Abstract
gp130-linked cytokines such as interleukin-6 (IL-6) stimulate the formation of tyrosine-phosphorylated signal transducer and activator of transcription 3 (P-STAT3), which activates many genes, including the STAT3 gene itself. The resulting increase in the concentration of unphosphorylated STAT3 (U-STAT3) drives a second wave of expression of genes such as RANTES, IL6, IL8, MET, and MRAS that do not respond directly to P-STAT3. Thus, U-STAT3 sustains cytokine-dependent signaling at late times through a mechanism completely distinct from that used by P-STAT3. Many U-STAT3-responsive genes have κB elements that are activated by a novel transcription factor complex formed when U-STAT3 binds to unphosphorylated NFκB (U-NFκB), in competition with IκB. The U-STAT3/U-NFκB complex accumulates in the nucleus with help from the nuclear localization signal of STAT3, activating a subset of κB-dependent genes. Additional genes respond to U-STAT3 through an NFκB-independent mechanism. The role of signal-dependent increases in U-STAT3 expression in regulating gene expression is likely to be important in physiological responses to gp130-linked cytokines and growth factors that activate STAT3, and in cancers that have constitutively active P-STAT3.
Keywords: Gene chip, gene transcription, IL-6, Jak–Stat, NFκB
Signal transducer and activator of transcription 3 (STAT3), one of seven STAT family members, is activated in response to interleukin-6 (IL-6) (Akira et al. 1994). Many cytokines use the common gp130 receptor to activate the phosphorylation of STAT3 on tyrosine residue 705, leading to the formation of dimers through reciprocal phosphotyrosine–SH2 domain interactions. Several growth factors also stimulate STAT3 activation. STAT3 dimers bind to specific γ-interferon activation sequence (GAS) elements (TTCNNNGAA) in the promoters of the induced genes (Seidel et al. 1995).
Constitutive activation of STAT3 is observed in many types of tumors. Thus, STAT3 is an oncogene, promoting cell proliferation and survival (Haura et al. 2005; Hodge et al. 2005). STAT3 is persistently phosphorylated in many human cancer cell lines and primary tumors, including hepatocellular carcinomas, breast cancers, prostate cancers, and head and neck cancers, and also in several hematological malignancies. Furthermore, STAT3 is necessary for v-src-induced transformation, and a constitutively active mutant of STAT3 can transform fibroblasts in cooperation with other transcription factors (Joo et al. 2004). Genes encoding proteins that regulate cell survival, including Bcl-2, Bcl-xL, mcl-1, and Fas, are direct targets of STAT3, as are genes encoding the cell cycle regulatory proteins cyclin D1, cyclin E1, and p21. In addition, other transcription factors, including c-Myc, c-Jun, and c-Fos, are themselves STAT3 targets (Hirano et al. 2000). STAT3 also functions as a transcriptional repressor of p53 expression: Blocking STAT3 in cancer cells up-regulates the expression of p53, leading to p53-mediated apoptosis (Niu et al. 2005). Major mechanisms of STAT3 activation in tumor cells are autocrine production of IL-6 and paracrine activation by IL-6 from stroma and infiltrating inflammatory cells. Indeed, circulating IL-6 levels are usually high in cancer patients (Giannitrapani et al. 2002). STAT3 activation provides an important link between inflammation and cancer. For example, Tebbutt et al. (2002) generated gp130757F/F mice, which carry a Y757F point mutation that disrupts the binding of the negative regulators SOCS3 and SHP2 to gp130. As a result, these mice show hyperactivation of STAT3, resulting in chronic gastric inflammation and distal stomach tumors.
There are several reports that STAT3 and NF-κB interact with each other (Battle and Frank 2002). For example, Hagihara et al. (2005) demonstrated that STAT3 forms a complex with the p65 subunit of NFκB following stimulation of cells with IL-1 plus IL-6, and that the bound STAT3 interacts with nonconsensus sequences near the κB element of the SAA promoter. Moreover, they showed that a complex that includes STAT3, p65, and p300 is essential for the synergistic induction of the SAA gene by IL-1 plus IL-6. Yu et al. (2002) reported a physical and functional interaction between STAT3 and p65 that inhibits transcriptional activation of the iNOS gene. Yoshida et al. (2004) showed that STAT3 and p65 physically interact in vivo and that p65 homodimers can cooperate with unphosphorylated STAT3 (U-STAT3) when bound to a specific type of κB motif. Reciprocally, this interaction appears to inhibit the function of GAS-binding sites for STAT3. In contrast, the p50 subunit of NFκB can cooperate with phosphorylated STAT3 (P-STAT3) bound to GAS sites (Yoshida et al. 2004).
Previous work from this laboratory has shown that STAT1 can drive gene expression even in the absence of tyrosine phosphorylation. For example, Chatterjee-Kishore et al. (2000) showed that unphosphorylated STAT1 binds to IRF1, forming a complex that activates a half GAS-half ICS element in the LMP2 promoter. In the case of STAT3, our recent work (Yang et al. 2005) showed that its high-level expression drives the transcription of many genes in the complete absence of tyrosine phosphorylation, a function quite distinct from the role of P-STAT3 in driving inducible gene expression. This activity of STAT3 is likely to have important physiological functions, since the STAT3 gene has a GAS element that drives a major increase in the concentration of STAT3 protein in response to signal-dependent tyrosine phosphorylation of STAT3 (Kojima et al. 1996; Narimatsu et al. 2001). We now address the mechanism through which U-STAT3 regulates gene expression, showing that, for many genes, it does so through its ability to interact with NFκB. To understand how U-STAT3 functions, we expressed the Y705F mutant of STAT3, which cannot be phosphorylated on residue 705, at a high level in untransformed human mammary epithelial hTERT-HME1 cells and used coimmunoprecipitation and chromatin immunoprecipitation (ChIP) assays to identify cofactors and DNA elements to which Y705F-STAT3 binds. Our data reveal that Y705F-STAT3 forms a complex with unphosphorylated NFκB (U-NFκB), binding to the κB elements of promoters, such as that of the RANTES (CCL5) gene, to induce their transcription.
Results
Promoters that bind to U-STAT3
To find the direct targets of U-STAT3, we used ChIP to clone the bound DNA sequences. Flag-tagged Y705F-STAT3 was precipitated with anti-Flag (M2) antibody. The coprecipitated DNA fragments were linked to adaptors, amplified by PCR, and inserted into a vector, followed by sequencing of individual clones. A BLAST search facilitated the identification of 12 fragments, seven of which (Table 1) had promoter activity in a luciferase reporter assay (data not shown). Five of the seven active promoter fragments corresponded to genes shown previously to be up-regulated by U-STAT3 (Yang et al. 2005). Computer-based analysis revealed several common elements that bind to transcription factors in these promoters (Ap1, CRE, C/EBP, ETS, and κB) (see Table 1). Interestingly, each of the seven promoters has a κB element.
Table 1.
Promoters that bind to USTAT3 have κB sites
A ChIP assay was performed with chromatin from YF cells (containing the Y705F mutant of STAT3) by using anti-Flag M2. The immunoprecipitated DNA was amplified by PCR and cloned, and 12 clones were sequenced. Five of the cloned fragments had no promoter activity in a luciferase reporter assay (data not shown). The seven fragments that did have promoter activity are shown. Five of these were from genes previously shown to be responsive to Y705F-STAT3 and U-STAT3 and two were not analyzed previously (N/A). The fold induction of each of these five genes in response to high-level expression of Y705F-STAT3 is shown, as are the positions of the κB elements and consensus sites for other transcription factors.
USTAT3 uses the κB element to induce RANTES gene expression
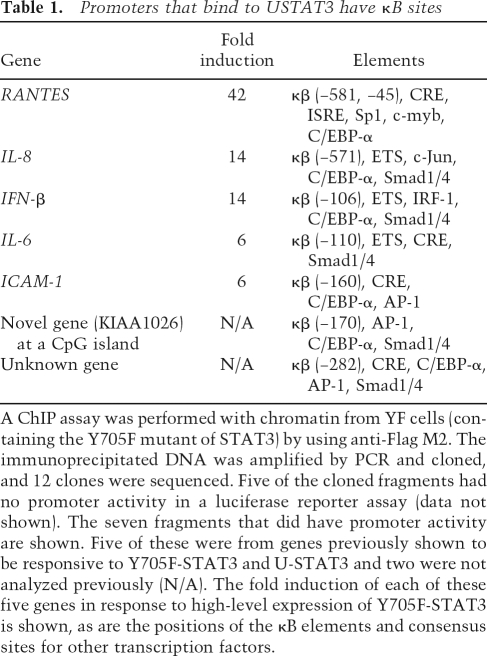
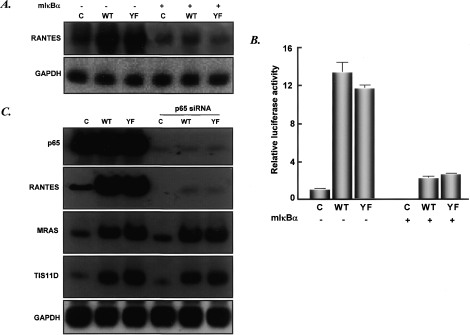
From previous work (Yang et al. 2005), we know that high-level expression of either U-STAT3 or Y705F-STAT3 drives the expression of many genes, including genes whose protein products are important in oncogenesis, cell cycle control, and the immune response (e.g., MET, MRAS, BCL2A1, IFNβ, and RANTES). Some of these genes are induced very substantially; i.e., RANTES was induced 28-fold by high levels of U-STAT3 and 42-fold by Y705F-STAT3. Since RANTES expression is induced so strongly, we studied its promoter to determine the role of the κB element. hTERT-HME1 cells were infected with retroviral vectors expressing wild-type or Y705F-STAT3, Flag-tagged at their C termini, and populations expressing 10- to 20-fold more STAT3 than wild-type cells were selected (Fig. 1A). These populations are named WT or YF, respectively. Note that this degree of increase corresponds well to the increase of STAT3 seen 36 h after exposure of hTERT-HME1 cells to IL-6, and, as seen before, RANTES mRNA accumulated substantially in response to high-level expression of either U-STAT3 or Y705F-STAT3 (Fig. 1A). To determine the responsible element in the RANTES promoter, we transfected hTERT-HME1 cells with constructs containing various fragments of this promoter fused to a luciferase reporter gene. Maximum transcriptional activity was observed with a −120 to −1 minimal fragment, and removal of this sequence from a longer fragment completely eliminated basal transcription (Fig. 1B). The activity induced in response to high-level expression of Y705F-STAT3 disappeared when the −120 to −1 region of the promoter was deleted (Fig. 1C). Sequence analysis showed that the human RANTES promoter contains several known elements; for example, CRE, ISRE, and κB. To test these for function, mutations were introduced into the −220 to −1 region, and the resulting fragments were linked to the pGL2-basic vector and tested in transient transfections of hTERT-HME1 and YF cells (Fig. 1D). Disruption of either the CRE or the ISRE element did not affect the activity of the promoter, but mutation of the κB element did. Therefore, the latter element plays an important role in mediating the response of RANTES to U-STAT3.
