Zhang and Yong Zhang for help with generation of atg5 cell lines; Petra Pham and Theresa Hyejeong Choi for assistance with the flow cytometry; Dr. Kane-Goldsmith for assistance with confocal microscopy; and Raj Patel for assistance with electron microscopy. We also thank Thomasina Sharkey for assistance with the preparation of the manuscript. This work was supported by a grant from the National Institutes of Health (R37 CA53370) and the Howard Hughes Medical Institute (to E.W).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1545107
References
- Aita V.M., Liang X.H., Murty V.V., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Liang X.H., Murty V.V., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Murty V.V., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Cayanis E., Kalachikov S., Gilliam T.C., Levine B., Kalachikov S., Gilliam T.C., Levine B., Gilliam T.C., Levine B., Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- Albertson D.G. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Albertson D.G., Collins C., McCormick F., Gray J.W., Collins C., McCormick F., Gray J.W., McCormick F., Gray J.W., Gray J.W. Chromosome aberrations in solid tumors. Nat. Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Anglade P., Vyas S., Javoy-Agid F., Herrero M.T., Michel P.P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E.C., Agid Y., Vyas S., Javoy-Agid F., Herrero M.T., Michel P.P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E.C., Agid Y., Javoy-Agid F., Herrero M.T., Michel P.P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E.C., Agid Y., Herrero M.T., Michel P.P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E.C., Agid Y., Michel P.P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E.C., Agid Y., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E.C., Agid Y., Mouatt-Prigent A., Ruberg M., Hirsch E.C., Agid Y., Ruberg M., Hirsch E.C., Agid Y., Hirsch E.C., Agid Y., Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol. Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- Baehrecke E.H. Autophagic programmed cell death in Drosophila. Cell Death Differ. 2003;10:940–945. doi: 10.1038/sj.cdd.4401280. [DOI] [PubMed] [Google Scholar]
- Barlogie B., Johnston D.A., Smallwood L., Raber M.N., Maddox A.M., Latreille J., Swartzendruber D.E., Drewinko B., Johnston D.A., Smallwood L., Raber M.N., Maddox A.M., Latreille J., Swartzendruber D.E., Drewinko B., Smallwood L., Raber M.N., Maddox A.M., Latreille J., Swartzendruber D.E., Drewinko B., Raber M.N., Maddox A.M., Latreille J., Swartzendruber D.E., Drewinko B., Maddox A.M., Latreille J., Swartzendruber D.E., Drewinko B., Latreille J., Swartzendruber D.E., Drewinko B., Swartzendruber D.E., Drewinko B., Drewinko B. Prognostic implications of ploidy and proliferative activity in human solid tumors. Cancer Genet. Cytogenet. 1982;6:17–28. doi: 10.1016/0165-4608(82)90017-6. [DOI] [PubMed] [Google Scholar]
- Brown E.J., Baltimore D., Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Cataldo A.M., Hamilton D.J., Nixon R.A., Hamilton D.J., Nixon R.A., Nixon R.A. Lysosomal abnormalities in degenerating neurons link neuronal compromise to senile plaque development in Alzheimer disease. Brain Res. 1994;640:68–80. doi: 10.1016/0006-8993(94)91858-9. [DOI] [PubMed] [Google Scholar]
- Degenhardt K., Sundararajan R., Lindsten T., Thompson C., White E., Sundararajan R., Lindsten T., Thompson C., White E., Lindsten T., Thompson C., White E., Thompson C., White E., White E. Bax and Bak independently promote cytochrome C release from mitochondria. J. Biol. Chem. 2002;277:14127–14134. doi: 10.1074/jbc.M109939200. [DOI] [PubMed] [Google Scholar]
- Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Anderson D., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Chen G., Mukherjee C., Shi Y., Gelinas C., Fan Y., Mukherjee C., Shi Y., Gelinas C., Fan Y., Shi Y., Gelinas C., Fan Y., Gelinas C., Fan Y., Fan Y., et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkman J. Angiogenesis and apoptosis. Semin. Cancer Biol. 2003;13:159–167. doi: 10.1016/s1044-579x(02)00133-5. [DOI] [PubMed] [Google Scholar]
