Abstract
Translation is a highly complex process that is regulated by a multitude of factors. Here, we show that the conserved kinase Ctk1 functions in translation by enhancing decoding fidelity. Ctk1 associates with translating ribosomes in vivo and is needed for efficient translation. Ctk1 phosphorylates Rps2, a protein of the small ribosomal subunit, on Ser 238. Importantly, Ctk1-depleted as well as rps2-S238A mutant cells show a defect in translation elongation through an increase in the frequency of miscoding. The role of Ctk1 in translation may be conserved as the mammalian homolog of Ctk1, CDK9, also associates with polysomes. Since Ctk1 interacts with the TREX (transcription and mRNA export) complex, which couples transcription to mRNA export, Ctk1/CDK9 might bind to correctly processed mRNPs during transcription and accompany the mRNP to the ribosomes in the cytoplasm, where Ctk1 enhances efficient and accurate translation of the mRNA.
Keywords: Ctk1, translation, phosphorylation, ribosome, translational fidelity, Rps2
Gene expression encompasses a multitude of different steps including transcription; 5′ capping, splicing, cleavage, and polyadenylation of the mRNA; packaging of the mRNA into a mature mRNP; nuclear export of the mRNP; and finally translation of the message into the encoded protein by the ribosome. It has been shown in recent years that the single steps of gene expression are physically and functionally linked (Maniatis and Reed 2002; Proudfoot et al. 2002).
The central coordinator of mRNA transcription and processing is the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (RNA Pol II). The CTD is highly conserved and consists of repeats of the consensus heptapeptide YSPTSPS. The CTD is differentially phosphorylated at Ser 2 (S2) and Ser 5 (S5) during transcription, which affects recruitment of mRNA processing factors.
Phosphorylation of S2 by the cyclin-dependent kinase (Cdk) Ctk1 is needed for regulation of transcription elongation (Cho et al. 2001). Ctk1 forms the active kinase complex CTDK-I together with Ctk2, its cyclin partner, and Ctk3, a protein of unknown function without homology with other proteins (Sterner et al. 1995). The potential human homolog of Ctk1 is CDK9, the kinase subunit of positive transcription elongation factor b (P-TEFb) (Zhu et al. 1997; Price 2000). Furthermore, it was shown that yeast CTDK-I promotes efficient transcription in HeLa nuclear cell extracts (Lee and Greenleaf 1997), providing a strong indication that the function of Ctk1 is conserved.
Recently, it has been shown that Ctk1 is also involved in RNA Pol I transcription (Bouchoux et al. 2004). Ctk1 interacts with RNA Pol I in vivo, and loss of Ctk1 causes a defect in RNA Pol I transcription. In addition, Ctk1 interacts genetically as well as biochemically with the TREX complex (Hurt et al. 2004). TREX couples transcription elongation to mRNA export (Jimeno et al. 2002; Strasser et al. 2002; Zenklusen et al. 2002). The TREX complex consists of the heterotetrameric THO complex, which plays a role in transcription elongation, the mRNA export factors Sub2 and Yra1, the SR-like proteins Gbp2 and Hrb1, and Tex1 (Strasser et al. 2002). According to our current model, the THO complex recruits Sub2, Yra1, Gbp2, Hrb1, and most likely also Tex1 to the nascent mRNA. These proteins in conjunction with other proteins such as Npl3 package the mRNA into a mature mRNP (Jensen et al. 2003). Yra1, in turn, interacts directly with the mRNA exporter Mex67-Mtr2, which can then facilitate the export of the mRNP through the nuclear pore complex. Importantly, most proteins described above for Saccharomyces cerevisiae are conserved in evolution, and an equivalent function has been shown in higher eukaryotes (Reed and Hurt 2002; Stutz and Izaurralde 2003).
Here, we show that Ctk1 enhances the decoding fidelity in translation. Sucrose density gradients reveal that Ctk1 associates with translating polysomes in vivo. Importantly, loss of Ctk1 greatly reduces translational activity as shown by in vitro translation assays. Ctk1 functions in translation fidelity as depletion of Ctk1 causes a higher sensitivity to translation inhibitors and an increase in the frequency of miscoding events. Interestingly, Ctk1 phosphorylates Rps2, a protein of the small ribosomal subunit known to be involved in translational accuracy, on S238, and rps2-S238A mutant cells show increased miscoding. The function of Ctk1 in translation seems to be conserved in evolution as the human homolog of Ctk1, CDK9, also associates with polysomes. As Ctk1 functions in transcription and associates with the TREX complex, this novel role of Ctk1 in translation might provide another connection between different steps in gene expression.