Figure 1.
The ability of U-STAT3 to regulate the RANTES promoter depends on a κB element. (A) Western and Northern analyses for STAT3 and RANTES expression in hTERT-HME1-derived cells. The cells were infected with retroviral constructs and stable pools were selected with G418. (C) hTERT-HME1 control cells; (WT) hTERT-HME1 cells expressing a high level of wild-type STAT3; (YF) hTERT-HME1 cells expressing a high level of Y705F-STAT3. Total cell lysates and total RNAs were analyzed. (B) Basal transcriptional activity of the human RANTES promoter in hTERT-HME1 cells. Luciferase constructs containing 5′- or 3′-deletions between bases −974 and −1 of the promoter were cotransfected with the pCH110 control plasmid and the cells were harvested 48 h later. The luciferase activity in each cell extract was normalized to the level of β-galactosidase activity (from pCH110) in the same extract. Values are means of triplicate determinations, and the bars show one standard error of the mean. (C) Inducible activity of human RANTES promoter fragments in YF cells. The reporter constructs were cotransfected with pCH110. The activities shown are relative to the activity of each fragment in hTERT-HME1 control cells. Values are means of triplicate determinations, and the bars show one standard error of the mean. (D) Inducible activity of promoter mutations in YF cells. The reporter constructs, containing mutations of individual promoter elements (marked by ×) of the 220-base-pair promoter fragment were transfected into the cells. Luciferase activities were determined and calculated relative to the values obtained in control cells as in C. (E) Y705F-STAT3 and p65 cooperate to drive the RANTES promoter. hTERT-HME1 cells were cotransfected with the pGL2-220 plasmid, in which the RANTES −1 to −220 promoter fragment drives luciferase expression, and pCH110, with or without pcDNA3.1-Y705F-STAT3 or pcDNA3.1-p65, expression plasmids for Y705F-STAT3 and p65, respectively. The cells, harvested 48 h later, were analyzed for luciferase activities, as described above. The reporter activities were normalized to activities in cells without cotransfection of p65 or Y705F-STAT3. Values are means of triplicate determinations, and the bars show one standard error of the mean. (F) U-STAT3 and p65 cooperate to activate the RANTES gene. hTERT-HME1 cells were transfected transiently with pcDNA3.1-p65 and/or pcDNA3.1-STAT3. After 48 h, total RNAs were isolated and analyzed by the Northern method.
RANTES expression is induced cooperatively by p65 and U-STAT3
Transient overexpression of either the p65 subunit of NFκB or U-STAT3 enhanced induction of the RANTES promoter by six- to sevenfold, whereas overexpression of both together led to a 15-fold increase (Fig. 1E). The accumulation of endogenous RANTES mRNA was also induced maximally when both p65 and STAT3 were overexpressed (Fig. 1F). Two-step ChIP assays, performed by using anti-STAT3 first and then anti-p65, confirmed that both proteins were bound simultaneously to the RANTES promoter (Supplementary Fig. 1). Further experiments showed that overexpression of either U-STAT3 or Y705F-STAT3 did not cause phosphorylation of Ser536 of p65 (Supplementary Fig. 2) and that conditioned medium from these cell populations did not activate expression of either RANTES or IL1β mRNA (Supplementary Fig. 3). Therefore, the increase in RANTES expression in response to U-STAT3 is not likely to be due to the phosphorylation of NFκB in the cytosol or to the secretion of factors that activate NFκB through cell-surface receptors.
A complex of U-STAT3 with the p65 and p50 subunits of NFκB binds to the κB element of the RANTES promoter
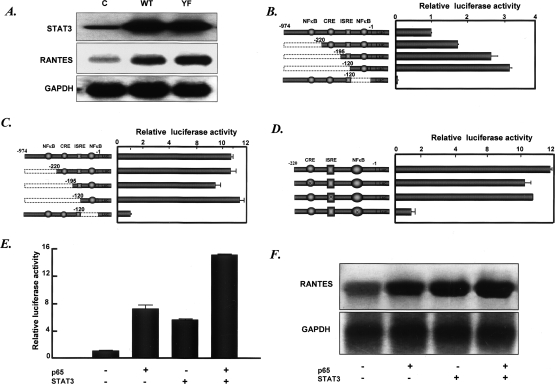
Electrophoretic mobility shift assays (EMSAs) were performed with an oligonucleotide that includes the complex κB element of the human RANTES promoter. Whole-cell lysates were prepared from hTERT-HME1, wild-type, and YF cells. Two major bands were detected (Fig. 2A, lanes 1–3). We interpret the upper band to include two species, one with p65 homodimers and one with p65–p50 heterodimers, and the lower band to represent species with p50 homodimers. In both wild-type and YF cells, all three bands increased (Fig. 2A, lanes 2,3). Only the upper two bands were supershifted by anti-p65 (Fig. 2A, lane 4), whereas anti-p50 supershifted both the lower band, which disappeared completely, and the lower part of the upper band (Fig. 2A, lane 5). Anti-STAT3 supershifted all three bands (Fig. 2A, lane 6), indicating that all three species of NFκB are bound to Y705F-STAT3. Neither anti-IκB nor anti-c-myc caused any of the bands to change (Fig. 2A, lanes 7,8). When the same cells were treated with TNF-α, EMSAs (Fig. 2A, lanes 9–16) showed that all of the complexes formed in cell extracts containing activated NFκB migrated more rapidly than those formed in extracts of untreated cells. Furthermore, the complexes formed with extracts of TNF-α-treated cells were no longer supershifted by anti-STAT3 (Fig. 2A, lane 14). Therefore, when cells are treated with TNF-α to activate NFκB, our EMSAs no longer revealed the binding of U-STAT3 to the RANTES κB element together with p65 and p50. However, as shown below, coimmunoprecipitation experiments still detected the association of NFκB and STAT3 when either transcription factor was phosphorylated.
Figure 2.
U-STAT3 binds to U-NFκB. (A) DNA-binding assays. The EMSAs shown were performed with whole-cell extracts. Assays with nuclear extracts (not shown) gave similar results. (C) hTERT-HME1 control cells. A DNA fragment of the human RANTES promoter, bases −58 to −29, containing a κB element, was used as the labeled probe. (Lanes 1–8) Extracts of untreated cells: control cells (lane 1), WT cells (lane 2), YF cells (lane 3), and supershifts obtained with extracts of YF cells following addition of antibodies directed against p65, p50, STAT3, IκB, or c-Myc (lanes 4–8). (Lanes 9–16) Same as lanes 1–8 except that the extracts are from cells treated with TNF-α for 4 h. (B) EMSAs. Whole-cell extracts were made from hTERT-HME1 cells, untreated or treated with IL-6. The probe was same as in A. (C) Northern analysis. Total RNAs (20 μg per lane) from hTERT-HME1 cells untreated or treated with IL-6 were analyzed by the Northern method. (D,E) STAT3 binds to p65, p50, and p105 but not to IκB. STAT3 was immunoprecipitated from whole-cell extracts of the cells shown in Figure 1A by using anti-Flag M2 beads. Western analyses were performed to detect p65, p50, and IκB.
From our previous work (Yang et al. 2005), we know that long-term treatment of hTERT-HME1 cells with IL-6 increases endogenous STAT3 expression by 20- to 30-fold and that the increased concentration of STAT3 induces a second wave of gene expression that includes the MET, M-RAS, and RANTES genes. To determine whether the increased concentration of STAT3 can still bind to and cooperate with p65 and p50 to induce gene expression, hTERT-HME1 cells were treated with IL-6 for 0, 2, 4, 8, 16, or 32 h and EMSAs were performed. As shown in Figure 2B, when STAT3 was induced strongly by IL-6 (Yang et al. 2005; data not shown) at late times, STAT3/p65/p50 complexes were detected in EMSAs. Furthermore, the level of RANTES mRNA parallels the level of STAT3 induced by IL-6 (Fig. 2C; Yang et al. 2005), indicating that induced endogenous STAT3, as well as exogenous STAT3 expressed at a high level, is capable of binding to NF-κB to drive gene expression.
U-STAT3 binds to NFκB in competition with IκB
To demonstrate the interaction between NFκB and U-STAT3 more directly, we performed coimmunoprecipitation assays, using wild-type and YF cells, which contain Flag-tagged STAT3 proteins (Fig. 1A). Anti-Flag beads were incubated with lysates of these cells to pull down U-STAT3 and Y705F-STAT3, and the presence of p65 and p50 was assayed in the immunoprecipitates (Fig. 2D,E). P65 and p50, as well as the p50 precursor p105, were pulled down with STAT3, but IκB was not. The levels of p65 and p50 were not affected by the level of STAT3 expression. In addition to its association with unphosphorylated p65, U-STAT3 can also bind to p65 that has been phosphorylated on Ser536 in response to TNF-α (Supplementary Fig. 2). In the EMSA assays of Figure 2A, an association between U-STAT3 or Y705F-STAT3 and NFκB activated in response to TNF-α was not observed, in contrast to the results of Supplementary Figure 2, possibly because complexes of STAT3 and P-NFκB do not bind well to DNA under the EMSA conditions that we have employed. In addition, in cells treated with IL-6, P-STAT3 can also be seen to bind to p65 and p50 (Supplementary Fig. 4; see also Fig. 3 in Yoshida et al. 2004). Therefore, in addition to the association of U-STAT3 and unphosphorylated p65, immunoprecipitation assays reveal that STAT3 and p65 also bind to each other when each is phosphorylated in response to IL-6 or TNF-α, respectively. Furthermore, pull-down experiments using extracts from untreated or IL-6-treated Hep3B cells indicate that both U-STAT3 and P-STAT3 interact primarily with the Rel-homology DNA-binding domain of p65 (Supplementary Fig. 5). The interaction appears to be stronger with the isolated Rel domain than with full-length p65, suggesting that opening of the interaction between the p65 transactivation domain and the Rel domain may facilitate binding. A similar phenomenon has been reported previously for the interaction between p65 and CBP (Zhong et al. 1998). In addition, as discussed below, several other laboratories have observed interactions between phosphorylated and unphosphorylated forms of p65 and STAT3.
