Abstract
Embryonal rhabdomyosarcoma (ERMS) is a devastating cancer with specific features of muscle differentiation that can result from mutational activation of RAS family members. However, to date, RAS pathway activation has not been reported in a majority of ERMS patients. Here, we have created a zebrafish model of RAS-induced ERMS, in which animals develop externally visible tumors by 10 d of life. Microarray analysis and cross-species comparisons identified two conserved gene signatures found in both zebrafish and human ERMS, one associated with tumor-specific and tissue-restricted gene expression in rhabdomyosarcoma and a second comprising a novel RAS-induced gene signature. Remarkably, our analysis uncovered that RAS pathway activation is exceedingly common in human RMS. We also created a new transgenic coinjection methodology to fluorescently label distinct subpopulations of tumor cells based on muscle differentiation status. In conjunction with fluorescent activated cell sorting, cell transplantation, and limiting dilution analysis, we were able to identify the cancer stem cell in zebrafish ERMS. When coupled with gene expression studies of this cell population, we propose that the zebrafish RMS cancer stem cell shares similar self-renewal programs as those found in activated satellite cells.
Keywords: Zebrafish, rhabdomyosarcoma, RAS, P53, transgenic
Rhabdomyosarcoma (RMS) is the most common soft-tissue sarcoma of childhood, affecting >250 new patients each year in the United States (Arndt and Crist 1999). Treatment is often very aggressive, involving local irradiation, chemotherapy, and tumor resection. For patients that present with metastatic disease at the time of diagnosis, prognosis is abysmal, with <25% of patients achieving 5-year survival (Arndt and Crist 1999). Embryonal RMS (ERMS), the most common subtype of pediatric RMS, is not only morphologically distinct from the alveolar subtype (ARMS) but is also associated with transformation by different molecular mechanisms (Xia et al. 2002). For example, 85% of ARMS have chromosomal translocations involving the PAX3 or PAX7 and the forkhead transcription factor (FKHR) gene loci (2;13 or 1;3, respectively). By contrast, ERMS do not have recurrent chromosomal translocations, but instead most exhibit allelic loss at 11p15.5, likely resulting in deregulation of the tumor suppressor gene, SCL22A18 (BWR1A) (Schwienbacher et al. 1998). Additionally, mutational activation of RAS family members has been reported in ERMS patients but is relatively infrequent (5%–35%) (Stratton et al. 1989; Chen et al. 2006). Inactivation of the P53 DNA damage pathway is common in both pediatric subtypes of RMS (Felix et al. 1992; Keleti et al. 1996). Finally, gene expression profiling and hierarchical clustering of human RMS fails to identify molecular signatures that distinguish disease subtypes based solely on morphological classification (Wachtel et al. 2004). In fact, these studies suggest that translocation-positive ARMS is molecularly distinct from both translocation-negative ARMS and ERMS, indicating that different molecular mechanisms govern the genesis of discrete disease subtypes.
Several murine models of RMS have been reported in the literature (Hahn et al. 1998; Sharp et al. 2002; Fleischmann et al. 2003; Nanni et al. 2003; Keller et al. 2004). A model of ARMS was developed in which a Pax3:Fkhr knock-in allele can be conditionally activated in muscle cells (Keller et al. 2004). Upon complete loss of the ARF locus, these transgenic mice develop malignancies that are histologically similar to human ARMS; however, tumor penetrance is low and latency is very long. A second mouse model of RMS utilizes transgenic animals that broadly misexpress the HGF/SF gene and, upon complete loss of the p16-INK4a locus, transgenic animals develop ERMS (Sharp et al. 2002). P53 inactivation, when coupled with HER-2/neu tyrosine kinase activation, can also lead to induction of RMS and salivary tumors (Nanni et al. 2003). Even more recently, a pleiomorphic RMS mouse model has been created in which RAS activation and P53 loss result in tumor formation in adult mice (Tsumura et al. 2006). Although these mouse models establish a clear role for P53 pathway disruption in the genesis of RMS and suggest that tyrosine kinase/RAS signaling pathway activation may be required for tumor initiation in translocation-negative RMS, these models require complex breeding strategies, multiple genetic perturbations, and a long latency for tumor development. Additionally, no comprehensive whole-genome approaches have been utilized to predict how well these mouse models accurately mimic human disease.
Cancer cells have the unique ability to recapitulate disease when introduced into transplant recipients, suggesting that self-renewal pathway acquisition is common in malignancy. In fact, recent studies have suggested that only a small portion of cells contained within the tumor mass have self-renewal capacity and are sufficient to cause disease. It is postulated that these rare cancer stem cells survive conventional treatment regimes, ultimately inducing secondary disease and relapse in patients. In solid tumors, such as brain (Singh et al. 2004) and breast tumors (Al-Hajj et al. 2003), the cancer stem cell has been identified; however, in most malignancies, including ERMS, the existence and characterization of the cancer stem cell have yet to be elucidated. Moreover, the mechanisms governing self-renewal are largely unknown and are now just beginning to emerge for diseases in which cancer stem cells have been identified (Krivtsov et al. 2006).
Here, we developed a robust zebrafish transgenic model of RAS-induced RMS in which nearly 50% of injected animals develop disease by 80 d of life. Zebrafish tumors express clinical diagnostic markers of human RMS and are morphologically similar to human ERMS. Microarray analysis and gene set enrichment analysis (GSEA) revealed that zebrafish RMS is similar to the human embryonal subtype of disease but not the alveolar subtype. Closer analysis of this evolutionarily conserved gene set identified a novel RAS signature up-regulated in human ERMS, pancreatic adenocarcinoma, and RAS-infected mammary epithelial cells. These results suggest that RAS pathway activation may be common in this subtype of disease. Next, we created dual fluorescently labeled RMS that allows for the identification of discrete subpopulations of cells within the tumor mass. Using fluorescence activated cell sorting (FACS), cell transplantation, and limiting dilution analysis, we identified the serially transplantable cancer stem cell in zebrafish RMS, a cell that shares a common gene expression signature with nontransformed muscle satellite cells. Microarray analysis of this population identified a unique transcriptional network that is likely associated with stem cell self-renewal in zebrafish ERMS (zERMS).
Results
A transgenic construct that drives gene expression in muscle-associated cells
The rag2 promoter is expressed in immature T- and B-cell lineages, olfactory rosettes, and sperm (Jessen et al. 2001; Langenau et al. 2004). Upon sectioning 7-, 10-, 21-, 28-, and 80-d-old rag2-EGFP-bcl2 and rag2-dsRED2 transgenic animals, transgene-expressing cells were also detected in the mononuclear component of the skeletal musculature, comprising mononuclear satellite cells, differentiating myoblasts, and the rare fusing myoblasts, but not multinucleated terminally differentiated muscle fibers (Supplementary Fig. S1; Supplemental Material). Similar results have been reported for rag1-GFP transgenic zebrafish, in which short promoter fragments drive GFP expression in muscle cells resulting from loss of an upstream negative regulatory element contained in both the rag1 and rag2 promoter loci (Jessen et al. 1999). In fact, promoter deletion analysis identified an E-box sequence (the binding site for MyoD family members) contained within the 6.5-kB rag2 promoter that is partially required for misexpression within satellite and myoblast cell populations (Supplementary Fig. S4). So although previous transgenic models utilizing the rag2 promoter driving expression of the mouse c-Myc gene develop T-cell acute lymphoblastic leukemia (Langenau et al. 2003), other tumor types may be predicted based on the aberrant activation of the rag2 promoter in nonlymphoid tissues.
Embryos injected with the rag2-kRASG12D construct develop RMS
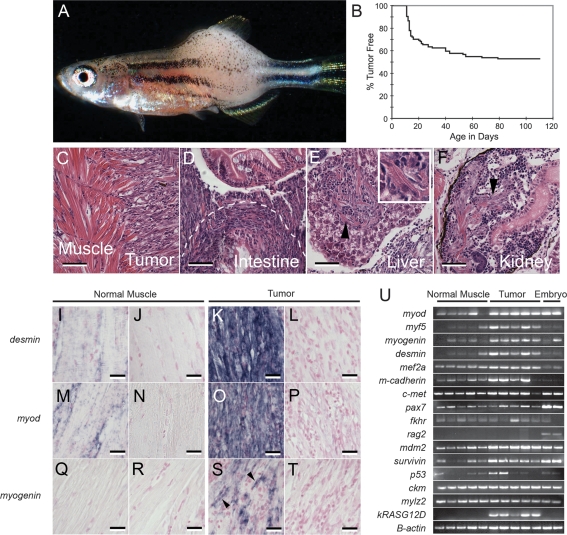
AB strain zebrafish embryos injected at the one-cell stage of development with a human kRASG12D-containing transgene (rag2-kRASG12D, 100 ng/μL) are phenotypically normal at 5 d post-fertilization (dpf) but develop externally visible tumors beginning at 10 dpf, with 47% of mosaic transgenic fish developing tumors by 80 dpf (n = 49 of 104) (Fig. 1A,B; Supplementary Fig. S2). Animals surviving past 80 d of life are likely mosaic for the rag2-kRASG12D transgene, but fail to express or have very low-level kRASG12D expression in satellite cell populations. Thus, tumor onset largely plateaus after 3 mo of age. Histological analysis revealed that tumor masses are composed of heterogeneous cell populations comprising undifferentiated muscle cells, multinucleated striated muscle fibers, and infiltrating blood cells (Fig. 1C; data not shown). Tumors were highly invasive, being found in the intestine (Fig. 1D), liver (Fig. 1E), kidney (Fig. 1F), and testes (data not shown). Striations lying within invasive tumors provided the first evidence that these malignancies were muscle in origin (Fig. 1E,F). Remarkably, lymphoid hyperplasia was observed in only one injected animal at 90 d of life (n = 1 in >1000 animals analyzed) (Supplementary Fig. S3). No other tumor types were observed in injected animals.
Figure 1.