Results
Ctk1 associates with translating polysomes
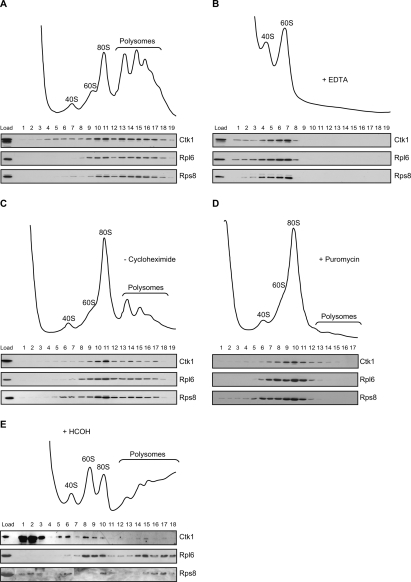
Kinases function in the regulation of a multitude of fundamental cellular processes. Interestingly, we found the kinase Ctk1, which has been shown to function in transcription (Cho et al. 2001; Bouchoux et al. 2004), to be associated with ribosomes. Whole-cell extracts were fractionated into soluble proteins, ribosomal subunits, monosomes, and polysomes on a sucrose density gradient, and each fraction of the gradient was assessed for the presence of Ctk1 by Western blotting. For comparison, the distribution of Rpl6, a ribosomal protein of the large subunit, and Rps8, a component of the small ribosomal subunit, was visualized. Ctk1 was present in fractions containing ribosomal subunits, monosomes, and polysomes (Fig. 1A).
Figure 1.
Ctk1 associates with polysomes. (A) Sedimentation behavior of Ctk1-TAP on sucrose density gradients. (Top panel) Ribosomal fractions (40S, 60S, 80S, and polysomes) were determined by OD254 nm measurement of the gradient fractions. (Bottom panel) The presence of Ctk1-TAP in each fraction was visualized by Western blotting using an anti-protein A antibody. (Bottom panel) For comparison, the sedimentation behavior of a ribosomal protein of the large ribosomal subunit (Rpl6) and of the small ribosomal subunit (Rps8) is shown. (B, top panel) Sedimentation behavior of Ctk1-TAP on sucrose density gradients containing EDTA, which disrupts mono- and polysomes. (Bottom panel) Ctk1 shifts to the fractions containing ribosomal subunits. (C) The distribution of Ctk1 shifts to the 80S fraction when cycloheximide is omitted. The sucrose density gradient is as in A, but lacking cycloheximide. Without cycloheximide the ribosomes run off the mRNA, and the amount of polysomes is specifically reduced. Compared with A, Ctk1, Rpl6, and Rps8 shift to the fractions of the 80S peak. (D) Ctk1 is lost from the high-density fractions in gradients with puromycin. The sucrose density gradient is as in C, but containing puromycin, which leads to a complete loss of polysomes. Ctk1 as well as Rpl6 and Rps8 are lost from the high-density fractions. (E) Ctk1 binds to ribosomes in vivo. Cells were cross-linked with formaldehyde prior to lysis of the cells in buffer containing 500 mM KCl and separation of the lysate on a 500 mM KCl sucrose density gradient. Ctk1, Rpl6, and Rps8 were visualized by Western blotting of each fraction.
The sedimentation of Ctk1 in the heavier fractions of the gradient is unlikely to represent Ctk1 in high-molecular-weight transcription complexes, because Rbp1, the largest subunit of RNA Pol II, and Paf1, a subunit of the Paf complex that is involved in transcription elongation, were present exclusively in the light fractions of a sucrose density gradient (data not shown). As Ctk1 has been implicated in RNA Pol I transcription (Bouchoux et al. 2004), Ctk1 could be associated with pre-40S, pre-60S, or 90S particles. In order to exclude this possibility, we assessed the sedimentation behavior of exemplary components of these preribosomal particles: Tsr1, Rix1, and Utp9. None of these three proteins shows a sedimentation behavior similar to that of Ctk1 (data not shown). Thus, it is unlikely that the sedimentation behavior of Ctk1 on sucrose density gradients reflects an association of Ctk1 with preribosomes or transcription complexes.