The SH2 and NLS domains of U-STAT3 are required for productive interaction with p65 and p50
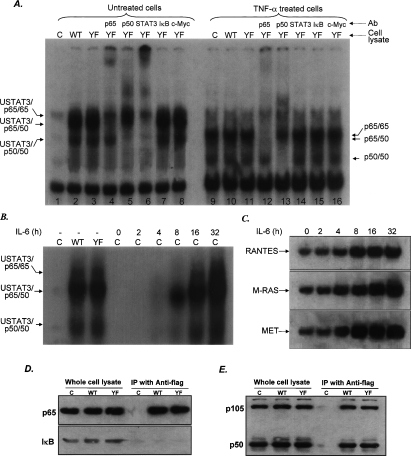
We transfected expression constructs for several different truncated GFP-tagged STAT3 proteins (Fig. 3A) transiently into PC3 cells, which have a very low level of endogenous STAT3 (Yuan et al. 2005). Full-length STAT3 (residues 1–770) was immunoprecipitated by both anti-p65 and anti-p50, as were two N-terminal truncations (150–770 and 162–770) and STAT3β (1–715), a naturally occurring C-terminal truncation of this protein. Variant STAT3 proteins with the C-terminal truncations 1–322 and 1–495 did not bind to p65 or p50 (Fig. 3B). These data indicate that the region of STAT3 from 495 to 715, which includes the SH2 domain, is essential for its interaction with NFκB. Functional analysis of these STAT3 deletions, using a RANTES promoter-driven luciferase construct in cotransfection experiments, showed that only full-length STAT3 and the 150–770 and 1–715 (STAT3β) truncated proteins were active (Fig. 3C), even though the 162–770 truncated protein still binds to NFκB (Fig. 3B). The 150–770 protein has a nuclear localization sequence (NLS) that the 162–770 protein lacks (Liu et al. 2005). Taken together, the results provide strong evidence that the region between residues 495 and 715, which includes the SH2 domain, is required for U-STAT3 to bind to U-NFκB, whereas the NLS (residues 150–162) is required for the bound protein to function in transcription.
Figure 3.
The SH2 and NLS domains of STAT3 are required for interaction with p65 and p50 and for up-regulation of the RANTES promoter. (A) STAT3 domains and deletion constructs. (B) PC3 cells were transfected with expression constructs for N- and C-terminal deletions of STAT3 and, 48 h later, whole-cell lysates were prepared and assayed by coimmunoprecipitation with anti-p65 or anti-p50 and by the Western method with anti-STAT3. Three different antibodies that react with the N-terminal, C-terminal, and middle portions of STAT3 were used. (C) Expression constructs for N- and C-terminal deletions of STAT3 were cotransfected into PC3 cells with pGL2-220 and pCH110, and the cells were harvested for luciferase assays 48 h later. (D) Northern analysis. Total RNAs (20 μg per lane) from hTERT-HME1 cells untreated or treated with IL-6 were analyzed by the Northern method. (E) DNA-binding assays. hTERT-HME1 cells expressing a high level of full-length STAT3 or 162–770 truncated STAT3 were untreated or treated with TNF-α for 4 h. EMSAs were performed with cytoplasmic and nuclear fractions. A DNA fragment of the human RANTES promoter, bases −58 to −29, containing a κB element, was used as the labeled probe. Assays were performed by adding equal amounts of proteins. (C) hTERT-HME1 control cells; (S3) hTERT-HME1 cells expressing a high level of STAT3. (F) hTERT-HME1 cells expressing a high level of full-length or 162–770 truncated STAT3 were grown on cover slips and stained with primary antibodies directed against STAT3 and p65. Following treatment with DAPI (blue nuclear stain) and fluorescent secondary antibodies for STAT3 (green) and p65 (red), the cells were examined by using confocal microscopy. The yellow pixels in the composite image demonstrate the close association of the two proteins.
Based on the above observations, it is logical to propose that 162–770 truncated STAT3 may inhibit NFκB-dependent signaling, for example, in response to TNF-α. To test this possibility, hTERT-HME1 cells in which full-length or 162–770 truncated STAT3 was stably overexpressed by 10-fold were treated with TNF-α for 4 h or were untreated. Northern analyses were performed to detect RANTES and GAPDH mRNAs. As we saw before, high-level expression of full-length STAT3 increases RANTES expression by 10- to 20-fold. TNF-α treatment also increases RANTES expression by 10- to 15-fold because the RANTES gene has a κB element that mediates the response to TNF-α. However, in cells expressing a high level of 162–770 truncated STAT3, untreated cells showed no increases of RANTES mRNA and the level in cells treated with TNF-α increased only slightly, by two- to threefold, much less than in control cells treated with TNF-α or in untreated cells expressing a high level of full-length STAT3 (Fig. 3D). This result indicates that 162–770 truncated STAT3 can inhibit NFκB-dependent signaling in response to an external stimulus.
We also performed EMSAs to determine whether truncated STAT3 affects the ability of the STAT3/p65/p50 complex to bind to a κB probe. hTERT-HME1 cells in which full-length or 162–770 truncated STAT3 was stably expressed at a high level were treated with TNF-α or left untreated. Four hours after treatment, the cells were harvested and cytoplasmic and nuclear fractions were prepared. As expected, full-length STAT3 increased NFκB-binding activity, and the activated complexes were translocated into the nucleus (Fig. 3E, top panel, lanes 1–5,11–15). These increased complexes could be supershifted by antibodies to p65, p50, or STAT3 (Fig. 3E, top panel, lanes 3–5,13–15). TNF-α treatment activates NFκB, and the complexes are located mainly in nucleus (Fig. 3E, top panel, lanes 6–10,16–20). However, these complexes could be supershifted only by antibodies to p65 or p50, but not by anti-STAT3 (Fig. 3E, top panel, lanes 8–10,18–20). 162–770 truncated STAT3 increases NFκB-binding activity only in the cytoplasm and not in the nucleus (Fig. 3E, bottom panel, lanes 1–5,11–15). These cytoplasmic complexes could be supershifted by antibodies to p65, p50, or STAT3 (Fig. 3E, bottom panel, lanes 3–5,13–15). TNF-α treatment is still capable of activating NFκB in these cells, but less than in cells expressing a high level of full-length STAT3 (Fig. 3E, top and bottom panels, lanes 16–20). In TNF-α-treated cells, activated NFκB was translocated into the nucleus completely (Fig. 3E, top and bottom panels, lanes 6,16) indicating that, although 162–770 truncated STAT3 still binds to NFκB, it fails to activate gene expression through a κB element. This observation is consistent with the data from the coimmunoprecipitation and luciferase assays (Fig. 3A–D).
We used immunocytochemistry to demonstrate that STAT3 binds to p65 and p50 in vivo. Full-length or 162–770 truncated STAT3 were expressed at a level fivefold to 10-fold higher than endogenous STAT3, which was detected with a secondary antibody tagged with a green label. Endogenous p65 was detected with a secondary antibody tagged with a red label. In control cells, STAT3 was distributed evenly between the cytoplasm and nucleus, while p65 was seen mainly in the cytoplasm (Fig. 3F, top panel). In cells with a high level of wild-type STAT3, the protein was distributed evenly between the cytoplasm and nucleus as before, but p65 was now seen predominantly in the nucleus (Fig. 3F, middle panel). Interestingly, in cells expressing a high level of 162–770 truncated STAT3, this protein was predominately in the cytoplasm, consistent with the results of others (Liu et al. 2005). As expected, p65 was seen primarily in the cytoplasm, as well (Fig. 3F, bottom panel). By double immunofluorescence we find that the two wild-type proteins are present simultaneously in the nucleus (Fig. 3F, middle panel), consistent with the possibility that STAT3 and p65 indeed do bind to each other in vivo.
Expression of IκBα superrepressor or ablation of p65 blocks RANTES expression in response to U-STAT3
In the absence of an activating signal, the steady-state equilibrium for NFκB localization is toward the cytosol, as a result of interaction with IκB (Birbach et al. 2002). Activating stimuli, such as the proinflammatory cytokines TNF-α and IL-1, liberate NFκB by inducing the phosphorylation of IκB, triggering its ubiquitination and degradation (Henkel et al. 1993; Palombella et al. 1994; Roff et al. 1996). Activation of IκB kinase leads to the phosphorylation of IκBα on Ser32 and Ser36, followed by its rapid proteasome-mediated degradation, allowing free NFκB to enter the nucleus. We were unable to overexpress exogenous wild-type IκB from a construct in these cells (data not shown), probably because it is too unstable when not complexed to p65 and p50. Therefore, we used the serine-to-alanine double mutant of IκBα, S32/36A (mIκBα), which is a superrepressor since it cannot be phosphorylated in response to activating signals. mIκBα was expressed transiently in hTERT-HME1-derived cells, at a level fivefold to 10-fold higher than that of IκB in control cells (data not shown), and mRNA expression from the endogenous RANTES gene (Fig. 4A) and RANTES-driven promoter (Fig. 4B) were analyzed. As expected, high-level expression of either U-STAT3 or Y705F-STAT3 induced the expression of endogenous RANTES mRNA strongly. The inductions were strongly suppressed by mIκBα (Fig. 4A). Similar results were obtained in the luciferase reporter assays (Fig. 4B). To assess the role of p65 in a functional assay, we used an small interfering RNA (siRNA) to cause an almost complete elimination of its expression in all three cell lines (Fig. 4C). The knock down of p65 eliminates both U-STAT3-induced and basal RANTES expression. These results provide strong support for a model in which an active transcription complex comprising U-STAT3 and U-NFκB is formed by competition between U-STAT3 and IκB for U-NFκB.