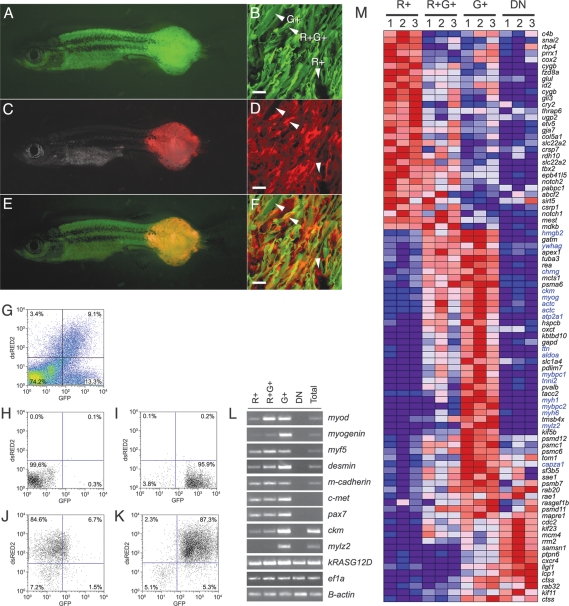
rag2-kRASG12D-injected animals develop RMS. (A) Bright-field image of a 30-d-old zebrafish with RMS. (B) Tumor onset in AB strain fish injected with rag2-kRASG12D (49 of 105 animals developed RMS by 80 dpf). (C–F) RMS cells invade into adjacent muscle (C), intestine (D), liver (E), and kidney (F). Dotted line in D outlines the outer edge of effaced intestine. Arrowheads denote striated muscle tumors. The boxed region in E is a magnified view of a striated cell in the liver. Bars: C–F, 50 μm. (I–T) kRASG12D-induced tumors express clinical diagnostic markers of RMS as determined by RNA in situ hybridization. (I,K,M,O,Q,S) Antisense probes. (J,L,N,P,R,T) Sense controls. Arrowheads in S denote multinucleated, myogenin-positive cells within RMS. Bars: I–T, 20 μm. (U) Semiquantitative RT–PCR comparing normal muscle and RMS from 30-d-old fish. Embryo cDNA served as a positive control in most samples (24 hpf). (mylz2) myosin light chain 2; (ckm) creatine kinase.
Tumors from 30-d-old animals expressed high RNA levels of clinical diagnostic markers of human RMS including desmin and myod within both the fibrous and small, round cell populations (n = 9) (Fig. 1I–P). In contrast, myogenin was expressed predominantly in the multinucleated fibers contained within the tumor mass and only rarely in mononuclear tumor cells (Fig. 1Q–T). Tumors also express high transcript levels for satellite cell markers (c-Met, m-cadherin, and myf5) and myoblast differentiation genes (myod, mef2a, and myogenin), again suggesting that RAS-induced RMS are highly heterogeneous (Fig. 1U). kRASG12D was expressed only in tumor while rag2 expression was absent in both tumor and normal muscle, indicating that promoter expression does not accurately recapitulate endogenous rag2 expression (Supplementary Fig. S4; Supplemental Material).
Conserved molecular pathways underlying RMS in zebrafish and humans
GSEA is a computational method for assessing whether a predefined gene set is statistically enriched in one biological state as compared with another (Subramanian et al. 2005). This method has been used in human (Ramaswamy et al. 2001) and mouse (Sweet-Cordero et al. 2005) to identify gene signatures associated with cancer and to classify zebrafish tumor types based on gene expression (Lam et al. 2006). In these latter experiments, chemically induced zebrafish liver tumors were shown to be most similar to human liver malignancies, but not prostate, lung, or gastric cancers, providing strong evidence that GSEA can be used to classify tumors based on expression profiling and cross-species comparisons. In our analysis, we used GSEA to assess whether conserved pathways are activated in both zebrafish and human RMS. The gene sets were determined experimentally by microarray analysis comparing zebrafish RAS-induced RMS (n = 8) to normal control muscle (n = 9) at 1.75-, 2.0-, 2.25-, 2.5-, and 3.0-fold change levels (for 2.25-fold change, see Supplementary Table S1). Several fold change cut-offs were utilized in our GSEA analysis to verify that differences between disease and normal states were reproducible and not due to arbitrary assignment of gene lists. The corresponding human and mouse homologs were identified on several array platforms (Supplementary Table S2) and analyzed for enrichment in data sets comprising a cancer state compared with a normal tissue state. Human pediatric ERMS and translocation-positive ARMS (Wachtel et al. 2004) were compared with normal juvenile muscle samples (Kang et al. 2005) using GSEA.
The zebrafish up-regulated gene sets were significantly associated with the human ERMS data set at all five fold changes but never with the ARMS data set (for 2.25-fold change, see Figs. 2A–C, 3A; for all other fold changes, see Supplementary Table S3). Rank-ordered gene lists for human ERMS compared with normal muscle at 2.25-fold change are provided in Supplementary Table S4. In contrast, the down-regulated gene sets identified in zebrafish RMS were not significantly down-regulated in either human ERMS or ARMS (Figs. 2D,E, 3A). Additional comparisons verified that our GSEA results could not be ascribed to differences in hybridization techniques utilized in normal muscle and RMS samples (Supplemental Material; Supplementary Table S3). In total, the GSEA analysis at the 2.25-fold change cut-off is representative of those completed at 1.75-, 2.0-, 2.5-, and 3.0-fold (Supplementary Table S3) and was subsequently used to define the up-regulated and down-regulated zERMS gene sets. The up-regulated zERMS gene set contains 329 probe sets, of which 166 known gene homologs can be identified in human, and the down-regulated gene set contains 314 probe sets, of which there are 130 homologous human genes (Supplementary Table S1). GSEA comparisons are graphically represented in Figure 2 (an associated gene list is provided in Supplementary Table S5). Finally, a subset of transcriptional targets identified in this analysis were validated by real-time quantitative RT–PCR comparing zebrafish RMS to normal muscle, establishing that array analysis identifies true transcriptionally regulated gene products in RMS (Supplementary Figure S5).
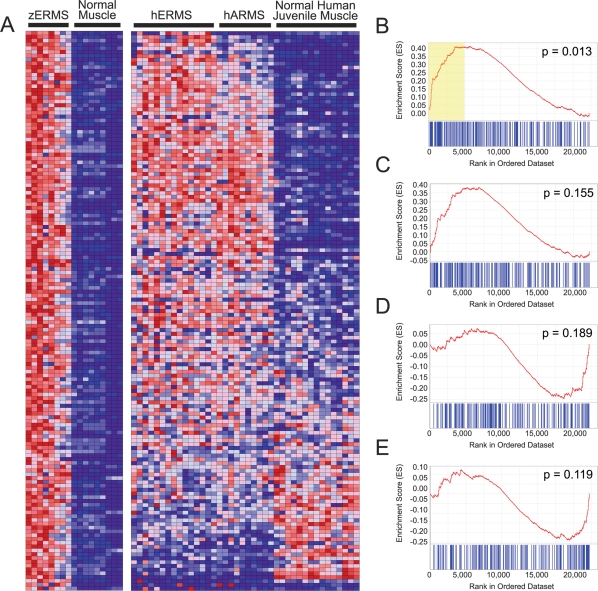
Figure 2.
GSEA identifies a conserved gene signature found in both zebrafish and human ERMS. (A) Heat map showing genes up-regulated in zERMS when compared with normal muscle at 2.25-fold change (left) and juxtaposed to the corresponding human orthologs in ERMS, ARMS, and normal juvenile muscle (right). (B–E) Graphical representation of the rank-ordered gene lists found when comparing human RMS to normal muscle. The up-regulated gene set identified in zebrafish RMS is significantly enriched in human ERMS (B; ES = 0.414, NES = 1.518, FDR q-val = 0.023, p = 0.013) but not the alveolar subtype (C; ES = 0.384, NES = 1.251, FDR q-val = 0.223, p = 0.155). The down-regulated gene set identified in zebrafish RMS is not significantly enriched in either ERMS (D) or ARMS (E). The yellow box in B defines the genes that contribute maximally to the GSEA score in human ERMS. (ES) Enrichment score.
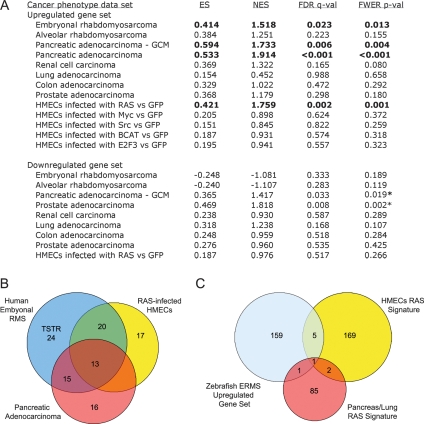
Figure 3.
GSEA identifies a novel evolutionarily conserved RAS signature and a tumor-specific signature associated with ERMS. (A) The up-regulated gene set identified in zERMS is significantly enriched in the human ERMS, pancreatic adenocarcinoma, and RAS-infected mammary epithelial cell (HMEC) data sets (denoted by bold lettering), while the down-regulated gene set is not significantly enriched in any data set. (ES) Enrichment score; (NES) Normalized Enrichment Score; (FDR) False discovery rate; (FWER p-val) FWER p-value. The asterisks denote samples that have discordant gene set enrichment, exhibiting up-regulation of a down-regulated gene set. (B) The genes from the up-regulated zERMS gene set that contribute maximally to the GSEA score in the pancreatic adenocarcinoma, human ERMS, and RAS-infected HMEC data sets differ. The 24 genes comprising the TSTR are marked. (C) Previously identified RAS signatures share few genes in common with the up-regulated gene set identified in the zebrafish transgenic model of RAS-induced RMS (2.25-fold change gene list). Genes contained within each overlapping group are noted in B and C.
Identification of a novel RAS signature and a RMS-specific signature
We questioned whether this up-regulated zERMS gene set was found in other tumor types, identifying a “cancer-associated” gene set rather than one specific to the ERMS phenotype. Unexpectedly, the zERMS up-regulated gene set was significantly associated with the human pancreatic adenocarcinoma data set (Iacobuzio-Donahue et al. 2003; Sweet-Cordero et al. 2005) but not the renal cell carcinoma, lung, colon, or prostate adenocarcinoma data sets (for 2.25-fold change, see Fig. 3A; for additional fold changes, see Supplementary Table S6) (Ramaswamy et al. 2001). Given that kRASG12D mutations are found in >90% of pancreatic adenocarcinomas and that the zERMS up-regulated gene list was identified using a zebrafish transgenic model of human kRASG12D-induced RMS, we questioned whether our up-regulated zebrafish gene set comprised a RAS-specific signature. To test this hypothesis, human mammary epithelial cells (HMECs) infected with activated RAS, MYC, SRC, B-CATENIN, or E2F3 were compared with cells infected with GFP using GSEA and our zERMS gene sets (Bild et al. 2006). The zERMS up-regulated gene set was significantly associated with RAS status (Fig. 3A; for a rank order gene list at 2.25-fold change, see Supplementary Table S4; for additional fold change analysis, see Supplementary Table S6) but not MYC, SRC, B-CATENIN, or E2F3. The down-regulated gene set was not associated with oncogene status (Fig. 3A). Next, we compared kRasG12D-induced mouse lung adenocarcinomas to normal lung using GSEA (Sweet-Cordero et al. 2005). The up-regulated zERMS gene set was significantly associated with mouse lung adenocarcinoma at multiple fold change differences (2.25-fold change, enrichment score [ES] = 0.406, normalized ES [NES] = 1.439, false discovery rate [FDR] = 0.047, p = 0.026; for additional fold changes, see Supplementary Table S6). Closer analysis of the genes contained within our data set revealed several known transcriptional targets of RAS including mcl-1 (Irvine et al. 2004), mdm2, dusp4 (Yip-Schneider et al. 2001), pim1 (Krumenacker et al. 2001), and g3bp (Irvine et al. 2004). Taken together these experiments validate that our zERMS up-regulated data set comprises a bona fide RAS signature.