To further corroborate that the sedimentation behavior of Ctk1 reflects an association with ribosomes, monosomes and polysomes were disrupted into ribosomal subunits by addition of EDTA. Under these conditions, Ctk1 was lost from the heavy fractions, and a higher amount of Ctk1 appeared in the fractions of the ribosomal subunits (Fig. 1B), indicating that the sedimentation behavior of Ctk1 depends on the presence of polysomes. However, since we cannot exclude that the addition of EDTA disrupts a complex other than polysomes, but dependent on divalent cations, we sought to disrupt the polysomes in a more specific way. First, cycloheximide was omitted from the extracts, which leads to run-off of the ribosomes from the message. The profile shows a reduced level of polysomes and a corresponding increase of the 80S peak (Fig. 1C). Concomitantly, the distribution of Ctk1—as well as that of the ribosomal proteins Rpl6 and Rps8—shifted from the heavier polysome fractions to the 80S fraction (Fig. 1C). In order to completely disrupt the polysomes, we added puromycin, an inhibitor that causes release of the nascent peptide and the mRNA from ribosomes and leads to a complete loss of polysomes from the gradient (Fig. 1D). Ctk1, Rpl6, and Rps8 shifted from the heavy fractions to the fractions corresponding to the 80S peak (Fig. 1D). All of the above sucrose density gradients show that Ctk1 is specifically associated with polysomes.
Ctk1 associates with polysomes in vivo
Since Ctk1 is predominantly nuclear at steady state (Lee and Greenleaf 1991), we wanted to show that Ctk1 associates with ribosomes in vivo. Thus, cells were cross-linked with formaldehyde prior to lysis. The extract was then prepared in buffer containing high salt and separated on a high salt sucrose density gradient to dissociate any proteins including Ctk1 from the ribosomes that had not been cross-linked. Under these conditions, a small amount of Ctk1 cosedimented with ribosomes, as is clearly visible by comparison with the sedimentation behavior of Rpl6 (Fig. 1E, lanes 8–10,14,15,17) and of Rps8 (Fig. 1E, especially lane 6). That only a small amount of Ctk1 remains associated with ribosomes in this experiment is expected for two reasons. First, the efficiency of the cross-link will not be 100%, and hence part of Ctk1 bound to a ribosome in vivo will be lost. Second, Ctk1 localizes mainly to the nucleus (Lee and Greenleaf 1991). Thus, it is expected that only a small fraction of Ctk1 associates with ribosomes at any given time. However, this experiment shows that Ctk1 is associated with translating ribosomes in vivo, consistent with a function of Ctk1 in translation.
Loss of Ctk1 function causes a decrease in translational activity
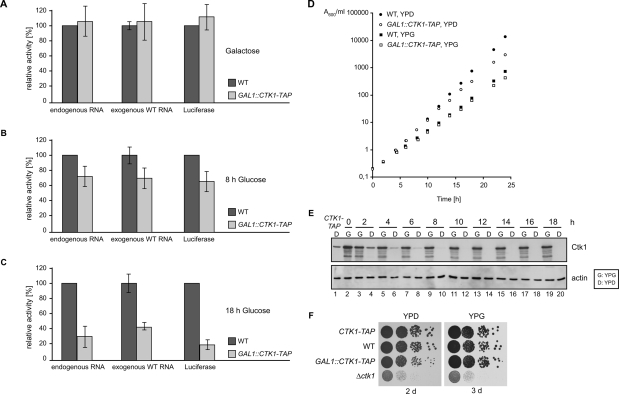
The association of Ctk1 with translating polysomes suggests a novel role of this kinase in translation. In order to test this hypothesis, in vitro translation assays were performed. Complete deletion of CTK1 causes a severe growth phenotype (Fig. 2F, Δctk1) that might lead to secondary effects. Hence, we constructed a strain carrying a C-terminal TAP-tagged version of Ctk1 driven by the GAL1 promoter in place of the endogenous CTK1 gene (GAL1∷CTK1-TAP) to be able to deplete Ctk1 by growth in glucose-containing medium. Expression of Ctk1 from the GAL1 promoter leads to slight overexpression of Ctk1 (Fig. 2E, cf. first two lanes) and a minor reduction in growth (Fig. 2D, cf. growth of wild type and GAL1∷CTK1-TAP in YPG). As control, translation-active extracts were prepared from wild-type and GAL1∷CTK1-TAP cells grown in galactose (Ctk1 expressed), and equal amounts of extract were tested for their activity to translate endogenous mRNAs. Both extracts have similar translational activities (Fig. 2A, left panel). In contrast, after 8 h of Ctk1 depletion—by growth of the GAL1∷CTK1-TAP strain in glucose-containing medium—the translational activity of these extracts drops to ∼70% compared with extracts from an isogenic wild-type strain (Fig. 2B, left panel). Importantly, the growth of the GAL1∷CTK1-TAP strain is identical, for the first 8 h after a shift from galactose- to glucose-containing medium, to the growth of the wild-type strain. Only later than 8 h after