Figure 4.
Inhibition of NFκB decreases RANTES gene expression in response to U-STAT3. (A) hTERT-HME1-derived cells were transfected transiently with the pcDNA3.1-mIκBα construct, which encodes the NFκB superrepressor and, 48 h later, total RNAs were isolated and analyzed. (B) The RANTES promoter-driven luciferase reporter construct pGL2-220 was transfected with pCH110, with or without pcDNA3.1-mIκBα, into hTERT-HME1-derived cells and, 48 h later, luciferase assays were performed. (C) hTERT-HME1-derived cells were transfected transiently with a siRNA directed against p65 and, 24 h later, the cells were transfected again as in A. The cells were harvested after 48 h more and total RNA was extracted. All of the mRNAs shown were assayed on the same Northern transfer.
Array-based expression analysis identifies three subsets of genes responsive to USTAT3 or TNF-α
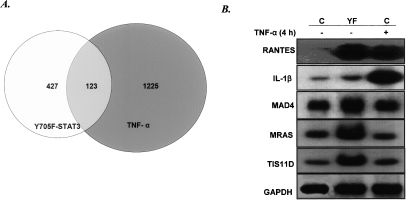
The levels of mRNAs isolated from TNF-α-treated or untreated hTERT-HME1-derived cells were analyzed (Fig. 5A). 1225 genes were induced more than threefold by TNF-α treatment, and 427 genes were induced more than threefold by high-level expression of Y705F-STAT3 (Supplementary Table 1). Of these, 123 genes were induced more than threefold by either TNF-α or by Y705F-STAT3. Therefore, most TNF-α-induced genes are not responsive to a high level of Y705F-STAT3, and most genes induced by Y705F-STAT3 do not respond to TNF-α. Typical genes from each of the three groups were analyzed by the Northern method: RANTES, which is induced by both TNF-α and Y705F-STAT3; IL1β, which is induced only by TNF-α; and MAD4, MRAS, and TIS11D, which are induced only by Y705F-STAT3 (Fig. 5B). The data clearly show that only a subset of the genes that respond to TNF-α respond also to Y705F-STAT3. Furthermore, many of the genes that respond to Y705F-STAT3 probably do not have functional κB elements, since they do not respond to TNF-α. This possibility was confirmed for MRAS and TIS11D since, in contrast to RANTES, their expression was not affected by eliminating p65 (Fig. 4C).
Figure 5.
Comparison of genes induced by high-level expression of Y705F-STAT3 with those induced by treatment with TNF-α. hTERT-HME1 control cells or YF cells were treated with 50 ng/mL TNF-α for 4 h or were untreated. Total RNAs were isolated and analyzed by using the CodeLink gene chip system. Genes with a more than threefold change in expression, compared with expression in untreated hTERT-HME1 cells, were scored. (A) Comparison of the genes expressed in response to a high level of Y705F-STAT3 or treatment with TNF-α. (B) Northern analysis of gene expression.
Discussion
The intracellular concentration of U-STAT3 increases when the STAT3 gene is activated in response to gp130-linked cytokines, allowing U-STAT3 to compete more effectively with IκB for U-NFκB to form a novel transcription factor that induces RANTES expression by binding to the proximal κB site of the promoter. Since the Y705F mutant of STAT3, which cannot be phosphorylated on tyrosine, also activates RANTES expression, this function of U-STAT3 is clearly distinct from the absolute requirement for tyrosine phosphorylation that enables STAT3 dimers to bind to GAS sequences (Wen et al. 1995; Kaptein et al. 1996; Zhang et al. 1999).
Studies of variant STAT3 proteins in which different domains have been deleted indicate that the region between residues 495 and 715, which includes the SH2 domain, is required for binding to p65 and p50. The small domain between residues 150 and 162 comprises an NLS sequence (Liu et al. 2005) that is necessary to activate gene expression in response to U-STAT3, suggesting that U-STAT3 contributes to the function of the ternary complex with p65 and p50 by facilitating the nuclear localization of the complex through interaction with importin-α3 (Fig. 6). Additional phosphorylation of P-STAT3 dimers on Ser727 is needed for maximal activation of transcription, but not for DNA binding (Wen and Darnell 1995, 1997). Neither residues 716–770, comprising the transactivation domain of STAT3 (and missing in STAT3β), nor Ser727 are absolutely required for the activity of U-STAT3 on the RANTES promoter (Fig. 3). However, it remains possible that the C-terminal domain, together with phosphorylated Ser727, might facilitate the transactivation function of the U-STAT3:U-NFκB complex on other promoters. For example, Ng et al. (2006) have shown that STAT3 is phosphorylated on Ser727 but not Tyr705 in response to activation of the TrkA receptor by nerve growth factor, and that serine-phosphorylated STAT3 is important in driving signal-dependent gene expression. The N-terminal domain of U-STAT3 is not required for binding to NFκB or for function, since the protein that includes residues 150–770 is fully active. Further work is required to delineate in more detail how the individual domains of U-STAT3 function in the ternary complex and to define the relevant domains of p65 and p50. The data of Supplementary Figure 5 already indicate that the Rel domain of p65 is required. Interestingly, the natural increase in the level of U-STAT3 in response to long-term treatment with IL-6 is capable of activating NFκB, and this activation drives the expression of the RANTES, MRAS, and MET genes (Fig. 2B,C). Note that 162–770 truncated STAT3 binds to NFκB but holds the complex in the cytosol, inhibiting the signal-dependent translocation of NFκB into the nucleus and target gene expression (Fig. 3D–F).
Figure 6.
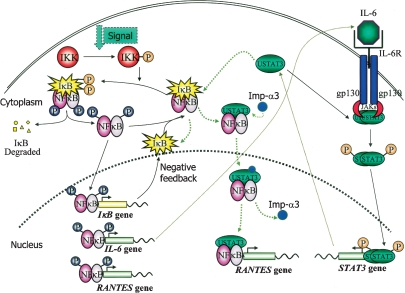
Interactions between the STAT3 and NFκB pathways. U-STAT3, induced to a high level due to activation of the STAT3 gene in response to ligands such as IL-6, competes with IκB for p65/p50. The U-STAT3:U-NFκB complex activates the RANTES promoter plus a subset of other promoters that have κB elements. U-STAT3 also drives the expression of some genes that do not have κB elements, by an unknown mechanism (not shown). The κB element of the IL6 gene is driven by canonical NFκB signaling in response to ligands such as TNF-α or IL-1, setting up the positive feedback loop that is driven by the activation of STAT3 in response to secreted IL-6, leading to an increased level of U-STAT3 that sustains the activation of genes such as RANTES. (Imp-α3) Importin-α3.
Although the binding of U-STAT3 to phosphorylated NFκB is not detected by EMSA under our conditions (Fig. 2A), immunoprecipitation experiments do detect the binding of these two proteins in TNF-α-treated cells (Supplementary Fig. 2). Conversely, the binding of P-STAT3 to U-NFκB is also detected by immunoprecipitation (Supplementary Figs. 4, 5). Interactions among phosphorylated and unphosphorylated forms of STAT3 and NFκB have been reported previously by several groups. U-STAT3 forms a complex with the p65 subunit of P-NFκB on a κB sequence in the human IL-8 promoter, inducing gene expression in response to IL-1β (Yoshida et al. 2004). U-STAT3 binds to both p65 and p50, and a specific type of κB sequence motif supports both the binding of p65 homodimers and cooperativity with U-STAT3 (Yoshida et al. 2004). Agrawal et al. (2003) showed that P-NFκB synergistically cooperates with P-STAT3 and C/EBPβ to enhance transcription of the C-reactive protein (CRP) gene. Hagihara et al. (2005) found that STAT3 plays an essential role in cytokine-driven expression of the serum amyloid A (SAA) gene, which does not have a typical STAT3 response element in its promoter. P-STAT3 and P-p65 form a complex following stimulation of cells with both IL-1 and IL-6, after which STAT3 interacts with nonconsensus sequences at the 3′ boundary of κB element of the SAA promoter to enhance transcription. Yu et al. (2002) found that U-STAT3, through direct interaction with p65, serves as a dominant-negative inhibitor of the ability of P-NFκB to induce cytokine-dependent induction of the iNOS promoter in mesangial cells.
In addition to its interactions with NFκB, STAT3 has been shown to bind to other transcription factors. For example, it forms a complex with the CRE-binding protein on the JUNB promoter (Kojima et al. 1996) and with c-Jun on the α2-macrogloblin APRE (Schaefer et al. 1995). Other reports show that STAT3 has an effect on CRE-like sites in the C/EBPB promoter (Niehof et al. 2001) and the glucocorticoid response element (Zhang and Fuller 1997; Zhang et al. 1999), which lack classical GAS sequences. We found that fewer than half of the genes that respond to high-level expression of Y705F-STAT3 respond also to TNF-α (Fig. 5). The Y705F-STAT3-responsive genes that do not respond to TNF-α probably do not have functional κB elements and, as shown in Figure 4C, two such genes do not need p65 in order to respond to Y705F-STAT3. Therefore, it is extremely likely that Y705F-STAT3 (or U-STAT3) interacts productively with one or more transcription factors different from NFκB to drive the expression of this class of genes. Identification of these factors and characterization of their interactions with U-STAT3 remain to be accomplished.