We questioned whether our zERMS up-regulated gene signature was specific to RAS status rather than identifying up-regulated genes involved in tumor-specific and tissue-restricted pathways (TSTR) associated with the ERMS phenotype. Thus, we defined the RMS-specific TSTR (24 genes in total), a subset of up-regulated genes contained within our zERMS up-regulated gene set that contribute maximally to the GSEA score in human ERMS but not pancreatic adenocarcinoma (Global Cancer Map [GCM] data set from Ramaswamy et al. 2001) or HMECs infected with RAS (Fig. 3B; Supplementary Table S7). The TSTR was significantly enriched in human RMS but not HMECs infected with activated RAS (Supplementary Table S8). In fact, muscle regulatory factor 5 (MYF5) is contained within the TSTR (MYF5 is the first probe set identified in Fig. 2A), suggesting that the expression of this gene likely identifies and participates in the lineage and stage-specific state of RMS cells. From this analysis, we conclude that the up-regulated zERMS gene set defines at least two distinct gene signatures, one being associated with RMS-specific pathway activation and a second associated with kRASG12D status.
ERMS onset can be modified by P53 pathway disruption
Given that P53 is mutationally inactivated in human ERMS (Xia et al. 2002), we questioned whether this pathway was altered in tumors arising in AB strain wild-type fish. RT–PCR analysis of zERMS revealed that p53 expression was variable in both normal muscle and ERMS (Fig. 1U), while sequencing of the p53 locus from wild-type tumors (n = 7 fish, exons 4–9) failed to identify inactivating mutations in this gene, suggesting that alternative mechanisms may exist to disrupt this pathway within zebrafish tumors. In fact, mdm2 and survivin expression were elevated in zERMS when compared with normal muscle (Fig. 1U). Both of these gene products suppress P53-dependent apoptosis and are overexpressed in ERMS (Keleti et al. 1996; Caldas et al. 2006).
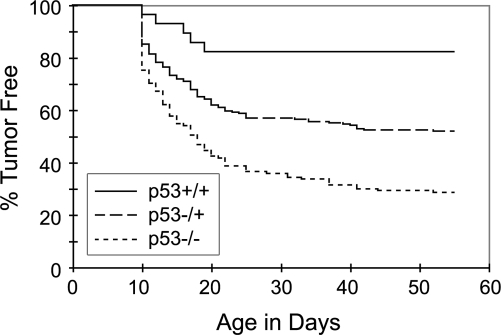
Given that P53 pathway modulators were up-regulated in our zERMS model, we wanted to assess whether p53 pathway disruption collaborates with tumor onset in mosaic transgenic animals. The rag2-kRASG12D transgene was injected into heterozygous and homozygous p53 loss of function (p53 LOF, Tu strain) (Berghmans et al. 2005) mutant incrosses at the one-cell stage of life. Heterozygous and homozygous p53 LOF fish have markedly increased tumor incidence compared with wild-type injected siblings (p = 0.0039 and p < 0.00001, respectively) (Fig. 4). Additionally, homozygous p53 LOF animals developed more tumors than heterozygous p53 LOF siblings (p = 0.00001) (Fig. 4). Interestingly, wild-type Tu strain animals (the strain in which P53-LOF studies were completed) developed fewer tumors than AB strain fish (cf. Figs. 4 and 1B), suggesting that as is seen in mouse, strain differences can affect tumor onset and development. Taken together, our experiments suggest that despite the rapidity of tumor onset in our model, tumor latency can be modified by altered p53 pathway deregulation.
Figure 4.
p53 pathway deregulation alters tumor onset in zebrafish RAS-induced ERMS. p53 LOF collaborates with kRASG12D to increase penetrance of RMS in animals injected with the rag2-kRASG12D transgene at the one-cell stage of life (wild-type vs. heterozygotes, p = 0.0039; wild-type vs. homozygotes, p < 0.00001; heterozygotes vs. homozygotes, p = 0.00001; n = 98 of 137 homozygous p53 LOF fish developed tumors, n = 103 of 217 in heterozygous fish, and n = 5 of 28 in wild-type fish).
Establishing a coinjection approach for labeling cell populations in zERMS
Transgenes integrate into the genome as concatamers (Houdebine and Chourrout 1991); thus, we reasoned that coinjection of two constructs into one-cell stage embryos may lead to cosegregation of transgenes within developing tumors. rag2-dsRED2 and rag2-kRASG12D constructs were coinjected into α-actin-GFP transgenic embryos at the one-cell stage of development (Fig. 5). Because α-actin-GFP is expressed in more mature muscle cells and is not expressed in satellite cells (Beauchamp et al. 2000), we anticipated that this strategy would allow for the differential labeling of RMS cell populations based on differentiation status. Most coinjected animals that developed RMS had dsRED2-labeled malignancy by 30 d of life (n = 60 of 62) (Fig. 5A–F), validating that coinjection of two trangenes leads to coexpression in a vast majority of developing tumors. GFP and dsRED2 expression was found in distinct cell populations as well as colocalized within developing tumors (Fig. 5A–F; Supplementary Figure S6). FACS analysis confirmed that the mononuclear component of the tumor was also comprised of four populations of cells (double negative [DN], GFP+/dsRED2− [G+], dsRED2+/GFP− [R+], and double positive [R+G+]) (Fig. 5G). Each of these four populations were isolated by FACS (purity ranging from 84.6% to 99.6%) (Fig. 5H–K) and assessed for expression of muscle markers as determined by semiquantitative RT–PCR analysis (Fig. 5L). All four cell populations expressed human kRASG12D, while the R+ population exhibited satellite cell marker expression (cMet+, m-cadherin+, and myf5+) and lower or undetectable levels of myoblast and mature muscle markers (myod, myogenin, creatine kinase, and myosin light chain 2 [mylz2]) when compared with either the R+G+ or G+ cell populations. In contrast, the DN population expressed lower levels of kRASG12D when compared with the other three populations and high levels of blood cell markers (data not shown).
Figure 5.
Coinjection strategies can be used to label distinct cell populations within zebrafish RMS. (A) GFP fluorescent image of RMS developing in a rag2-dsRED2+/α-actin-GFP+-injected animal. (B) GFP fluorescence in cryostat section. (C) dsRED2 fluorescence image of injected fish shown in A. (D) dsRED2 fluorescence in cryostat section. (E,F) Merged images. Arrowheads in B, D, and F denote cells that expresses GFP (G+), dsRED2+ (R+) or both (R+G+) within zebrafish RMS. Bars: B,D,F, 20 μm. (G) FACS profile of α-actin-GFP transgenic animal injected at the one-cell stage with rag2-dsRED2 and rag2-kRASG12D. (H–K) The four cell populations can be isolated to relative purity by FACS. (L) Semiquantitative RT–PCR analysis confirms that expression of dsRED2+ and GFP+ can be used to identify discrete populations of tumor cells based on their stage of muscle differentiation. Total refers to total cells isolated from RMS by FACS based on cell viability. (M) Microarray analysis of sorted cell populations from three tumors (numbered 1–3 at top of heat map). Gene symbols are at right, with blue denoting genes expressed in normal differentiating muscle cells.
To confirm that our transgenic approach labels tumor cells based on differentiation status, we performed microarray analysis on sorted cell populations. From this analysis, we find that the four tumor cell populations are molecularly distinct from one another (Fig. 5M; associated Affymetrix identifiers provided in Supplementary Table S5). In fact, the R+ population contains genes expressed in early, undifferentiated muscle populations. For example, cox2 has been hypothesized to regulate satellite cell proliferation, differentiation, and fusion (Mendias et al. 2004) and is expressed in rapidly proliferating C2C12 cells grown in high serum but subsequently down-regulated during differentiation (Tomczak et al. 2004). Additionally, id2, gli3, and notch1 are highly expressed in the R+ cell population and are rapidly up-regulated in C2C12 cells after withdrawal of serum, indicating that these genes are expressed in early, dividing muscle progenitor populations (Delgado et al. 2003). By contrast, the R+G+ population transcribes genes known to be expressed later in muscle differentiation, including creatine kinase (ckm), myogenin (myog), α-actin (actc), high mobility group box 2 (hmgb2), and acetylcholine receptor γ subunit (chrng), whereas the G+ population transcribes genes that are expressed even later in muscle development including myosin heavy chains (myh1 and myh6), mylz2, and troponin I (tnni2) (Tomczak et al. 2004). Finally, the DN population is comprised of blood cells, expressing the macrophage markers l-plastin (lcp1) and cathepsin S (ctss). From our RT–PCR and microarray analyses, we conclude that our cell labeling procedures identify RMS cells based on differentiation status.