the shift does the GAL1∷CTK1-TAP strain start to grow more slowly than a wild-type strain (Fig. 2D). Consistently, Ctk1 is detectable by Western blotting during the first 8 h of depletion (Fig. 2E, lanes 4,6,8,10). Thus, a defect in translational activity is evident at a time point when the cells still grow as well as a wild-type strain, indicating that the observed translational defect is not a secondary effect. After 18 h of depletion of Ctk1, the translational activity of the Ctk1-depleted cells (GAL1∷CTK1-TAP) drops to ∼30% of the activity of wild-type cells (Fig. 2C, left panel). At this time point, the Ctk1-depleted cells (GAL1∷CTK1-TAP, YPD) grow a bit more slowly than the wild-type strain (Fig. 2D, WT, YPD). However, this growth defect is not severe, as Ctk1-depleted cells grow almost like wild-type cells for 2 d on plates (Fig. 2F, left panel). Taken together, depletion of Ctk1 causes a defect in translation, which is most likely direct as it occurs prior to an impairment of growth.
Figure 2.
Ctk1 is necessary for efficient translation. (A) Translation-active extracts were prepared from wild-type (WT) and GAL1∷CTK1-TAP cells grown in galactose-containing medium (Ctk1 expressed), and equal amounts of each extract were tested in an in vitro translation assay with endogenous mRNA (left panel), exogenous total RNA (middle panel), and in vitro transcribed mRNA coding for luciferase (right panel). Both extracts have the same translational activity. (B) Depletion of Ctk1 for 8 h leads to a translation defect of ∼30%. Wild-type and GAL1∷CTK1-TAP cells were shifted to glucose-containing medium for 8 h before translation-active extracts were prepared. Assays were performed as in A. (C) After 18 h of depletion of Ctk1, translation activity drops to ∼30%. Experiments were performed as in A after an 18-h shift to glucose-containing medium. For each panel in A–C, the activity of the wild-type extracts was set to 100%. Values and error bars represent the results of two independent experiments of three independent extracts each. (D) Growth curves of wild-type (WT, filled circle/square) and GAL1∷CTK1-TAP (open circle/square) cultures in galactose-containing medium (YPG, squares) and after shift to glucose-containing medium (YPD, circles) by measuring their optical density at 600 nm. (E) Depletion of Ctk1 after shift to glucose-containing medium. The same amount of whole-cell extracts from GAL1∷CTK1-TAP cells grown in galactose-containing medium (G: YPG) or shifted to glucose-containing medium (D: YPD) for the indicated time points was analyzed for the amount of Ctk1-TAP by SDS-PAGE and Western blotting with PAP antibody. As loading control, the amount of actin was assessed. (Lane 1) For comparison with the physiological Ctk1 expression, an extract from a CTK1-TAP strain, which expresses Ctk1 under its endogenous promoter, is shown. (F) Growth of CTK1-TAP, wild-type (WT), GAL1∷CTK1-TAP, and Δctk1 cells on plates. Tenfold dilutions of cells were spotted on glucose-containing medium (YPD) or galactose-containing medium (YPG) and grown at 30°C for 2 and 3 d, respectively.
Since Ctk1 is involved in RNA Pol I and II transcription (Cho et al. 2001; Bouchoux et al. 2004), a possible explanation for the low translation activity of Ctk1-depleted extracts could be that they contain fewer ribosomes and/or mRNAs. We thus assessed the level of ribosomal proteins and the translatability of endogenous mRNAs present in equal amounts of wild-type and Ctk1-depleted extracts. First, the levels of two ribosomal proteins representative of the small and the large ribosomal subunit, Rps8 and Rpl6, respectively, are equal as assessed by Western blotting (data not shown). Second, the mRNA extracted from Ctk1-depleted extracts was translated as well as the mRNA of wild-type extracts when used as template in in vitro translation assays with wild-type extracts that had been treated with nuclease to destroy any endogenous mRNAs (data not shown). Taken together, the levels of ribosomal proteins and the translatability of mRNAs of wild-type and Ctk1-depleted extracts used in the in vitro translation assay are the same and thus most likely do not account for the loss of translational activity in extracts lacking Ctk1.