Interconnections between signaling pathways that use activated NFκB and those that use activated STAT3 are shown in Figure 6. The current work reveals the importance of U-STAT3 in connections between these two important classes of pathways. IL-1 is an important mediator of the inflammatory response since it induces other proinflammatory cytokines, chemokines, and acute phase proteins (Dinarello 1996). From the work presented here, we can now appreciate that the expression of IL-6 in response to activation of NFκB by IL-1 initiates a positive feedback loop in which secreted IL-6 stimulates the tyrosine phosphorylation of STAT3, leading secondarily to an increase in U-STAT3, which then drives the expression of a subset of NFκB-activated genes, including RANTES. Thus, the κB element of the RANTES promoter can function to give strong expression in two ways, directly in response to TNF-α or IL-1 or indirectly in response to IL-6 (Fig. 6). This dual regulation of RANTES transcription may be important in regulating its physiological functions, with short-term expression in response to IL-1 controlled by P-NFκB and more sustained expression, indirectly in response to IL-6, regulated by U-STAT3:P-NFκB (Fig. 6).
RANTES is an important mediator of acute and chronic inflammation, with genetic evidence indicating its involvement in immunopathological disorders (Kim et al. 2004; Simeoni et al. 2004; Boger et al. 2005; Wang et al. 2005; Charo and Ransohoff 2006). Its wide spectrum of biological activities is transduced through three distinct chemokine receptors, CCR1, CCR3, and CCR5. These targets of RANTES are present on a diversity of leukocytes, including memory T cells, eosinophils, and monocytes (Fujisawa et al. 2000; Luther and Cyster 2001). Depending on the cellular context, RANTES can deliver chemoattractant or activating signals, with the latter inducing responses of dendritic cells that range from eosinophil degranulation to production of cytokines. The levels of RANTES mRNA (Fig. 2C) and the mRNAs encoding MET, MRAS, and TIS11D (Fig. 2C; Yang et al. 2005) are increased coordinately with U-STAT3 levels in cells treated with IL-6 for long times (32–48 h). Sustained RANTES expression, as might be driven by increased expression of U-STAT3 following exposure of cells to gp130-linked cytokines, may be highly significant biologically. For example, elevated RANTES levels can impair the entry into cells of macrophage-tropic HIV-1 via CCR5 (Simmons et al. 2000). Also, micromolar concentration of RANTES can deliver costimulatory signals to T cells, augmenting responses through the T-cell receptor (Bacon et al. 1995). Elegant structural studies indicate that these concentrations may be achievable in vivo through formation of multimeric RANTES aggregates on a glycosaminoglycan substrate (Johnson et al. 2004; Shaw et al. 2004; Proudfoot 2006). Although the expression of RANTES was first thought to be limited to active T cells, recent data have shown that it is produced by a variety of tissue types in response to specific stimuli. RANTES mRNA is expressed late (3–5 d) after activation of resting T cells, whereas in fibroblasts, renal epithelial, and mesangial cells, RANTES mRNA is quickly up-regulated by TNF-α stimulation (Hirano et al. 2003; Ogura et al. 2005).
The full biological relevance of the ability of P-STAT3 to increase the intracellular concentration of U-STAT3 remains to be established. In the context of cancer, the constitutive tyrosine phosphorylation of STAT3 in many different tumors is likely to lead to increased expression of U-STAT3, which in turn drives the expression of oncogenes such as MET and MRAS (Yang et al. 2005). In cell culture system, long-term treatment with IL-6 to increase total U-STAT3, the expression levels for RANTES, as well as MET, MRAS, and TIS11D (Fig. 2C; Yang et al. 2005) are increased coordinately with U-STAT3 levels at late IL-6-treated time points (32–48 h). The biological role of U-STAT3-driven gene expression in normal physiology is best addressed by experiments with genetically altered mice. An important attempt to do this was reported by Narimatsu et al. (2001), who mutated the GAS element of the endogenous STAT3 promoter. The ability of IL-6 to increase STAT3 expression was abrogated in some tissues but not in others, probably because STAT3-dependent expression of the STAT3 gene can be regulated through additional elements that were not recognized and therefore were not mutated. Incomplete suppression of the response of the STAT3 gene to IL-6 might well account for the observed mild phenotype of the promoter knock-in mouse. Since complete deletion of STAT3 is embryonic lethal (Takeda et al. 1997), it remains to be seen whether mice with complete loss of the STAT3-dependent induction of U-STAT3 expression would have severe defects, as might be expected if the up-regulation of U-STAT3 is important for the full physiological functions of the many cytokines that use the common gp130 receptor subunit to phosphorylate STAT3.
Materials and methods
Cells and reagents
hTERT-HME1 cells (Clontech) were grown in MCDB 170 medium with supplements of bovine pituitary extract, hydrocortisone, insulin, gentamycin, human epidermal growth factor, and amphotericin-B, all from Clonetics. PC3 cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin (100 U/mL) and streptomycin (100 μg/mL) (GIBCO-BRL). TNF-α was used at a concentration of 50 ng/mL. Antibodies against STAT3 (C-20, K-15, and H-190), p65, p50, IκBα, and c-Myc were from Santa Cruz Biotechnologies, and antibodies against Tyr705-phosphorylated STAT3 (pTyr-STAT3) and Ser536-phosphorylated p65 were from Cell Signaling Technology. WT and YF cells, expressing a high level of wild-type STAT3 or Y705F-STAT3, respectively, were described previously (Yang et al. 2005). The construct for truncated 162–770 wild-type STAT3 and cells expressing a high level of this protein were generated as described before (Yang et al. 2005). Plasmids encoding GFP-tagged STAT3 N- and C-terminal deletion mutants were generous gifts from Nancy C. Reich, State University of New York, Stony Brook, Stony Brook, NY (Liu et al. 2005). The p65 expression plasmid was described by Yoshida et al. (2004). NFκB siRNA was from Cell Signaling Technology.
Luciferase reporter plasmid
A 1.5-kb DNA fragment containing the human RANTES promoter (−1426 to +128), obtained from a ChIP experiment, was inserted to the pcDNA3.1 vector. RANTES promoter deletion and mutation reporter constructs were gifts from Dr. Antonella Casola, University of Texas Medical Branch, Galveston, TX (Casola et al. 2002). Further deletions of these luciferase reporter constructs were performed by PCR, by introducing KpnI and NheI sites, followed by subcloning to the same restriction sites of the pGL2-basic vector, to generate pGL2-974 (−974 ∼ −1), pGL2-220 (−220 ∼ −1), pGL2-195 (−195 ∼ −1), pGL2-120 (−120 ∼ −1), pGL2-del (−974 ∼ −120), pGL2-CRE-m (5′-AAACT GATGAGCTCACTCTA-3′ to 5′-AAACTtcTtAtagacCgCTA -3′), pGL2-ISRE-m (5′-TTTCAGTTTTCTTTTCC-3′ to 5′-TT TCAGTaaaCTaaaCC-3′), and pGL2-NFκB-m (5′-TTTTGGAAA CTCCCCTTAGGGGATGCCCT-3′ to 5′-TTTTGGcAcCTtaa CgTA cGCCATGCatT-3′), respectively (Casola et al. 2002). Note that the RANTES promoter sequence used has two κB sites and that both were mutated. To guard against PCR-associated incorporation errors, the integrity of all the constructs generated was confirmed by sequencing.
Transfection and luciferase assays
RANTES promoter–luciferase reporter (Luc) constructs were transfected into hTERT-HME1 cells by using the Fugene 6 reagent (Roche). Cells were plated and cultured in 12-well plates to 40% confluence before transfection. After a change to fresh media, 1 μg/well luciferase plasmid plus 0.5 μg/well pCH110 (β-galactoside plasmid for internal control) were cotransfected. Forty hours later, the cells were harvested and the cell pellets were lysed in 200 μL of buffer (Reporter lysis buffer, Promega), mixed by vortexing for 5 sec, and spun at 2000g for 5 min at room temperature. Cell lysate (60 μL) was mixed with 60 μL of luciferase assay buffer (Promega) for activity measurements in an Auto Lumat BG-P luminometer (MGM Instruments). For the β-galactosidase activity assay, the luminescent β-galactosidase detection Kit II (Clontech) was used.
p65 siRNA transfection
hTERT-HME1-derived cells were grown in 60-mm plates to 40% confluence before transfection. Media were aspirated from the cells, which were washed twice with sterile phosphate-buffered saline. Then, 5 mL of fresh medium were added to each plate, with 10% serum and without antibiotics. Twenty microliters of 10 μM p65 siRNA (Cell Signaling Technology) were added to 300 μL of siRNA Transfection Medium (Santa Cruz Biotechnology), mixed gently, kept at room temperature for 20 min, and added drop-wise to the plates with gentle rocking. After incubation for 24 h at 37°C, the transfection media were removed and the cells were transfected again, following the same protocol. After another 48 h, the cells were harvested and total RNA or protein was extracted for analysis.
Coimmunoprecipitations
The protocol provided by Sigma-Aldrich was followed, with slight modifications. For immunoprecipitation of p65, cells were lysed in buffer (50 mM Tris HCl at pH 8.0, 150 mM NaCl, 1% NP-40) and Sepharose G beads were used. For immunoprecipitation of STAT3, the EZview Red ANTI-FLAG M2 Affinity Gel system (Sigma-Aldrich) was used, exactly as in the protocol provided by the manufacturer.
Western and Northern analyses
These procedures were carried out essentially as described before (Yang et al. 2005). For Western analyses, membranes were probed with primary antibodies specific for STAT3 (Santa Cruz Biotechnology, 482), Tyr705-STAT3, p65 (Santa Cruz Biotechnology, 109), p50 (Santa Cruz Biotechnology, 114), or Ser536-p65. For Northern analyses, 20 μg of total RNA were used. Human cDNA probes for RANTES, MET, IL1β, MAD4, MRAS, and TIS11D were cut from I.M.A.G.E. clones (Invitrogen or the American Type Culture Collection). Templates for the human GAPDH cDNA were obtained by RT–PCR. Signals were normalized for loading by comparing the intensities of GAPDH mRNA on the same membranes.
EMSAs
This procedure was performed as reported previously (Yang et al. 2005). hTERT-HME1-derived cells, untreated or treated with TNF-α for 4 h, were lysed in EMSA lysis buffer, supplemented with protease inhibitors. The probe was the NFκB consensus sequence (top strand, 5′-TTTTGGAAACTCCCCTTAGGGGA TGCCCCT-3′) from the RANTES promoter (Nelson et al. 1993; Genin et al. 2000). Labeled probe (104 DPM) was used in each binding reaction. For supershift analyses, whole-cell extracts were preincubated for 20 min at room temperature with polyclonal antibodies specific for STAT3, p65, p50 (see above), IκB (Santa Cruz Biotechnology, 371), or c-Myc (Santa Cruz Biotechnology, 788) before adding radiolabeled probe.