Identifying the serially transplantable cell population in ERMS
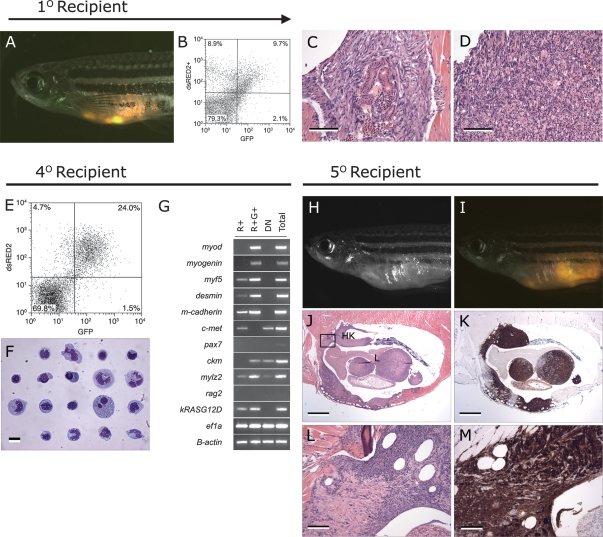
To assess whether zERMS are transplantable, unsorted primary tumor cells were isolated from rag2-dsRED2+/α-actin-GFP+ RMS and injected intraperitoneally into sublethally irradiated primary recipients (2 × 104 to 4 × 103 cells per animal, n = 10 tumors analyzed). Recipient fish developed small bundles of dsRED2+/GFP− cells within the peritoneal cavity by 7 d post-transplant and then progressed to having dsRED2+/GFP+ masses near the site of injection by 14 d (Fig. 6A). Tumor heterogeneity was largely similar to that found in primary tumors, with transplanted RMS having spindled cell components (Fig. 6C) and/or large cell aggregates (Fig. 6D). FACS analysis also confirmed heterogeneity of engrafted tumor cells with transplant animals containing three distinct populations of mononuclear cells (n = 3, R+, R+G+, and DN) (Fig. 6B). Remarkably, the G+ cell populations are severely diminished or absent in primary transplant recipients. These results, in conjunction with RT–PCR results (Fig. 5L), likely indicate that G+ cells found in primary tumors contain both RAS-affected cells and residual normal cell populations, and that over time, RAS-expressing cell populations may acquire additional genetic or epigenetic alterations that further perturb the muscle differentiation program.
Figure 6.
The dsRED2+ cell population from double-transgenic rag2-dsRED2/α-actin-GFP animals contains the serially transplantable cancer stem cell in zERMS. (A–D) Primary transplanted tumors from α-actin-GFP+/rag2-dsRED2+ fish (1° Recipient). (A) Merged image of GFP fluorescent, dsRED2 fluorescent, and brightfield images. (B) FACS analysis of primary recipient engrafted with ERMS. (C,D) Histological analysis reveals heterogeneity in transplant animals, with some fish having masses of spindled cells (C) or round cell aggregates (D), or both. Bars: C,D, 100 μm. (E–G) Cells isolated from serially transplanted animals, in this case a quaternary recipient animal (4° Recipient). (E) FACS plot of tumor cells isolated from a 4° recipient. (F) Wright-Giemsa-stained cytospins of FACS-sorted R+ cells from quaternary tumor. (G) Semiquantitative RT–PCR analysis of FACS-sorted cell populations. Total refers to total cells isolated from quaternary transplanted RMS isolated by FACS based on cell viability and that serve as an input control. (H–M) Fish transplanted with 50 R+ cells defined in E–G (5° Recipient). (H) Bright-field image of transplant recipient animal. (I) Merged image of a dsRED2+/GFP+ tumor in same animal. Hematoxylin and eosin-stained (J) and anti-GFP-immunostained (K) section of transplanted fish showing that RMS cells infiltrate the liver (L), head kidney (HK), and skeletal muscle. (L,M) High-power magnification of boxed region in J. Bars: J,K, 1 mm; L,M, 100 μm.
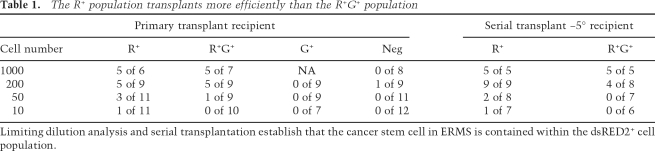
To define the population of cells that contains the ERMS transplantable cell population, FACS-sorted cells were obtained from primary rag2-dsRED2+/α-actin-GFP+ RMS and injected into irradiated recipient fish at limiting dilution (2 × 104, 4 × 103, 1 × 103, 200, 50, and 10 cells). In the example depicted in Table 1, the FACS-sorted cell populations were enriched after the first FACS procedure (R+, 83.3%; R+G+, 82.1%; G+, 52.2%; and DN, 98.9%). In the four tumors analyzed by limiting dilution analysis, the R+ gated cell population exhibited superior engraftment potential when compared with either the R+G+, G+, or the DN cell populations (Table 1). Primary recipients transplanted with R+ cells developed ERMS that were indistinguishable from animals transplanted with unsorted cell populations (data not shown).
Table 1.
The R+ population transplants more efficiently than the R+G+ population
Limiting dilution analysis and serial transplantation establish that the cancer stem cell in ERMS is contained within the dsRED2+ cell population.
To further confirm that the R+ population contains the ERMS transplantable cell population, FACS-sorted cell populations from serially transplanted tumors were assessed for engraftment potential. In the example shown (Fig. 6), ERMS cells were serially propagated from primary tumor-bearing animals to irradiated recipients, allowed to engraft, and then were reisolated and again introduced into irradiated hosts. In total, the cells were serially transplanted four times, and then subpopulations of ERMS cells were isolated from quaternary recipient animals using FACS (R+, 97% purity; G+R+, 85% purity) (Fig. 6E–G). Despite the R+ population being very pure after sorting, this population remains morphologically heterogeneous (Fig. 6F), suggesting that tumor cells contained in this sorted cell population comprise a mixture of muscle cell types. However, semiquantitative RT–PCR analysis of FACS-sorted quaternary transplant populations revealed that the R+ cells have an expression signature similar to that of activated satellite cells (cMet+, m-cadherin+, myf5+, and myod−) (Fig. 6G; Cornelison and Wold 1997). In contrast, pax7, a marker of quiescent satellite cells, is not expressed at high levels in this population, nor was it differentially expressed in whole tumor when compared with normal muscle. In contrast, the R+G+ cells comprise more differentiated mononuclear cells, expressing higher levels of myod, myogenin, ckm, and mylz2. Taken together, these results suggest that the R+ ERMS cancer stem cell is molecularly similar to normal muscle satellite cells, and that like satellite cell populations (Dhawan and Rando 2005), heterogeneity may also be observed in the ERMS cancer stem cell population.
FACS-sorted cell populations were introduced into irradiated, 5° recipients at limiting dilution (Fig. 6H–M), confirming that as was observed in primary tumors, the R+ cell population is enriched for tumor initiating cells (Table 1). As few as 10 R+ cells were required for transplantation of disease into irradiated 5° recipient fish by 14 d post-transplantation (n = 1 of 7). Serially transplanted ERMS in 5° recipient animals largely recapitulated the heterogeneity observed in both primary tumors (Fig. 1C–F) and transplanted tumors arising in primary recipients (Fig. 6C–M). By contrast, the R+G+ population was less-efficient at inducing disease (n = 0 of 7 and n = 0 of 6 animals injected with 50 and 10 cells, respectively), while the DN population from this tumor failed to engraft in secondary and tertiary recipient animals (DN, 99% purity; n = 0 of 8 and n = 0 of 7 fish, 2 × 104 cells per animal, respectively). Similar results were obtained for two other serially transplanted ERMS (Supplementary Table S9) and additional experiments validated that the R+ stem cell was serially transplantable (Supplemental Material).
Discussion
Conserved molecular pathways in ERMS
The molecular pathways underlying zebrafish development are remarkably conserved when compared with mouse and human (Thisse and Zon 2002); however, it is largely unknown whether cancer pathways are also conserved in the zebrafish. In the case of zebrafish RAS-induced RMS, tumors express clinical diagnostic markers of human disease and appear morphologically similar to human ERMS. Additionally, although p53 is not mutationally inactivated in zebrafish RAS-induced RMS, p53 pathway disruption significantly alters tumor onset, suggesting that suppression of the p53 response pathway may also be important for RMS tumor initiation in our model. In fact, survivin and mdm2 are up-regulated in zebrafish RMS and are known suppressors of the p53 pathway (Xia et al. 2002; Caldas et al. 2006). MDM2 is commonly amplified and overexpressed in human ERMS (Keleti et al. 1996) and SURVIVIN overexpression is observed in human RMS, with suppression of this protein within established tumors leading to tumor regression (Caldas et al. 2006). Finally, microarray and GSEA uncovered gene expression signatures that are up-regulated in both human ERMS and zebrafish RMS, underscoring the molecular similarity of these diseases. When comparing our up-regulated zebrafish RMS gene signature to human RMS, the most striking finding was that MYF5 is differentially up-regulated in both zebrafish and human ERMS, but not ARMS, suggesting that MYF5-expressing cells are likely overrepresented in the ERMS subtype of disease. In fact, we find that the cancer stem cell in zebrafish RMS is most similar to nontransformed, activated satellite cell populations, which express myf5. Taken together, our data strongly supports the conservation of molecular pathways involved in the genesis of ERMS in both zebrafish and human.
One unexpected finding of our microarray and cross-species comparisons was the identification of a RAS signature in human ERMS. This signature was also up-regulated in human pancreatic adenocarcinoma, a malignancy in which 90% of patients have kRASG12D mutations, and HMECs infected with RAS but not MYC, SRC, B-CATENIN, or E2F3. By contrast, the down-regulated gene signature identified in zERMS is not enriched in any of these data sets and does not correlate with RAS status. Similar results were reported for RAS signatures identified in a mouse model of kRasG12D-induced lung adenocarcinoma where the RAS signature comprised only up-regulated genes (Sweet-Cordero et al. 2005). Although it is formally possible that genes down-regulated by RAS are not conserved between species, it is far more likely that RAS may preferentially activate gene transcription.
The up-regulated gene sets found in zERMS contain direct targets of the RAS pathway. For example, G3BP has been shown to be a downstream effector of RAS signaling (Parker et al. 1996; Irvine et al. 2004) and is significantly up-regulated in human breast cancers (Barnes et al. 2002) and ERMS. Similarly, the anti-apoptotic gene, MDM2, is regulated by RAS signaling pathways (Ashcroft et al. 2002), as is Dusp4/MKP2, a gene that is involved in repressing the ERK pathway and that ultimately compensates for high levels of RAS activation within transformed cells (Yip-Schneider et al. 2001). Remarkably, although our up-regulated zERMS gene set contains transcriptional targets of RAS, the RAS signature we identified in zebrafish RMS is different from those previously identified (Sweet-Cordero et al. 2005; Bild et al. 2006). Moreover, the RAS signatures identified by Sweet-Cordero et al. (2005) and Bild et al. (2006) also differ from one another. This result can be explained by two possibilities. First, microarray and bioinformatics approaches define gene sets based on the identification of the most differentially regulated gene transcripts between two data sets. Those genes that are differentially regulated, but expressed at lower transcript levels, are excluded from these gene sets. Second, transcriptional targets of the RAS pathway likely differ based on cellular context. This may reflect the use of different tissue-specific RAS pathways or the differential activation of pathways downstream from RAS. Because each of these three signatures is up-regulated in multiple RAS-affected tumors and tissues (Sweet-Cordero et al. 2005; Bild et al. 2006), our data suggest that RAS regulates a larger set of transcriptional targets than previously appreciated.