- Gebhart E. Double minutes, cytogenetic equivalents of gene amplification, in human neoplasia—A review. Clin. Transl. Oncol. 2005;7:477–485. doi: 10.1007/BF02717000. [DOI] [PubMed] [Google Scholar]
- Gomez-Santos C., Ferrer I., Santidrian A.F., Barrachina M., Gil J., Ambrosio S., Ferrer I., Santidrian A.F., Barrachina M., Gil J., Ambrosio S., Santidrian A.F., Barrachina M., Gil J., Ambrosio S., Barrachina M., Gil J., Ambrosio S., Gil J., Ambrosio S., Ambrosio S. Dopamine induces autophagic cell death and α-synuclein increase in human neuroblastoma SH-SY5Y cells. J. Neurosci. Res. 2003;73:341–350. doi: 10.1002/jnr.10663. [DOI] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Yokoyama M., Mishima K., Saito I., Okano H., Mishima K., Saito I., Okano H., Saito I., Okano H., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hellman A., Zlotorynski E., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Zlotorynski E., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Smith D.I., Trakhtenbrot L., Kerem B., Trakhtenbrot L., Kerem B., Kerem B. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- Hennessy B.T., Smith D.L., Ram P.T., Lu Y., Mills G.B., Smith D.L., Ram P.T., Lu Y., Mills G.B., Ram P.T., Lu Y., Mills G.B., Lu Y., Mills G.B., Mills G.B. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- Jin S. Autophagy, mitochondrial quality control, and oncogenesis. Autophagy. 2006;2:80–84. doi: 10.4161/auto.2.2.2460. [DOI] [PubMed] [Google Scholar]
- Jin S., White E., White E. Role of autophagy in cancer: Management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O.P., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Kallioniemi O.P., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Gray J.W., Waldman F., Pinkel D., Waldman F., Pinkel D., Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O.P., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Kallioniemi O.P., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Pinkel D., Gray J.W., Waldman F.M., Gray J.W., Waldman F.M., Waldman F.M. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc. Natl. Acad. Sci. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Taylor M.P., Jackson W.T., Taylor M.P., Jackson W.T., Jackson W.T. Cellular autophagy: Surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Emr S.D., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Cregg J.M., Dunn W.A., Jr., Emr S.D., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Cregg J.M., Dunn W.A., Jr., Emr S.D., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Dunn W.A., Jr., Emr S.D., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Emr S.D., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Sibirny A., Subramani S., Thumm M., Veenhuis M., Subramani S., Thumm M., Veenhuis M., Thumm M., Veenhuis M., Veenhuis M., et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Mizushima N., Ohsumi Y., Uchiyama Y., Ohsumi Y., Uchiyama Y., Uchiyama Y., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Ueno T., Koike M., Uchiyama Y., Kominami E., Koike M., Uchiyama Y., Kominami E., Uchiyama Y., Kominami E., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., Ohsumi Y., Tokuhisa T., Mizushima N., Tokuhisa T., Mizushima N., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J., Klionsky D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Lin C.T., Lyu Y.L., Xiao H., Lin W.H., Whang-Peng J., Lyu Y.L., Xiao H., Lin W.H., Whang-Peng J., Xiao H., Lin W.H., Whang-Peng J., Lin W.H., Whang-Peng J., Whang-Peng J. Suppression of gene amplification and chromosomal DNA integration by the DNA mismatch repair system. Nucleic Acids Res. 2001;29:3304–3310. doi: 10.1093/nar/29.16.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little K.C., Chartrand P., Chartrand P. Genomic DNA is captured and amplified during double-strand break (DSB) repair in human cells. Oncogene. 2004;23:4166–4172. doi: 10.1038/sj.onc.1207570. [DOI] [PubMed] [Google Scholar]