ChIP analyses
The protocol is from previous publications (Weinmann and Farnham 2002; Li et al. 2003). Briefly, 108 cells were cross-linked in 1% formaldehyde for 10 min before adding 0.125 M glycine to terminate the reaction. The cells were trypsinized and resuspended in 6 mL of cell lysis buffer (5 mM PIPES at pH 8.0, 85 mM KCl, 0.5% Nonidet P-40, 10 μL/mL PMSF, 1 μL/mL aprotinin, 1 μL/mL leupeptin). After incubation for 10 min on ice, nuclei were collected and resuspended in 1 mL of nuclear lysis buffer (50 mM Tris HCl at pH 8.1, 10 mM EDTA, 1% SDS) plus protease inhibitors to obtain chromatin preparations, which were then sonicated to an average length of ∼0.5–2 kb by using 15 pulses of 30 sec each with 2-min rests at setting 5 of a Fisher Model 60 sonic dismembranator. Sonicated samples were immunoprecipitated with anti-M2 (anti-Flag), which distinguishes exogenous (tagged) from endogenous (untagged) U-STAT3. The cross-links were then reversed in 0.3 M NaCl in the presence of RNaseA (Roche), 10 mg/mL, for 4–5 h at 65°C. DNA fragments were purified by ethanol precipitation. The immunoprecipitated DNA was amplified by a ligation-mediated PCR (LM–PCR) procedure in which the samples were pretreated with T4 DNA polymerase to blunt the ends of the DNA. Then a linker was ligated to the DNA fragments, allowing them to be amplified by PCR, using primers located in the linker. The PCR products were ligated into a pcDNA3.1-based vector by using Rapid Ligation Kit (Roche). Inserts were sequenced by using a vector-specific primer and T7 or Sp6 polymerase.
CodeLink expression array experiments
Total RNAs were analyzed by using CodeLink arrays (GenUs Biosystems). Data were analyzed by using GenUs software. Expression was normalized against the levels of GAPDH and ACTIN mRNAs in the all samples. The levels of mRNAs in TNF-α-treated cells or untreated YF cells were compared with the levels in hTERT-HME1 control cells. The data are presented in Supplementary Table 1.
Immunocytochemistry
hTERT-HME1 cells stably expressing a high level of full-length wild-type STAT3 or 162–770 truncated STAT3 (Yang et al. 2005) were grown on glass cover slips for 24 h before fixation. Cells at ∼50% confluency were fixed in 4% paraformaldehyde for 15 min and absolute methanol for 5 min at room temperature and then treated with blocking buffer (1× PBS + 0.3% Triton X-100 + 10% FBS). STAT3 was detected with mouse anti-human STAT3 (Cell Signaling Technology) and rabbit anti-human p65 (Santa Cruz Biotechnology). Signals were visualized by Alexa Fluor 488 goat anti-mouse (green fluorescent) and Alexa Fluor 494 goat anti-rabbit (red fluorescent) secondary antibodies. Images were captured with a Zeiss Axioskop fluorescence microscope.
Acknowledgments
We thank Dr. Judith Drazba, Cleveland Clinic, for help with confocal microscopy; Dr. Antonella Casola, University of Texas Medical Branch, for human RANTES promoter constructs; and Dr. Nancy C. Reich, Stony Brook University, for STAT3 constructs. This work was supported by grant P01 CA 62220 to G.R.S. from the National Cancer Institute and RO1 A144122 to P.E.A. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. We acknowledge Dr. Arvind Kumar for help with supplemental experiments, and we are especially grateful to Drs. Ian M. Kerr, Richard Ransohoff, and Anette van Boxel-Dezaire for many helpful discussions and suggestions.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1553707
References
- Agrawal A., Cha-Molstad H., Samols D., Kushner I., Cha-Molstad H., Samols D., Kushner I., Samols D., Kushner I., Kushner I. Overexpressed nuclear factor-κB can participate in endogenous C-reactive protein induction, and enhances the effects of C/EBPβ and signal transducer and activator of transcription-3. Immunology. 2003;108:539–547. doi: 10.1046/j.1365-2567.2003.01608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Nishio Y., Inoue M., Wang X.J., Wei S., Matsusaka T., Yoshida K., Sudo T., Naruto M., Kishimoto T., Nishio Y., Inoue M., Wang X.J., Wei S., Matsusaka T., Yoshida K., Sudo T., Naruto M., Kishimoto T., Inoue M., Wang X.J., Wei S., Matsusaka T., Yoshida K., Sudo T., Naruto M., Kishimoto T., Wang X.J., Wei S., Matsusaka T., Yoshida K., Sudo T., Naruto M., Kishimoto T., Wei S., Matsusaka T., Yoshida K., Sudo T., Naruto M., Kishimoto T., Matsusaka T., Yoshida K., Sudo T., Naruto M., Kishimoto T., Yoshida K., Sudo T., Naruto M., Kishimoto T., Sudo T., Naruto M., Kishimoto T., Naruto M., Kishimoto T., Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- Bacon K.B., Premack B.A., Gardner P., Schall T.J., Premack B.A., Gardner P., Schall T.J., Gardner P., Schall T.J., Schall T.J. Activation of dual T cell signaling pathways by the chemokine RANTES. Science. 1995;269:1727–1730. doi: 10.1126/science.7569902. [DOI] [PubMed] [Google Scholar]
- Battle T.E., Frank D.A., Frank D.A. The role of STATs in apoptosis. Curr. Mol. Med. 2002;2:381–392. doi: 10.2174/1566524023362456. [DOI] [PubMed] [Google Scholar]
- Birbach A., Gold P., Binder B.R., Hofer E., de Martin R., Schmid J.A., Gold P., Binder B.R., Hofer E., de Martin R., Schmid J.A., Binder B.R., Hofer E., de Martin R., Schmid J.A., Hofer E., de Martin R., Schmid J.A., de Martin R., Schmid J.A., Schmid J.A. Signaling molecules of the NF-κB pathway shuttle constitutively between cytoplasm and nucleus. J. Biol. Chem. 2002;277:10842–10851. doi: 10.1074/jbc.M112475200. [DOI] [PubMed] [Google Scholar]
- Boger C.A., Fischereder M., Deinzer M., Aslanidis C., Schmitz G., Stubanus M., Banas B., Kruger B., Riegger G.A., Kramer B.K., Fischereder M., Deinzer M., Aslanidis C., Schmitz G., Stubanus M., Banas B., Kruger B., Riegger G.A., Kramer B.K., Deinzer M., Aslanidis C., Schmitz G., Stubanus M., Banas B., Kruger B., Riegger G.A., Kramer B.K., Aslanidis C., Schmitz G., Stubanus M., Banas B., Kruger B., Riegger G.A., Kramer B.K., Schmitz G., Stubanus M., Banas B., Kruger B., Riegger G.A., Kramer B.K., Stubanus M., Banas B., Kruger B., Riegger G.A., Kramer B.K., Banas B., Kruger B., Riegger G.A., Kramer B.K., Kruger B., Riegger G.A., Kramer B.K., Riegger G.A., Kramer B.K., Kramer B.K. RANTES gene polymorphisms predict all-cause and cardiac mortality in type 2 diabetes mellitus hemodialysis patients. Atherosclerosis. 2005;183:121–129. doi: 10.1016/j.atherosclerosis.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Casola A., Henderson A., Liu T., Garofalo R.P., Brasier A.R., Henderson A., Liu T., Garofalo R.P., Brasier A.R., Liu T., Garofalo R.P., Brasier A.R., Garofalo R.P., Brasier A.R., Brasier A.R. Regulation of RANTES promoter activation in alveolar epithelial cells after cytokine stimulation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;283:L1280–L1290. doi: 10.1152/ajplung.00162.2002. [DOI] [PubMed] [Google Scholar]
- Charo I.F., Ransohoff R.M., Ransohoff R.M. The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 2006;354:610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- Chatterjee-Kishore M., Wright K.L., Ting J.P., Stark G.R., Wright K.L., Ting J.P., Stark G.R., Ting J.P., Stark G.R., Stark G.R. How Stat1 mediates constitutive gene expression: A complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C.A. Biologic basis for Interleukin-1 in disease. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- Fujisawa T., Kato Y., Nagase H., Atsuta J., Terada A., Iguchi K., Kamiya H., Morita Y., Kitaura M., Kawasaki H., Kato Y., Nagase H., Atsuta J., Terada A., Iguchi K., Kamiya H., Morita Y., Kitaura M., Kawasaki H., Nagase H., Atsuta J., Terada A., Iguchi K., Kamiya H., Morita Y., Kitaura M., Kawasaki H., Atsuta J., Terada A., Iguchi K., Kamiya H., Morita Y., Kitaura M., Kawasaki H., Terada A., Iguchi K., Kamiya H., Morita Y., Kitaura M., Kawasaki H., Iguchi K., Kamiya H., Morita Y., Kitaura M., Kawasaki H., Kamiya H., Morita Y., Kitaura M., Kawasaki H., Morita Y., Kitaura M., Kawasaki H., Kitaura M., Kawasaki H., Kawasaki H., et al. Chemokines induce eosinophil degranulation through CCR-3. J. Allergy Clin. Immunol. 2000;106:507–513. doi: 10.1067/mai.2000.108311. [DOI] [PubMed] [Google Scholar]
- Genin P., Algarte M., Roof P., Lin R., Hiscott J., Algarte M., Roof P., Lin R., Hiscott J., Roof P., Lin R., Hiscott J., Lin R., Hiscott J., Hiscott J. Regulation of RANTES chemokine gene expression requires cooperativity between NF-κ B and IFN-regulatory factor transcription factors. J. Immunol. 2000;164:5352–5361. doi: 10.4049/jimmunol.164.10.5352. [DOI] [PubMed] [Google Scholar]