RAS family members are mutationally activated infrequently in pediatric ERMS (Stratton et al. 1989; Chen et al. 2006). Additionally, Chen and colleagues have reported that SHP2 (PTPN11) mutations were relatively uncommon in human RMS (Chen et al. 2006) and mutations in BRAF are absent in RMS (Miao et al. 2004). Both of these genes are mutated in human cancer and exert their oncogenic effects through activation of the RAS pathway. Our data imply that RAS pathway activation is common in human ERMS. We suggest that RAS pathway activation resulting from as-yet-unidentified molecular mechanisms may be involved in the genesis of a majority of human ERMS, possibly occurring through mutational activation of kinases or other positive regulators of RAS. Alternatively, it is also possible that suppressors of RAS activity may be deleted or inactivated in muscle malignancies.
Identifying the cancer stem cell in zERMS
The cancer stem cell in zERMS was identified by utilizing fluorescent transgenic approaches in conjunction with FACS and cell transplantation. Specifically, the rag2-kRASG12D transgene was coinjected with rag2-dsRED2 into α-actin-GFP transgenic animals. Primary RMS contain four distinct populations of cells, all of which express human kRASG12D. By transplanting each sorted cell population into irradiated recipients, we establish that the rag2-dsRED+/α-actin-GFP− cell population (R+) is better at engrafting disease than any other tumor subpopulation. Moreover, most tumors that developed in transplanted animals contained R+, rag2-dsRED2+/α-actin-GFP+ (R+G+), and DN cell populations, while the α-actin-GFP+ (G+) population was lost or severely reduced upon serial passage. These data suggest that RAS is sufficient to induce RMS in our zebrafish transgenic model but that tumor cells likely acquire additional perturbations that disrupt normal muscle developmental programs, ultimately causing a full block in differentiation. In human patients, early tumor development is difficult to detect and thus, these events are necessarily difficult to study, yet it is likely that genetic perturbations that block differentiation are also found in human tumors. Additional gene expression studies comparing differentiation status of zERMS upon serial transplantation will likely uncover molecular pathways required in this process.
The rag2-dsRED2+/α-actin-GFP− cancer stem cell population in zebrafish RAS-induced RMS is molecularly similar to nontransformed, satellite cells. Both of these cell types express early muscle markers including myf5, m-cadherin, c-MET, and desmin. Moreover, microarray analysis of the RMS cancer stem cell population identifies a set of genes that is both differentially up-regulated when compared with other ERMS tumor cell types and yet exclusively expressed in early normal, mononuclear muscle cell populations. For example, id2, gli3, cox2, and notch1 are transcriptionally activated in dividing C2C12 cells and upon transfer to low-serum conditions, cells repress expression of these genes and begin to differentiate (Delgado et al. 2003; Tomczak et al. 2004). Gli3 is required for Myf5 transcription in a subset of muscle progenitor cells (McDermott et al. 2005) and Id2 is a potent inhibitor of the MyoD transcription factor (Benezra et al. 1990). Thus, it is likely that these two gene products act in concert to induce myf5 while actively repressing myod activity in the zebrafish RMS stem cell. Finally, notch signaling is important for asymmetric cell division occurring in activated satellite cells, with dividing cells producing a committed myogenic precursor and a quiescent satellite cell (Conboy and Rando 2002). Thus, it is possible that notch activity in both the ERMS cancer stem cell and the rag2-dsRED2+/α-actin-GFP+ differentiated tumor cell populations reflects use of this signaling pathway in division of the ERMS cancer stem cell. Taken together, microarray analysis of the ERMS cancer stem cell population suggests that a subset of molecular pathways involved in normal satellite cell self-renewal may also regulate proliferation and self-renewal pathways in RMS.
The cancer stem cell and the tumor-initiating cell have not been previously identified in either ERMS or ARMS; however, mouse models of ARMS suggest that the cancer-initiating cell in this subtype of disease may be a committed progenitor cell rather than a satellite cell (Keller et al. 2004). In such a setting, committed-muscle progenitors that develop into the ARMS cancer-initiating cell must reacquire self-renewal capacity. Our data suggests a different self-renewal mechanism in ERMS. In our model, the cancer stem cell population is more similar to normal, activated satellite cells rather than committed, differentiating progenitors. These results suggest that the tumor initiating cell and cancer stem cell may be one in the same in ERMS: an activated satellite cell. Such a finding would explain why ERMS is predominantly a pediatric disease and is more common than the alveolar subtype. For example, P53 pathway disruption and activating mutations contained within RAS family members may be sufficient to induce disease when occurring within muscle satellite cells. In such a setting, mutations that confer reacquisition of self-renewal would not be required. In fact, studies in which oncogenic N-ras or H-ras were introduced into myoblast cells results in suppression of both differentiation and fusion (Olson et al. 1987); however, studies linking RAS activation to the retention or stimulation of self-renewal in muscle have yet to be reported. It will be interesting in subsequent experiments to delineate the contribution of RAS to differentiation arrest and/or self-renewal in the ERMS stem cell. Taken together, our results suggest that common molecular pathways are found in both satellite cells and the ERMS cancer stem cell, raising the interesting possibility that the satellite cell may be the cell of origin in this disease and that the molecular pathways regulating self-renewal may be similar between these cell types.
In conclusion, our experiments in the zebrafish have led to the identification of evolutionarily conserved pathways in ERMS and the isolation of the cancer stem cell in this disease, opening new avenues of investigation to better understand the molecular pathways governing rhabdomyosarcomagenesis and cancer stem cell self-renewal.
Materials and methods
Animals
Zebrafish maintenance and developmental staging were conducted as described previously (Langenau et al. 2003).
Vectors
The human kRASG12D and dsRED2 ORFs were amplified by PCR, digested with BamHI and HindIII, and cloned into the rag2-GFP vector (Jessen et al. 2001; Langenau et al. 2003).
RT–PCR fragments generated from 24-h wild-type AB embryo cDNA were cloned into the pGEMT-easy vector (Promega). Plasmids containing myod, desmin, and myogenin were obtained and used to generate in situ probes. All PCR primers are available in Supplementary Table S10.
Microinjection and creation of stable transgenic lines
The rag2-kRASG12D and rag2-dsRED2 constructs were linearized with XhoI, phenol:chloroform-extracted, ethanol-precipitated, resuspended in 0.5× TE + 100 mM KCl, and injected into one-cell stage AB strain, α-actin-GFP transgenic (Higashijima et al. 1997), or p53 LOF (p53M214K) (Berghmans et al. 2005) animals.
Immunohistochemistry and RNA in situ hybridization
Paraffin embedding and sectioning, in situ hybridization, cryostat sectioning, and immunohistochemical analysis were performed essentially as described (Guyon et al. 2003; Langenau et al. 2005).
RT–PCR analysis
RNA was isolated from whole tumors, wild-type muscle, and FACS-sorted cell populations (Trizol, GIBCO-BRL). RNA was treated with DNaseI prior to reverse transcription, and RT–PCR was performed. PCR primers and thermocycling conditions are described in detail (Supplementary Table S10; Supplemental Material).
Statistical analysis
The Fisher exact test was used to compare the numbers of fish with and without tumor at day 55 in the three genotypic groups (p53+/+, p53+/−, and p53−/−). Pairwise comparisons were performed; there was no adjustment for multiple comparisons. Age at tumor onset was presented graphically using the method of Kaplan and Meier (1958).
Sequencing of the p53 locus
Sequencing of the p53 gene locus in tumors was completed as previously described (Langenau et al. 2005).
Microarray analysis and GSEA
RNA was collected from eight zebrafish RMS and nine normal muscle samples (AB strain, 30 dpf). Complementary RNA was prepared from 5 μg of RNA and hybridized to zebrafish Affymetrix microarrays. Probe cell intensity (CEL) files were imported into D-chip, normalized in batches against an invariant set, and filtered according to the following criteria: B/E or E/B (fold change) > 1.5 using a 90% confidence bound of fold change, E − B or B − E > 100, p value for testing E = B < 0.05. To account for multiple comparisons, 100 permutations of the data were completed. From this analysis, 1817 differentially regulated probe sets were obtained (1094 up-regulated and 723 down-regulated). The median false discovery was 0.1% and the 90% FDR was 1.8%. Subsequently, various fold change lists (1.75, 2.0, 2.25, 2.5, and 3.0) were compiled based on the lower confidence bound, a more conservative estimate of fold change.
Annotation of the zebrafish probe sets was completed using the both the Affymetrix NetAffx Analysis Center (http://www.affymetrix.com/analysis/index.affx) and the Zon Laboratory/Children’s Hospital Zebrafish Project Initiative homepage (http://134.174.23.167/ZonRHmapper).
For published microarray data sets, original CEL files were obtained (Supplementary Table S2; Ramaswamy et al. 2001; Wachtel et al. 2004; Sweet-Cordero et al. 2005; Bild et al. 2006). Files were imported into D-chip and normalized against an invariant set, and gene cluster text (GCT) files containing all probes were generated. For the second pancreatic adenocarcinoma and normal pancreas data set, GCT files were obtained directly (Supplementary Table S2; Iacobuzio-Donahue et al. 2003). GSEA analysis was completed using phenotype permutation with a weighted enrichment statistic and a Singal2Noise Metric for ranking genes. One-thousand permutations of the data were completed to obtain an FDR q-val. Significance was defined as having an FDR q-val < 0.25 and a FWER p-value of <0.05 (FWERP p-val). Human and mouse gene sets used in our analysis are described in more detail online (Supplemental Material).
For microarray analysis performed on FACS-sorted RMS cell populations, RNA was extracted from 1.5 × 104 to 4.5 × 104 sorted cells and amplified twice to obtain enough probe to hybridize to arrays.
FACS
Kidney and muscle cells from transgenic rag2-EGFP-bcl-2 and α-actin-GFP fish and tumor cells from kRASG12D-induced RMS were isolated. For muscle and tumor preparations, samples were minced in 10 mL of 0.9× PBS and treated with 100 μL of liberase III (25 μg/mL; Roche) at room temperature for 30 min. Subsequently, 500 μL of FBS was added to inactivate liberase enzymes. Samples were filtered twice (40 μm), centrifuged at 1000g for 5 min, and resuspended in 500 μL of 0.9× PBS + 5% FBS containing propidium iodide. FACS was completed as previously described (Traver et al. 2003). All samples were double sorted to obtain highly enriched cell populations and doublets were excluded based on size.