In addition to endogenous mRNA, we also tested the translation activity of extracts from Ctk1-depleted cells with exogenously added mRNA and with in vitro transcribed luciferase mRNA as a substrate. For exogenous mRNA, the total RNA was extracted from a wild-type strain and added to in vitro translation assays with wild-type and Ctk1-depleted extracts that had been treated with nuclease to destroy the endogenous mRNA. With exogenous wild-type mRNA as template, extracts from cells depleted for Ctk1 for 8 and 18 h also show a significantly reduced translational activity as compared with wild-type extracts (Fig. 2A–C, middle panel). When in vitro transcribed luciferase mRNA was used as template in the in vitro translation reaction and the luciferase activity was measured, Ctk1-depleted extracts had a significantly reduced translation activity, which was similar to the effects observed with the other two templates (Fig. 2A–C, right panel). Taken together, loss of Ctk1 function leads to a decrease in translational activity with three different mRNAs as template. Thus, we conclude that Ctk1 exhibits a positive and most likely direct function in translation (also see below), establishing a completely new function for Ctk1.
Ctk1 functions in translation elongation and decoding fidelity
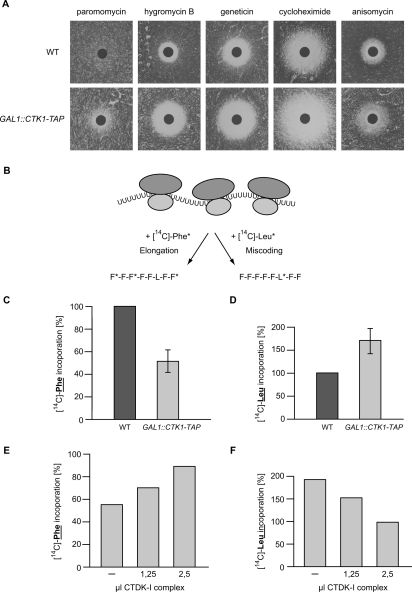
As a first attempt to specify the function of Ctk1 in translation, we tested whether Ctk1-depleted cells are sensitive to different translation inhibitors. Depletion of Ctk1 renders cells significantly more sensitive to paromomycin, hygromycin B, geneticin, cycloheximide, and anisomycin than an isogenic wild-type strain (Fig. 3A). This hypersensitivity of Ctk1-depleted cells to inhibitors of translation elongation is consistent with a positive function of Ctk1 in translation. Ctk1-depleted cells display the greatest sensitivity to paromomycin, hygromycin, and geneticin—aminoglycoside antibiotics that bind to the decoding region of the ribosomal A site. These drugs decrease the translational accuracy by stimulating the stable association of near-cognate aminoacyl tRNA to the ribosomal A site (Palmer et al. 1979; Singh et al. 1979; Moazed and Noller 1987). Hypersensitivity of the mutant strain toward these antibiotics, especially paromomycin, has often been linked to loss of translational accuracy, suggesting that Ctk1 might be necessary for correct decoding of the mRNA.
Figure 3.