- Giannitrapani L., Cervello M., Soresi M., Notarbartolo M., La Rosa M., Virruso L., D’Alessandro N., Montalto G., Cervello M., Soresi M., Notarbartolo M., La Rosa M., Virruso L., D’Alessandro N., Montalto G., Soresi M., Notarbartolo M., La Rosa M., Virruso L., D’Alessandro N., Montalto G., Notarbartolo M., La Rosa M., Virruso L., D’Alessandro N., Montalto G., La Rosa M., Virruso L., D’Alessandro N., Montalto G., Virruso L., D’Alessandro N., Montalto G., D’Alessandro N., Montalto G., Montalto G. Circulating IL-6 and sIL-6R in patients with hepatocellular carcinoma. Ann. N. Y. Acad. Sci. 2002;963:46–52. doi: 10.1111/j.1749-6632.2002.tb04093.x. [DOI] [PubMed] [Google Scholar]
- Hagihara K., Nishikawa T., Sugamata Y., Song J., Isobe T., Taga T., Yoshizaki K., Nishikawa T., Sugamata Y., Song J., Isobe T., Taga T., Yoshizaki K., Sugamata Y., Song J., Isobe T., Taga T., Yoshizaki K., Song J., Isobe T., Taga T., Yoshizaki K., Isobe T., Taga T., Yoshizaki K., Taga T., Yoshizaki K., Yoshizaki K. Essential role of STAT3 in cytokine-driven NF-κB-mediated serum amyloid A gene expression. Genes Cells. 2005;10:1051–1063. doi: 10.1111/j.1365-2443.2005.00900.x. [DOI] [PubMed] [Google Scholar]
- Haura E.B., Turkson J., Jove R., Turkson J., Jove R., Jove R. Mechanisms of disease: Insights into the emerging role of signal transducers and activators of transcription in cancer. Nat. Clin. Pract. Oncol. 2005;2:315–324. doi: 10.1038/ncponc0195. [DOI] [PubMed] [Google Scholar]
- Henkel T., Machleidt T., Alkalay I., Kronke M., Ben-Neriah Y., Baeuerle P.A., Machleidt T., Alkalay I., Kronke M., Ben-Neriah Y., Baeuerle P.A., Alkalay I., Kronke M., Ben-Neriah Y., Baeuerle P.A., Kronke M., Ben-Neriah Y., Baeuerle P.A., Ben-Neriah Y., Baeuerle P.A., Baeuerle P.A. Rapid proteolysis of IκBα is necessary for activation of transcription factor NF-κB. Nature. 1993;365:182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- Hirano T., Ishihara K., Hibi M., Ishihara K., Hibi M., Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19:2548–2556. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- Hirano F., Komura K., Fukawa E., Makino I., Komura K., Fukawa E., Makino I., Fukawa E., Makino I., Makino I. Tumor necrosis factor α TNF-α-induced RANTES chemokine expression via activation of NF-κB and p38 MAP kinase: Roles of TNF-α in alcoholic liver diseases. J. Hepatol. 2003;38:483–489. doi: 10.1016/s0168-8278(02)00456-7. [DOI] [PubMed] [Google Scholar]
- Hodge D.R., Hurt E.M., Farrar W.L., Hurt E.M., Farrar W.L., Farrar W.L. The role of IL-6 and STAT3 in inflammation and cancer. Eur. J. Cancer. 2005;41:2502–2512. doi: 10.1016/j.ejca.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Johnson Z., Kosco-Vilbois M.H., Herren S., Cirillo R., Muzio V., Zaratin P., Carbonatto M., Mack M., Smailbegovic A., Rose M., Kosco-Vilbois M.H., Herren S., Cirillo R., Muzio V., Zaratin P., Carbonatto M., Mack M., Smailbegovic A., Rose M., Herren S., Cirillo R., Muzio V., Zaratin P., Carbonatto M., Mack M., Smailbegovic A., Rose M., Cirillo R., Muzio V., Zaratin P., Carbonatto M., Mack M., Smailbegovic A., Rose M., Muzio V., Zaratin P., Carbonatto M., Mack M., Smailbegovic A., Rose M., Zaratin P., Carbonatto M., Mack M., Smailbegovic A., Rose M., Carbonatto M., Mack M., Smailbegovic A., Rose M., Mack M., Smailbegovic A., Rose M., Smailbegovic A., Rose M., Rose M., et al. Interference with heparin binding and oligomerization creates a novel anti-inflammatory strategy targeting the chemokine system. J. Immunol. 2004;173:5776–5785. doi: 10.4049/jimmunol.173.9.5776. [DOI] [PubMed] [Google Scholar]
- Joo A., Aburatani H., Morii E., Iba H., Yoshimura A., Aburatani H., Morii E., Iba H., Yoshimura A., Morii E., Iba H., Yoshimura A., Iba H., Yoshimura A., Yoshimura A. STAT3 and MITF cooperatively induce cellular transformation through upregulation of c-fos expression. Oncogene. 2004;23:726–734. doi: 10.1038/sj.onc.1207174. [DOI] [PubMed] [Google Scholar]
- Kaptein A., Paillard V., Saunders M., Paillard V., Saunders M., Saunders M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak–STAT signal transduction. J. Biol. Chem. 1996;271:5961–5964. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- Kim J.J., Lee J.H., Jang C.H., Kim Y.S., Chae S.C., Chung H.T., Choi T.W., Lee J.H., Lee J.H., Jang C.H., Kim Y.S., Chae S.C., Chung H.T., Choi T.W., Lee J.H., Jang C.H., Kim Y.S., Chae S.C., Chung H.T., Choi T.W., Lee J.H., Kim Y.S., Chae S.C., Chung H.T., Choi T.W., Lee J.H., Chae S.C., Chung H.T., Choi T.W., Lee J.H., Chung H.T., Choi T.W., Lee J.H., Choi T.W., Lee J.H., Lee J.H. Chemokine RANTES promoter polymorphisms in allergic rhinitis. Laryngoscope. 2004;114:666–669. doi: 10.1097/00005537-200404000-00013. [DOI] [PubMed] [Google Scholar]
- Kojima H., Nakajima K., Hirano T., Nakajima K., Hirano T., Hirano T. IL-6-inducible complexes on an IL-6 response element of the junB promoter contain Stat3 and 36 kDa CRE-like site binding proteins. Oncogene. 1996;12:547–554. [PubMed] [Google Scholar]
- Li Z., Van Calcar S., Qu C., Cavenee W.K., Zhang M.Q., Ren B., Van Calcar S., Qu C., Cavenee W.K., Zhang M.Q., Ren B., Qu C., Cavenee W.K., Zhang M.Q., Ren B., Cavenee W.K., Zhang M.Q., Ren B., Zhang M.Q., Ren B., Ren B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., McBride K.M., Reich N.C., McBride K.M., Reich N.C., Reich N.C. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc. Natl. Acad. Sci. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther S.A., Cyster J.G., Cyster J.G. Chemokines as regulators of T cell differentiation. Nat. Immunol. 2001;2:102–107. doi: 10.1038/84205. [DOI] [PubMed] [Google Scholar]
- Narimatsu M., Maeda H., Itoh S., Atsumi T., Ohtani T., Nishida K., Itoh M., Kamimura D., Park S.J., Mizuno K., Maeda H., Itoh S., Atsumi T., Ohtani T., Nishida K., Itoh M., Kamimura D., Park S.J., Mizuno K., Itoh S., Atsumi T., Ohtani T., Nishida K., Itoh M., Kamimura D., Park S.J., Mizuno K., Atsumi T., Ohtani T., Nishida K., Itoh M., Kamimura D., Park S.J., Mizuno K., Ohtani T., Nishida K., Itoh M., Kamimura D., Park S.J., Mizuno K., Nishida K., Itoh M., Kamimura D., Park S.J., Mizuno K., Itoh M., Kamimura D., Park S.J., Mizuno K., Kamimura D., Park S.J., Mizuno K., Park S.J., Mizuno K., Mizuno K., et al. Tissue-specific autoregulation of the stat3 gene and its role in interleukin-6-induced survival signals in T cells. Mol. Cell. Biol. 2001;21:6615–6625. doi: 10.1128/MCB.21.19.6615-6625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P.J., Kim H.T., Manning W.C., Goralski T.J., Krensky A.M., Kim H.T., Manning W.C., Goralski T.J., Krensky A.M., Manning W.C., Goralski T.J., Krensky A.M., Goralski T.J., Krensky A.M., Krensky A.M. Genomic organization and transcriptional regulation of the RANTES chemokine gene. J. Immunol. 1993;151:2601–2612. [PubMed] [Google Scholar]
- Ng Y.P., Cheung Z.H., Ip N.Y., Cheung Z.H., Ip N.Y., Ip N.Y. STAT3 as a downstream mediator of Trk signaling and functions. J. Biol. Chem. 2006;281:15636–15644. doi: 10.1074/jbc.M601863200. [DOI] [PubMed] [Google Scholar]
- Niehof M., Streetz K., Rakemann T., Bischoff S.C., Manns M.P., Horn F., Trautwein C., Streetz K., Rakemann T., Bischoff S.C., Manns M.P., Horn F., Trautwein C., Rakemann T., Bischoff S.C., Manns M.P., Horn F., Trautwein C., Bischoff S.C., Manns M.P., Horn F., Trautwein C., Manns M.P., Horn F., Trautwein C., Horn F., Trautwein C., Trautwein C. Interleukin-6-induced tethering of STAT3 to the LAP/C/EBPβ promoter suggests a new mechanism of transcriptional regulation by STAT3. J. Biol. Chem. 2001;276:9016–9027. doi: 10.1074/jbc.M009284200. [DOI] [PubMed] [Google Scholar]