Transplantation
Fluorescently labeled single-cell preparations isolated by FACS were injected into irradiated AB strain adult fish (23 Gy, 2 d prior to transplant). For limiting dilution analysis, 2 × 104 RBCs were used as carrier cells along with RMS cells. Transplantation was completed essentially as described (Langenau et al. 2003; Traver et al. 2003).
To assess tumor engraftment of cells from double-transgenic rag2-dsRED2, α-actin-GFP animals, transplant fish were analyzed for dsRED2 and GFP fluorescence using a dissecting microscope at 7, 11, 14 or 18 d post-injection. Tumor-positive fish are defined as having either dsRED2+/GFP− or dsRED2+/GFP+ tumor masses. A subset of transplant fish were sacrificed to verify that (1) fluorescence equated with tumor formation, and (2) fish scored as negative for tumors by fluorescent microscopy were negative for tumors. Fluorescent microscopic analysis and sectioning yielded similar results.
Acknowledgments
We thank Scott Armstrong, Carla Kim, Alejandro Sweet-Cordero, Yariv Houvras, Craig Ceol, Richard White, Cicely Jette-Spaghetti, and Erik Langenau for critical review of this manuscript, and Anthony DiBase, Anna Burrows, Alan Flint, and Peter Schrow for expert technical support. D.M.L is the Edmond J. Safra Foundation Fellow from the Irvington Institute. L.M.K. and L.I.Z are investigators of the Howard Hughes Medical Institute. Funding for this work was provided by the Bernard F. and Alva B. Gimbel Foundation (L.M.K.) and NIH grant 5R01 CA103846-02 (L.I.Z). The Children’s Flow Cytometry Core is supported by a grant to Dr. Michael E. Greenberg (P30HD018655).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1545007
References
- Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F., Benito-Hernandez A., Morrison S.J., Clarke M.F., Morrison S.J., Clarke M.F., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt C.A., Crist W.M., Crist W.M. Common musculoskeletal tumors of childhood and adolescence. N. Engl. J. Med. 1999;341:342–352. doi: 10.1056/NEJM199907293410507. [DOI] [PubMed] [Google Scholar]
- Ashcroft M., Ludwig R.L., Woods D.B., Copeland T.D., Weber H.O., MacRae E.J., Vousden K.H., Ludwig R.L., Woods D.B., Copeland T.D., Weber H.O., MacRae E.J., Vousden K.H., Woods D.B., Copeland T.D., Weber H.O., MacRae E.J., Vousden K.H., Copeland T.D., Weber H.O., MacRae E.J., Vousden K.H., Weber H.O., MacRae E.J., Vousden K.H., MacRae E.J., Vousden K.H., Vousden K.H. Phosphorylation of HDM2 by Akt. Oncogene. 2002;21:1955–1962. doi: 10.1038/sj.onc.1205276. [DOI] [PubMed] [Google Scholar]
- Barnes C.J., Li F., Mandal M., Yang Z., Sahin A.A., Kumar R., Li F., Mandal M., Yang Z., Sahin A.A., Kumar R., Mandal M., Yang Z., Sahin A.A., Kumar R., Yang Z., Sahin A.A., Kumar R., Sahin A.A., Kumar R., Kumar R. Heregulin induces expression, ATPase activity, and nuclear localization of G3BP, a Ras signaling component, in human breast tumors. Cancer Res. 2002;62:1251–1255. [PubMed] [Google Scholar]
- Beauchamp J.R., Heslop L., Yu D.S., Tajbakhsh S., Kelly R.G., Wernig A., Buckingham M.E., Partridge T.A., Zammit P.S., Heslop L., Yu D.S., Tajbakhsh S., Kelly R.G., Wernig A., Buckingham M.E., Partridge T.A., Zammit P.S., Yu D.S., Tajbakhsh S., Kelly R.G., Wernig A., Buckingham M.E., Partridge T.A., Zammit P.S., Tajbakhsh S., Kelly R.G., Wernig A., Buckingham M.E., Partridge T.A., Zammit P.S., Kelly R.G., Wernig A., Buckingham M.E., Partridge T.A., Zammit P.S., Wernig A., Buckingham M.E., Partridge T.A., Zammit P.S., Buckingham M.E., Partridge T.A., Zammit P.S., Partridge T.A., Zammit P.S., Zammit P.S. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J. Cell Biol. 2000;151:1221–1234. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R., Davis R.L., Lockshon D., Turner D.L., Weintraub H., Davis R.L., Lockshon D., Turner D.L., Weintraub H., Lockshon D., Turner D.L., Weintraub H., Turner D.L., Weintraub H., Weintraub H. The protein Id: A negative regulator of helix–loop–helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Berghmans S., Murphey R.D., Wienholds E., Neuberg D., Kutok J.L., Fletcher C.D., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P., Murphey R.D., Wienholds E., Neuberg D., Kutok J.L., Fletcher C.D., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P., Wienholds E., Neuberg D., Kutok J.L., Fletcher C.D., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P., Neuberg D., Kutok J.L., Fletcher C.D., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P., Kutok J.L., Fletcher C.D., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P., Fletcher C.D., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P., Liu T.X., Schulte-Merker S., Kanki J.P., Schulte-Merker S., Kanki J.P., Kanki J.P., et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild A.H., Yao G., Chang J.T., Wang Q., Potti A., Chasse D., Joshi M.B., Harpole D., Lancaster J.M., Berchuck A., Yao G., Chang J.T., Wang Q., Potti A., Chasse D., Joshi M.B., Harpole D., Lancaster J.M., Berchuck A., Chang J.T., Wang Q., Potti A., Chasse D., Joshi M.B., Harpole D., Lancaster J.M., Berchuck A., Wang Q., Potti A., Chasse D., Joshi M.B., Harpole D., Lancaster J.M., Berchuck A., Potti A., Chasse D., Joshi M.B., Harpole D., Lancaster J.M., Berchuck A., Chasse D., Joshi M.B., Harpole D., Lancaster J.M., Berchuck A., Joshi M.B., Harpole D., Lancaster J.M., Berchuck A., Harpole D., Lancaster J.M., Berchuck A., Lancaster J.M., Berchuck A., Berchuck A., et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Caldas H., Holloway M.P., Hall B.M., Qualman S.J., Altura R.A., Holloway M.P., Hall B.M., Qualman S.J., Altura R.A., Hall B.M., Qualman S.J., Altura R.A., Qualman S.J., Altura R.A., Altura R.A. Survivin-directed RNA interference cocktail is a potent suppressor of tumour growth in vivo. J. Med. Genet. 2006;43:119–128. doi: 10.1136/jmg.2005.034686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Takita J., Hiwatari M., Igarashi T., Hanada R., Kikuchi A., Hongo T., Taki T., Ogasawara M., Shimada A., Takita J., Hiwatari M., Igarashi T., Hanada R., Kikuchi A., Hongo T., Taki T., Ogasawara M., Shimada A., Hiwatari M., Igarashi T., Hanada R., Kikuchi A., Hongo T., Taki T., Ogasawara M., Shimada A., Igarashi T., Hanada R., Kikuchi A., Hongo T., Taki T., Ogasawara M., Shimada A., Hanada R., Kikuchi A., Hongo T., Taki T., Ogasawara M., Shimada A., Kikuchi A., Hongo T., Taki T., Ogasawara M., Shimada A., Hongo T., Taki T., Ogasawara M., Shimada A., Taki T., Ogasawara M., Shimada A., Ogasawara M., Shimada A., Shimada A., et al. Mutations of the PTPN11 and RAS genes in rhabdomyosarcoma and pediatric hematological malignancies. Genes Chromosomes Cancer. 2006;45:583–591. doi: 10.1002/gcc.20322. [DOI] [PubMed] [Google Scholar]
- Conboy I.M., Rando T.A., Rando T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Cornelison D.D., Wold B.J., Wold B.J. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 1997;191:270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- Delgado I., Huang X., Jones S., Zhang L., Hatcher R., Gao B., Zhang P., Huang X., Jones S., Zhang L., Hatcher R., Gao B., Zhang P., Jones S., Zhang L., Hatcher R., Gao B., Zhang P., Zhang L., Hatcher R., Gao B., Zhang P., Hatcher R., Gao B., Zhang P., Gao B., Zhang P., Zhang P. Dynamic gene expression during the onset of myoblast differentiation in vitro. Genomics. 2003;82:109–121. doi: 10.1016/s0888-7543(03)00104-6. [DOI] [PubMed] [Google Scholar]
- Dhawan J., Rando T.A., Rando T.A. Stem cells in postnatal myogenesis: Molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005;15:666–673. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Felix C.A., Kappel C.C., Mitsudomi T., Nau M.M., Tsokos M., Crouch G.D., Nisen P.D., Winick N.J., Helman L.J., Kappel C.C., Mitsudomi T., Nau M.M., Tsokos M., Crouch G.D., Nisen P.D., Winick N.J., Helman L.J., Mitsudomi T., Nau M.M., Tsokos M., Crouch G.D., Nisen P.D., Winick N.J., Helman L.J., Nau M.M., Tsokos M., Crouch G.D., Nisen P.D., Winick N.J., Helman L.J., Tsokos M., Crouch G.D., Nisen P.D., Winick N.J., Helman L.J., Crouch G.D., Nisen P.D., Winick N.J., Helman L.J., Nisen P.D., Winick N.J., Helman L.J., Winick N.J., Helman L.J., Helman L.J. Frequency and diversity of p53 mutations in childhood rhabdomyosarcoma. Cancer Res. 1992;52:2243–2247. [PubMed] [Google Scholar]
- Fleischmann A., Jochum W., Eferl R., Witowsky J., Wagner E.F., Jochum W., Eferl R., Witowsky J., Wagner E.F., Eferl R., Witowsky J., Wagner E.F., Witowsky J., Wagner E.F., Wagner E.F. Rhabdomyosarcoma development in mice lacking Trp53 and Fos: Tumor suppression by the Fos protooncogene. Cancer Cell. 2003;4:477–482. doi: 10.1016/s1535-6108(03)00280-0. [DOI] [PubMed] [Google Scholar]