- Livingstone L.R., White A., Sprouse J., Livanos E., Jacks T., Tlsty T.D., White A., Sprouse J., Livanos E., Jacks T., Tlsty T.D., Sprouse J., Livanos E., Jacks T., Tlsty T.D., Livanos E., Jacks T., Tlsty T.D., Jacks T., Tlsty T.D., Tlsty T.D. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ. 2005;12:1535–1541. doi: 10.1038/sj.cdd.4401728. (Suppl. 2) [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T., Tokuhisa T., Ohsumi Y., Yoshimori T., Ohsumi Y., Yoshimori T., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondello C., Guasconi V., Giulotto E., Nuzzo F., Guasconi V., Giulotto E., Nuzzo F., Giulotto E., Nuzzo F., Nuzzo F. γ-Ray and hydrogen peroxide induction of gene amplification in hamster cells deficient in DNA double strand break repair. DNA Repair (Amst.) 2002;1:483–493. doi: 10.1016/s1568-7864(02)00035-6. [DOI] [PubMed] [Google Scholar]
- Narayanan V., Mieczkowski P.A., Kim H.M., Petes T.D., Lobachev K.S., Mieczkowski P.A., Kim H.M., Petes T.D., Lobachev K.S., Kim H.M., Petes T.D., Lobachev K.S., Petes T.D., Lobachev K.S., Lobachev K.S. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell. 2006;125:1283–1296. doi: 10.1016/j.cell.2006.04.042. [DOI] [PubMed] [Google Scholar]
- Nelson D.A., Tan T.T., Rabson A.B., Anderson D., Degenhardt K., White E., Tan T.T., Rabson A.B., Anderson D., Degenhardt K., White E., Rabson A.B., Anderson D., Degenhardt K., White E., Anderson D., Degenhardt K., White E., Degenhardt K., White E., White E. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes & Dev. 2004;18:2095–2107. doi: 10.1101/gad.1204904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. Centrosome aberrations: Cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Pua H.H., Dzhagalov I., Chuck M., Mizushima N., He Y.W., Dzhagalov I., Chuck M., Mizushima N., He Y.W., Chuck M., Mizushima N., He Y.W., Mizushima N., He Y.W., He Y.W. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 2006;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Eskelinen E.L., Mizushima N., Ohsumi Y., Mizushima N., Ohsumi Y., Ohsumi Y., et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D.W., Fojo A., Chin J.E., Roninson I.B., Richert N., Pastan I., Gottesman M.M., Fojo A., Chin J.E., Roninson I.B., Richert N., Pastan I., Gottesman M.M., Chin J.E., Roninson I.B., Richert N., Pastan I., Gottesman M.M., Roninson I.B., Richert N., Pastan I., Gottesman M.M., Richert N., Pastan I., Gottesman M.M., Pastan I., Gottesman M.M., Gottesman M.M. Human multidrug-resistant cell lines: Increased mdr1 expression can precede gene amplification. Science. 1986;232:643–645. doi: 10.1126/science.3457471. [DOI] [PubMed] [Google Scholar]
- Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y., Mizuta T., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y., Arakawa-Kobayashi S., Thompson C.B., Tsujimoto Y., Thompson C.B., Tsujimoto Y., Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Snijders A.M., Nowak N., Segraves R., Blackwood S., Brown N., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., Nowak N., Segraves R., Blackwood S., Brown N., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., Segraves R., Blackwood S., Brown N., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., Blackwood S., Brown N., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., Brown N., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., Hamilton G., Hindle A.K., Huey B., Kimura K., Hindle A.K., Huey B., Kimura K., Huey B., Kimura K., Kimura K., et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat. Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- Symington L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.E., Livanos E.M., Tlsty T.D., Livanos E.M., Tlsty T.D., Tlsty T.D. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes & Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- Yu L., Alva A., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E.H., Lenardo M.J., Alva A., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E.H., Lenardo M.J., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E.H., Lenardo M.J., Dutt P., Freundt E., Welsh S., Baehrecke E.H., Lenardo M.J., Freundt E., Welsh S., Baehrecke E.H., Lenardo M.J., Welsh S., Baehrecke E.H., Lenardo M.J., Baehrecke E.H., Lenardo M.J., Lenardo M.J. Regulation of an ATG7–beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A.J., Heintz N., Jin S., Yang C., Levine A.J., Heintz N., Yang C., Levine A.J., Heintz N., Levine A.J., Heintz N., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]