- Niu G., Wright K.L., Ma Y., Wright G.M., Huang M., Irby R., Briggs J., Karras J., Cress W.D., Pardoll D., Wright K.L., Ma Y., Wright G.M., Huang M., Irby R., Briggs J., Karras J., Cress W.D., Pardoll D., Ma Y., Wright G.M., Huang M., Irby R., Briggs J., Karras J., Cress W.D., Pardoll D., Wright G.M., Huang M., Irby R., Briggs J., Karras J., Cress W.D., Pardoll D., Huang M., Irby R., Briggs J., Karras J., Cress W.D., Pardoll D., Irby R., Briggs J., Karras J., Cress W.D., Pardoll D., Briggs J., Karras J., Cress W.D., Pardoll D., Karras J., Cress W.D., Pardoll D., Cress W.D., Pardoll D., Pardoll D., et al. Role of Stat3 in regulating p53 expression and function. Mol. Cell. Biol. 2005;25:7432–7440. doi: 10.1128/MCB.25.17.7432-7440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura N., Tobe M., Sakamaki H., Nagura H., Abiko Y., Kondoh T., Tobe M., Sakamaki H., Nagura H., Abiko Y., Kondoh T., Sakamaki H., Nagura H., Abiko Y., Kondoh T., Nagura H., Abiko Y., Kondoh T., Abiko Y., Kondoh T., Kondoh T. Tumor necrosis factor-α increases chemokine gene expression and production in synovial fibroblasts from human temporomandibular joint. J. Oral Pathol. Med. 2005;34:357–363. doi: 10.1111/j.1600-0714.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- Palombella V.J., Rando O.J., Goldberg A.L., Maniatis T., Rando O.J., Goldberg A.L., Maniatis T., Goldberg A.L., Maniatis T., Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-κB precursor protein and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Proudfoot A.E. The biological relevance of chemokine–proteoglycan interactions. Biochem. Soc. Trans. 2006;34:422–426. doi: 10.1042/BST0340422. [DOI] [PubMed] [Google Scholar]
- Roff M., Thompson J., Rodriguez M.S., Jacque J.M., Baleux F., Arenzana-Seisdedos F., Hay R.T., Thompson J., Rodriguez M.S., Jacque J.M., Baleux F., Arenzana-Seisdedos F., Hay R.T., Rodriguez M.S., Jacque J.M., Baleux F., Arenzana-Seisdedos F., Hay R.T., Jacque J.M., Baleux F., Arenzana-Seisdedos F., Hay R.T., Baleux F., Arenzana-Seisdedos F., Hay R.T., Arenzana-Seisdedos F., Hay R.T., Hay R.T. Role of IκBα ubiquitination in signal-induced activation of NFκB in vivo. J. Biol. Chem. 1996;271:7844–7850. doi: 10.1074/jbc.271.13.7844. [DOI] [PubMed] [Google Scholar]
- Schaefer T.S., Sanders L.K., Nathans D., Sanders L.K., Nathans D., Nathans D. Cooperative transcriptional activity of Jun and Stat3 β, a short form of Stat3. Proc. Natl. Acad. Sci. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H.M., Milocco L.H., Lamb P., Darnell J.E., Jr., Stein R.B., Rosen J., Milocco L.H., Lamb P., Darnell J.E., Jr., Stein R.B., Rosen J., Lamb P., Darnell J.E., Jr., Stein R.B., Rosen J., Darnell J.E., Jr., Stein R.B., Rosen J., Stein R.B., Rosen J., Rosen J. Spacing of palindromic half sites as a determinant of selective STAT signal transducers and activators of transcription DNA binding and transcriptional activity. Proc. Natl. Acad. Sci. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J.P., Johnson Z., Borlat F., Zwahlen C., Kungl A., Roulin K., Harrenga A., Wells T.N., Proudfoot A.E., Johnson Z., Borlat F., Zwahlen C., Kungl A., Roulin K., Harrenga A., Wells T.N., Proudfoot A.E., Borlat F., Zwahlen C., Kungl A., Roulin K., Harrenga A., Wells T.N., Proudfoot A.E., Zwahlen C., Kungl A., Roulin K., Harrenga A., Wells T.N., Proudfoot A.E., Kungl A., Roulin K., Harrenga A., Wells T.N., Proudfoot A.E., Roulin K., Harrenga A., Wells T.N., Proudfoot A.E., Harrenga A., Wells T.N., Proudfoot A.E., Wells T.N., Proudfoot A.E., Proudfoot A.E. The X-ray structure of RANTES: Heparin-derived disaccharides allows the rational design of chemokine inhibitors. Structure. 2004;12:2081–2093. doi: 10.1016/j.str.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Simeoni E., Winkelmann B.R., Hoffmann M.M., Fleury S., Ruiz J., Kappenberger L., Marz W., Vassalli G., Winkelmann B.R., Hoffmann M.M., Fleury S., Ruiz J., Kappenberger L., Marz W., Vassalli G., Hoffmann M.M., Fleury S., Ruiz J., Kappenberger L., Marz W., Vassalli G., Fleury S., Ruiz J., Kappenberger L., Marz W., Vassalli G., Ruiz J., Kappenberger L., Marz W., Vassalli G., Kappenberger L., Marz W., Vassalli G., Marz W., Vassalli G., Vassalli G. Association of RANTES G-403A gene polymorphism with increased risk of coronary arteriosclerosis. Eur. Heart J. 2004;25:1438–1446. doi: 10.1016/j.ehj.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Hibbitts S., Stine J.T., Gray P.W., Proudfoot A.E., Clapham P.R., Reeves J.D., Hibbitts S., Stine J.T., Gray P.W., Proudfoot A.E., Clapham P.R., Hibbitts S., Stine J.T., Gray P.W., Proudfoot A.E., Clapham P.R., Stine J.T., Gray P.W., Proudfoot A.E., Clapham P.R., Gray P.W., Proudfoot A.E., Clapham P.R., Proudfoot A.E., Clapham P.R., Clapham P.R. Co-receptor use by HIV and inhibition of HIV infection by chemokine receptor ligands. Immunol. Rev. 2000;177:112–126. doi: 10.1034/j.1600-065x.2000.17719.x. [DOI] [PubMed] [Google Scholar]
- Takeda K., Noguchi K., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., Akira S., Noguchi K., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., Akira S., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., Akira S., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., Akira S., Matsumoto M., Yoshida N., Kishimoto T., Akira S., Yoshida N., Kishimoto T., Akira S., Kishimoto T., Akira S., Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbutt N.C., Giraud A.S., Inglese M., Jenkins B., Waring P., Clay F.J., Malki S., Alderman B.M., Grail D., Hollande F., Giraud A.S., Inglese M., Jenkins B., Waring P., Clay F.J., Malki S., Alderman B.M., Grail D., Hollande F., Inglese M., Jenkins B., Waring P., Clay F.J., Malki S., Alderman B.M., Grail D., Hollande F., Jenkins B., Waring P., Clay F.J., Malki S., Alderman B.M., Grail D., Hollande F., Waring P., Clay F.J., Malki S., Alderman B.M., Grail D., Hollande F., Clay F.J., Malki S., Alderman B.M., Grail D., Hollande F., Malki S., Alderman B.M., Grail D., Hollande F., Alderman B.M., Grail D., Hollande F., Grail D., Hollande F., Hollande F., et al. Reciprocal regulation of gastrointestinal homeostasis by SHP2 and STAT-mediated trefoil gene activation in gp130 mutant mice. Nat. Med. 2002;8:1089–1097. doi: 10.1038/nm763. [DOI] [PubMed] [Google Scholar]
- Wang C.R., Guo H.R., Liu M.F., Guo H.R., Liu M.F., Liu M.F. RANTES promoter polymorphism as a genetic risk factor for rheumatoid arthritis in the Chinese. Clin. Exp. Rheumatol. 2005;23:379–384. [PubMed] [Google Scholar]
- Weinmann A.S., Farnham P.J., Farnham P.J. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- Wen Z., Darnell J.E., Jr., Darnell J.E., Jr. Mapping of Stat3 serine phosphorylation to a single residue 727 and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res. 1997;25:2062–2067. doi: 10.1093/nar/25.11.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z., Zhong Z., Darnell J.E., Jr., Zhong Z., Darnell J.E., Jr., Darnell J.E., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- Yang J., Chatterjee-Kishore M., Staugaitis S.M., Nguyen H., Schlessinger K., Levy D.E., Stark G.R., Chatterjee-Kishore M., Staugaitis S.M., Nguyen H., Schlessinger K., Levy D.E., Stark G.R., Staugaitis S.M., Nguyen H., Schlessinger K., Levy D.E., Stark G.R., Nguyen H., Schlessinger K., Levy D.E., Stark G.R., Schlessinger K., Levy D.E., Stark G.R., Levy D.E., Stark G.R., Stark G.R. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- Yoshida Y., Kumar A., Koyama Y., Peng H., Arman A., Boch J.A., Auron P.E., Kumar A., Koyama Y., Peng H., Arman A., Boch J.A., Auron P.E., Koyama Y., Peng H., Arman A., Boch J.A., Auron P.E., Peng H., Arman A., Boch J.A., Auron P.E., Arman A., Boch J.A., Auron P.E., Boch J.A., Auron P.E., Auron P.E. Interleukin 1 activates STAT3/nuclear factor-κB cross-talk via a unique TRAF6- and p65-dependent mechanism. J. Biol. Chem. 2004;279:1768–1776. doi: 10.1074/jbc.M311498200. [DOI] [PubMed] [Google Scholar]
- Yu Z., Zhang W., Kone B.C., Zhang W., Kone B.C., Kone B.C. Signal transducers and activators of transcription 3 STAT3 inhibits transcription of the inducible nitric oxide synthase gene by interacting with nuclear factor κB. Biochem. J. 2002;367:97–105. doi: 10.1042/BJ20020588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z.L., Guan Y.J., Chatterjee D., Chin Y.E., Guan Y.J., Chatterjee D., Chin Y.E., Chatterjee D., Chin Y.E., Chin Y.E. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Fuller G.M., Fuller G.M. The competitive binding of STAT3 and NF-κB on an overlapping DNA binding site. Biochem. Biophys. Res. Commun. 1997;237:90–94. doi: 10.1006/bbrc.1997.7082. [DOI] [PubMed] [Google Scholar]
- Zhang X., Wrzeszczynska M.H., Horvath C.M., Darnell J.E., Jr., Wrzeszczynska M.H., Horvath C.M., Darnell J.E., Jr., Horvath C.M., Darnell J.E., Jr., Darnell J.E., Jr. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell. Biol. 1999;19:7138–7146. doi: 10.1128/mcb.19.10.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H., Voll R.E., Ghosh S., Voll R.E., Ghosh S., Ghosh S. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]