- Guyon J.R., Mosley A.N., Zhou Y., O’Brien K.F., Sheng X., Chiang K., Davidson A.J., Volinski J.M., Zon L.I., Kunkel L.M., Mosley A.N., Zhou Y., O’Brien K.F., Sheng X., Chiang K., Davidson A.J., Volinski J.M., Zon L.I., Kunkel L.M., Zhou Y., O’Brien K.F., Sheng X., Chiang K., Davidson A.J., Volinski J.M., Zon L.I., Kunkel L.M., O’Brien K.F., Sheng X., Chiang K., Davidson A.J., Volinski J.M., Zon L.I., Kunkel L.M., Sheng X., Chiang K., Davidson A.J., Volinski J.M., Zon L.I., Kunkel L.M., Chiang K., Davidson A.J., Volinski J.M., Zon L.I., Kunkel L.M., Davidson A.J., Volinski J.M., Zon L.I., Kunkel L.M., Volinski J.M., Zon L.I., Kunkel L.M., Zon L.I., Kunkel L.M., Kunkel L.M. The dystrophin associated protein complex in zebrafish. Hum. Mol. Genet. 2003;12:601–615. [PubMed] [Google Scholar]
- Hahn H., Wojnowski L., Zimmer A.M., Hall J., Miller G., Zimmer A., Wojnowski L., Zimmer A.M., Hall J., Miller G., Zimmer A., Zimmer A.M., Hall J., Miller G., Zimmer A., Hall J., Miller G., Zimmer A., Miller G., Zimmer A., Zimmer A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat. Med. 1998;4:619–622. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- Higashijima S., Okamoto H., Ueno N., Hotta Y., Eguchi G., Okamoto H., Ueno N., Hotta Y., Eguchi G., Ueno N., Hotta Y., Eguchi G., Hotta Y., Eguchi G., Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev. Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- Houdebine L.M., Chourrout D., Chourrout D. Transgenesis in fish. Experientia. 1991;47:891–897. doi: 10.1007/BF01929879. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue C.A., Maitra A., Olsen M., Lowe A.W., van Heek N.T., Rosty C., Walter K., Sato N., Parker A., Ashfaq R., Maitra A., Olsen M., Lowe A.W., van Heek N.T., Rosty C., Walter K., Sato N., Parker A., Ashfaq R., Olsen M., Lowe A.W., van Heek N.T., Rosty C., Walter K., Sato N., Parker A., Ashfaq R., Lowe A.W., van Heek N.T., Rosty C., Walter K., Sato N., Parker A., Ashfaq R., van Heek N.T., Rosty C., Walter K., Sato N., Parker A., Ashfaq R., Rosty C., Walter K., Sato N., Parker A., Ashfaq R., Walter K., Sato N., Parker A., Ashfaq R., Sato N., Parker A., Ashfaq R., Parker A., Ashfaq R., Ashfaq R., et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am. J. Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine K., Stirling R., Hume D., Kennedy D., Stirling R., Hume D., Kennedy D., Hume D., Kennedy D., Kennedy D. Rasputin, more promiscuous than ever: A review of G3BP. Int. J. Dev. Biol. 2004;48:1065–1077. doi: 10.1387/ijdb.041893ki. [DOI] [PubMed] [Google Scholar]
- Jessen J.R., Willett C.E., Lin S., Willett C.E., Lin S., Lin S. Artificial chromosome transgenesis reveals long-distance negative regulation of rag1 in zebrafish. Nat. Genet. 1999;23:15–16. doi: 10.1038/12609. [DOI] [PubMed] [Google Scholar]
- Jessen J.R., Jessen T.N., Vogel S.S., Lin S., Jessen T.N., Vogel S.S., Lin S., Vogel S.S., Lin S., Lin S. Concurrent expression of recombination activating genes 1 and 2 in zebrafish olfactory sensory neurons. Genesis. 2001;29:156–162. doi: 10.1002/gene.1019. [DOI] [PubMed] [Google Scholar]
- Kang P.B., Kho A.T., Sanoudou D., Haslett J.N., Dow C.P., Han M., Blasko J.M., Lidov H.G., Beggs A.H., Kunkel L.M., Kho A.T., Sanoudou D., Haslett J.N., Dow C.P., Han M., Blasko J.M., Lidov H.G., Beggs A.H., Kunkel L.M., Sanoudou D., Haslett J.N., Dow C.P., Han M., Blasko J.M., Lidov H.G., Beggs A.H., Kunkel L.M., Haslett J.N., Dow C.P., Han M., Blasko J.M., Lidov H.G., Beggs A.H., Kunkel L.M., Dow C.P., Han M., Blasko J.M., Lidov H.G., Beggs A.H., Kunkel L.M., Han M., Blasko J.M., Lidov H.G., Beggs A.H., Kunkel L.M., Blasko J.M., Lidov H.G., Beggs A.H., Kunkel L.M., Lidov H.G., Beggs A.H., Kunkel L.M., Beggs A.H., Kunkel L.M., Kunkel L.M. Variations in gene expression among different types of human skeletal muscle. Muscle Nerve. 2005;32:483–491. doi: 10.1002/mus.20356. [DOI] [PubMed] [Google Scholar]
- Kaplan E.L., Meier P., Meier P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- Keleti J., Quezado M.M., Abaza M.M., Raffeld M., Tsokos M., Quezado M.M., Abaza M.M., Raffeld M., Tsokos M., Abaza M.M., Raffeld M., Tsokos M., Raffeld M., Tsokos M., Tsokos M. The MDM2 oncoprotein is overexpressed in rhabdomyosarcoma cell lines and stabilizes wild-type p53 protein. Am. J. Pathol. 1996;149:143–151. [PMC free article] [PubMed] [Google Scholar]
- Keller C., Arenkiel B.R., Coffin C.M., El-Bardeesy N., DePinho R.A., Capecchi M.R., Arenkiel B.R., Coffin C.M., El-Bardeesy N., DePinho R.A., Capecchi M.R., Coffin C.M., El-Bardeesy N., DePinho R.A., Capecchi M.R., El-Bardeesy N., DePinho R.A., Capecchi M.R., DePinho R.A., Capecchi M.R., Capecchi M.R. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: Cooperativity of Ink4a/ARF and Trp53 loss of function. Genes & Dev. 2004;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., Wang J., Hahn W.C., Gilliland D.G., Hahn W.C., Gilliland D.G., Gilliland D.G., et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Krumenacker J.S., Narang V.S., Buckley D.J., Buckley A.R., Narang V.S., Buckley D.J., Buckley A.R., Buckley D.J., Buckley A.R., Buckley A.R. Prolactin signaling to pim-1 expression: A role for phosphatidylinositol 3-kinase. J. Neuroimmunol. 2001;113:249–259. doi: 10.1016/s0165-5728(00)00430-6. [DOI] [PubMed] [Google Scholar]
- Lam S.H., Wu Y.L., Vega V.B., Miller L.D., Spitsbergen J., Tong Y., Zhan H., Govindarajan K.R., Lee S., Mathavan S., Wu Y.L., Vega V.B., Miller L.D., Spitsbergen J., Tong Y., Zhan H., Govindarajan K.R., Lee S., Mathavan S., Vega V.B., Miller L.D., Spitsbergen J., Tong Y., Zhan H., Govindarajan K.R., Lee S., Mathavan S., Miller L.D., Spitsbergen J., Tong Y., Zhan H., Govindarajan K.R., Lee S., Mathavan S., Spitsbergen J., Tong Y., Zhan H., Govindarajan K.R., Lee S., Mathavan S., Tong Y., Zhan H., Govindarajan K.R., Lee S., Mathavan S., Zhan H., Govindarajan K.R., Lee S., Mathavan S., Govindarajan K.R., Lee S., Mathavan S., Lee S., Mathavan S., Mathavan S., et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat. Biotechnol. 2006;24:73–75. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- Langenau D.M., Traver D., Ferrando A.A., Kutok J.L., Aster J.C., Kanki J.P., Lin S., Prochownik E., Trede N.S., Zon L.I., Traver D., Ferrando A.A., Kutok J.L., Aster J.C., Kanki J.P., Lin S., Prochownik E., Trede N.S., Zon L.I., Ferrando A.A., Kutok J.L., Aster J.C., Kanki J.P., Lin S., Prochownik E., Trede N.S., Zon L.I., Kutok J.L., Aster J.C., Kanki J.P., Lin S., Prochownik E., Trede N.S., Zon L.I., Aster J.C., Kanki J.P., Lin S., Prochownik E., Trede N.S., Zon L.I., Kanki J.P., Lin S., Prochownik E., Trede N.S., Zon L.I., Lin S., Prochownik E., Trede N.S., Zon L.I., Prochownik E., Trede N.S., Zon L.I., Trede N.S., Zon L.I., Zon L.I., et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- Langenau D.M., Ferrando A.A., Traver D., Kutok J.L., Hezel J.P., Kanki J.P., Zon L.I., Look A.T., Trede N.S., Ferrando A.A., Traver D., Kutok J.L., Hezel J.P., Kanki J.P., Zon L.I., Look A.T., Trede N.S., Traver D., Kutok J.L., Hezel J.P., Kanki J.P., Zon L.I., Look A.T., Trede N.S., Kutok J.L., Hezel J.P., Kanki J.P., Zon L.I., Look A.T., Trede N.S., Hezel J.P., Kanki J.P., Zon L.I., Look A.T., Trede N.S., Kanki J.P., Zon L.I., Look A.T., Trede N.S., Zon L.I., Look A.T., Trede N.S., Look A.T., Trede N.S., Trede N.S. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl. Acad. Sci. 2004;101:7369–7374. doi: 10.1073/pnas.0402248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau D.M., Feng H., Berghmans S., Kanki J.P., Kutok J.L., Look A.T., Feng H., Berghmans S., Kanki J.P., Kutok J.L., Look A.T., Berghmans S., Kanki J.P., Kutok J.L., Look A.T., Kanki J.P., Kutok J.L., Look A.T., Kutok J.L., Look A.T., Look A.T. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. 2005;102:6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott A., Gustafsson M., Elsam T., Hui C.C., Emerson C.P., Jr., Borycki A.G., Gustafsson M., Elsam T., Hui C.C., Emerson C.P., Jr., Borycki A.G., Elsam T., Hui C.C., Emerson C.P., Jr., Borycki A.G., Hui C.C., Emerson C.P., Jr., Borycki A.G., Emerson C.P., Jr., Borycki A.G., Borycki A.G. Gli2 and Gli3 have redundant and context-dependent function in skeletal muscle formation. Development. 2005;132:345–357. doi: 10.1242/dev.01537. [DOI] [PubMed] [Google Scholar]
- Mendias C.L., Tatsumi R., Allen R.E., Tatsumi R., Allen R.E., Allen R.E. Role of cyclooxygenase-1 and -2 in satellite cell proliferation, differentiation, and fusion. Muscle Nerve. 2004;30:497–500. doi: 10.1002/mus.20102. [DOI] [PubMed] [Google Scholar]
- Miao J., Kusafuka T., Fukuzawa M., Kusafuka T., Fukuzawa M., Fukuzawa M. Hotspot mutations of BRAF gene are not associated with pediatric solid neoplasms. Oncol. Rep. 2004;12:1269–1272. [PubMed] [Google Scholar]
- Nanni P., Nicoletti G., De Giovanni C., Croci S., Astolfi A., Landuzzi L., Di Carlo E., Iezzi M., Musiani P., Lollini P.L., Nicoletti G., De Giovanni C., Croci S., Astolfi A., Landuzzi L., Di Carlo E., Iezzi M., Musiani P., Lollini P.L., De Giovanni C., Croci S., Astolfi A., Landuzzi L., Di Carlo E., Iezzi M., Musiani P., Lollini P.L., Croci S., Astolfi A., Landuzzi L., Di Carlo E., Iezzi M., Musiani P., Lollini P.L., Astolfi A., Landuzzi L., Di Carlo E., Iezzi M., Musiani P., Lollini P.L., Landuzzi L., Di Carlo E., Iezzi M., Musiani P., Lollini P.L., Di Carlo E., Iezzi M., Musiani P., Lollini P.L., Iezzi M., Musiani P., Lollini P.L., Musiani P., Lollini P.L., Lollini P.L. Development of rhabdomyosarcoma in HER-2/neu transgenic p53 mutant mice. Cancer Res. 2003;63:2728–2732. [PubMed] [Google Scholar]
- Olson E.N., Spizz G., Tainsky M.A., Spizz G., Tainsky M.A., Tainsky M.A. The oncogenic forms of N-ras or H-ras prevent skeletal myoblast differentiation. Mol. Cell. Biol. 1987;7:2104–2111. doi: 10.1128/mcb.7.6.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker F., Maurier F., Delumeau I., Duchesne M., Faucher D., Debussche L., Dugue A., Schweighoffer F., Tocque B., Maurier F., Delumeau I., Duchesne M., Faucher D., Debussche L., Dugue A., Schweighoffer F., Tocque B., Delumeau I., Duchesne M., Faucher D., Debussche L., Dugue A., Schweighoffer F., Tocque B., Duchesne M., Faucher D., Debussche L., Dugue A., Schweighoffer F., Tocque B., Faucher D., Debussche L., Dugue A., Schweighoffer F., Tocque B., Debussche L., Dugue A., Schweighoffer F., Tocque B., Dugue A., Schweighoffer F., Tocque B., Schweighoffer F., Tocque B., Tocque B. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol. Cell. Biol. 1996;16:2561–2569. doi: 10.1128/mcb.16.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S., Tamayo P., Rifkin R., Mukherjee S., Yeang C.H., Angelo M., Ladd C., Reich M., Latulippe E., Mesirov J.P., Tamayo P., Rifkin R., Mukherjee S., Yeang C.H., Angelo M., Ladd C., Reich M., Latulippe E., Mesirov J.P., Rifkin R., Mukherjee S., Yeang C.H., Angelo M., Ladd C., Reich M., Latulippe E., Mesirov J.P., Mukherjee S., Yeang C.H., Angelo M., Ladd C., Reich M., Latulippe E., Mesirov J.P., Yeang C.H., Angelo M., Ladd C., Reich M., Latulippe E., Mesirov J.P., Angelo M., Ladd C., Reich M., Latulippe E., Mesirov J.P., Ladd C., Reich M., Latulippe E., Mesirov J.P., Reich M., Latulippe E., Mesirov J.P., Latulippe E., Mesirov J.P., Mesirov J.P., et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl. Acad. Sci. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienbacher C., Sabbioni S., Campi M., Veronese A., Bernardi G., Menegatti A., Hatada I., Mukai T., Ohashi H., Barbanti-Brodano G., Sabbioni S., Campi M., Veronese A., Bernardi G., Menegatti A., Hatada I., Mukai T., Ohashi H., Barbanti-Brodano G., Campi M., Veronese A., Bernardi G., Menegatti A., Hatada I., Mukai T., Ohashi H., Barbanti-Brodano G., Veronese A., Bernardi G., Menegatti A., Hatada I., Mukai T., Ohashi H., Barbanti-Brodano G., Bernardi G., Menegatti A., Hatada I., Mukai T., Ohashi H., Barbanti-Brodano G., Menegatti A., Hatada I., Mukai T., Ohashi H., Barbanti-Brodano G., Hatada I., Mukai T., Ohashi H., Barbanti-Brodano G., Mukai T., Ohashi H., Barbanti-Brodano G., Ohashi H., Barbanti-Brodano G., Barbanti-Brodano G., et al. Transcriptional map of 170-kb region at chromosome 11p15.5: Identification and mutational analysis of the BWR1A gene reveals the presence of mutations in tumor samples. Proc. Natl. Acad. Sci. 1998;95:3873–3878. doi: 10.1073/pnas.95.7.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp R., Recio J.A., Jhappan C., Otsuka T., Liu S., Yu Y., Liu W., Anver M., Navid F., Helman L.J., Recio J.A., Jhappan C., Otsuka T., Liu S., Yu Y., Liu W., Anver M., Navid F., Helman L.J., Jhappan C., Otsuka T., Liu S., Yu Y., Liu W., Anver M., Navid F., Helman L.J., Otsuka T., Liu S., Yu Y., Liu W., Anver M., Navid F., Helman L.J., Liu S., Yu Y., Liu W., Anver M., Navid F., Helman L.J., Yu Y., Liu W., Anver M., Navid F., Helman L.J., Liu W., Anver M., Navid F., Helman L.J., Anver M., Navid F., Helman L.J., Navid F., Helman L.J., Helman L.J., et al. Synergism between INK4a/ARF inactivation and aberrant HGF/SF signaling in rhabdomyosarcomagenesis. Nat. Med. 2002;8:1276–1280. doi: 10.1038/nm787. [DOI] [PubMed] [Google Scholar]
- Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B., Henkelman R.M., Cusimano M.D., Dirks P.B., Cusimano M.D., Dirks P.B., Dirks P.B. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Stratton M.R., Fisher C., Gusterson B.A., Cooper C.S., Fisher C., Gusterson B.A., Cooper C.S., Gusterson B.A., Cooper C.S., Cooper C.S. Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reaction. Cancer Res. 1989;49:6324–6327. [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Pomeroy S.L., Golub T.R., Lander E.S., Golub T.R., Lander E.S., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet-Cordero A., Mukherjee S., Subramanian A., You H., Roix J.J., Ladd-Acosta C., Mesirov J., Golub T.R., Jacks T., Mukherjee S., Subramanian A., You H., Roix J.J., Ladd-Acosta C., Mesirov J., Golub T.R., Jacks T., Subramanian A., You H., Roix J.J., Ladd-Acosta C., Mesirov J., Golub T.R., Jacks T., You H., Roix J.J., Ladd-Acosta C., Mesirov J., Golub T.R., Jacks T., Roix J.J., Ladd-Acosta C., Mesirov J., Golub T.R., Jacks T., Ladd-Acosta C., Mesirov J., Golub T.R., Jacks T., Mesirov J., Golub T.R., Jacks T., Golub T.R., Jacks T., Jacks T. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat. Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- Thisse C., Zon L.I., Zon L.I. Organogenesis—Heart and blood formation from the zebrafish point of view. Science. 2002;295:457–462. doi: 10.1126/science.1063654. [DOI] [PubMed] [Google Scholar]
- Tomczak K.K., Marinescu V.D., Ramoni M.F., Sanoudou D., Montanaro F., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Marinescu V.D., Ramoni M.F., Sanoudou D., Montanaro F., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Ramoni M.F., Sanoudou D., Montanaro F., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Sanoudou D., Montanaro F., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Montanaro F., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Kunkel L.M., Kohane I.S., Beggs A.H., Kohane I.S., Beggs A.H., Beggs A.H. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 2004;18:403–405. doi: 10.1096/fj.03-0568fje. [DOI] [PubMed] [Google Scholar]
- Traver D., Paw B.H., Poss K.D., Penberthy W.T., Lin S., Zon L.I., Paw B.H., Poss K.D., Penberthy W.T., Lin S., Zon L.I., Poss K.D., Penberthy W.T., Lin S., Zon L.I., Penberthy W.T., Lin S., Zon L.I., Lin S., Zon L.I., Zon L.I. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- Tsumura H., Yoshida T., Saito H., Imanaka-Yoshida K., Suzuki N., Yoshida T., Saito H., Imanaka-Yoshida K., Suzuki N., Saito H., Imanaka-Yoshida K., Suzuki N., Imanaka-Yoshida K., Suzuki N., Suzuki N. Cooperation of oncogenic K-ras and p53 deficiency in pleomorphic rhabdomyosarcoma development in adult mice. Oncogene. 2006;25:7673–7679. doi: 10.1038/sj.onc.1209749. [DOI] [PubMed] [Google Scholar]
- Wachtel M., Dettling M., Koscielniak E., Stegmaier S., Treuner J., Simon-Klingenstein K., Buhlmann P., Niggli F.K., Schafer B.W., Dettling M., Koscielniak E., Stegmaier S., Treuner J., Simon-Klingenstein K., Buhlmann P., Niggli F.K., Schafer B.W., Koscielniak E., Stegmaier S., Treuner J., Simon-Klingenstein K., Buhlmann P., Niggli F.K., Schafer B.W., Stegmaier S., Treuner J., Simon-Klingenstein K., Buhlmann P., Niggli F.K., Schafer B.W., Treuner J., Simon-Klingenstein K., Buhlmann P., Niggli F.K., Schafer B.W., Simon-Klingenstein K., Buhlmann P., Niggli F.K., Schafer B.W., Buhlmann P., Niggli F.K., Schafer B.W., Niggli F.K., Schafer B.W., Schafer B.W. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64:5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- Xia S.J., Pressey J.G., Barr F.G., Pressey J.G., Barr F.G., Barr F.G. Molecular pathogenesis of rhabdomyosarcoma. Cancer Biol. Ther. 2002;1:97–104. doi: 10.4161/cbt.51. [DOI] [PubMed] [Google Scholar]
- Yip-Schneider M.T., Lin A., Marshall M.S., Lin A., Marshall M.S., Marshall M.S. Pancreatic tumor cells with mutant K-ras suppress ERK activity by MEK-dependent induction of MAP kinase phosphatase-2. Biochem. Biophys. Res. Commun. 2001;280:992–997. doi: 10.1006/bbrc.2001.4243. [DOI] [PubMed] [Google Scholar]