Ctk1 plays a role in decoding fidelity. (A) Ctk1-depleted cells are sensitive to translation inhibitors. GAL1∷CTK1-TAP and wild-type (WT) cells were grown for 18 h in YPD and then plated on YPD plates, on which a filter containing paromomycin, hygromycin B, geneticin, cycloheximide, or anisomycin was placed. The size of the halo indicates the sensitivity of the strain toward the drug. (B) Schematic showing the poly(U) assay. The rate of translation elongation was determined by measuring the incorporation of radioactively labeled phenylalanine (F*; left arrow), and the rate of miscoding was measured by the incorporation of radioactively labeled leucine (L*; right arrow) when poly(U) RNA was used as a template in the translation assay. (C) Depletion of Ctk1 leads to a decrease in the efficiency of translation elongation. The incorporation of [14C]-labeled phenylalanine into the polypeptide chain of nuclease-treated extracts prepared from wild-type (WT) and GAL1∷CTK1-TAP cells after 18 h of depletion in glucose-containing medium was measured. The activity of wild-type cells was set to 100%. (D) Loss of Ctk1 function leads to an increase in the frequency of miscoding. Cells depleted for Ctk1 by growth in glucose-containing medium (YPD) for 18 h show an increase in the frequency of miscoding as measured by the incorporation of [14C]-labeled leucine. (E,F) The defect of Ctk1-depleted cells in translation elongation (E) and translational accuracy (F) can be rescued by reconstitution with purified CTDK-I complex in a dose-dependent manner. Purified CTDK-I complex was added to extracts depleted for Ctk1 for 18 h, and the efficiency of translation elongation and the miscoding frequency was measured as in C and D, respectively. As negative control, an eluate from a mock purification of a nontagged wild-type strain was added to the translation assays, and the activities of the CTDK-I-treated extracts were calculated relative to the mock-treated ones. One representative experiment of three independent experiments is shown.
In order to test specifically if translation elongation is impaired in cells lacking Ctk1, we compared the translation rate of poly(U) into polyphenylalanine; that is, under conditions in which peptide formation is independent of translation initiation factors (see Fig. 3B, left arrow; Wyers et al. 2000). Ctk1-depleted cells exhibit ∼50% of correct Phe* incorporation as compared with wild-type cells (Fig. 3C), indicating that loss of Ctk1 function leads to a defect in translation elongation. This decrease in translation elongation efficiency could be due to a decrease in the accuracy of translation, as suggested by the increased sensitivity of Ctk1-depleted cells toward drugs affecting the maintenance of accuracy. Therefore, we tested the misreading capacity of wild-type and Ctk1-depleted cells. To do this, the incorporation of leucine instead of phenylalanine in the poly(U) assay was measured. Leucine is incorporated into the polyphenylalanine product due to pairing of the UUU codon with the near-cognate UAA or CAA anti-codons of Leu-tRNAs (Fig. 3B, right arrow). Ctk1-depleted extracts show a 1.5-fold increase in leucine incorporation compared with extracts from wild-type cells (Fig. 3D). Thus, depletion of Ctk1 leads to an increase in the rate of miscoding events. Since loss of Ctk1 function leads to an increase in sensitivity toward translational inhibitors, a reduced incorporation of the correct amino acid during elongation, and an increase in the frequency of miscoding, we conclude that Ctk1 is necessary for a high decoding fidelity in translation elongation.
In order to show that the observed effect of Ctk1 depletion on translational accuracy is a direct one, reconstitution experiments were performed to assess whether the elongation and decoding defects of extracts depleted for Ctk1 can be rescued by addition of purified CTDK-I complex. The CTDK-I complex containing Ctk1, Ctk2, and Ctk3 was purified from yeast via TAP-tagged Ctk1 with buffer containing 1 M salt to exclude copurifying proteins other than Ctk2 and Ctk3. Addition of increasing amounts of CTDK-I complex to the Ctk1-depleted extracts restored the translation elongation defect (Fig. 3E) as well as the defect in correct decoding of the message (Fig. 3F) in a dose-dependent manner. That both translation defects observed in Ctk1-depleted extracts can be rescued by addition of purified CTDK-I complex indicates that the observed effects are specific. Thus, Ctk1 contributes directly to a higher rate of decoding fidelity during translation elongation.
Ctk1 is required for correct decoding in vivo
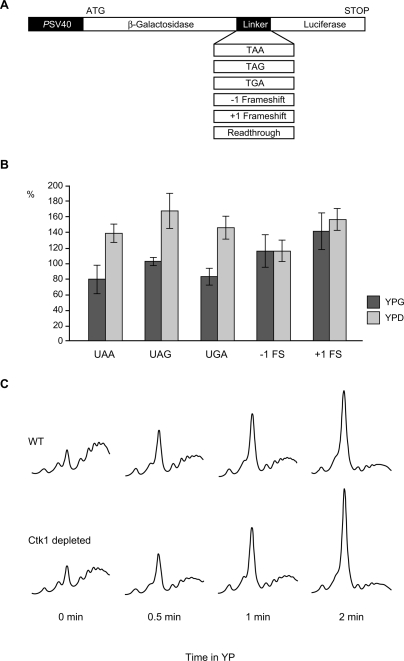
In order to test whether Ckt1 is also required for accurate translation in vivo, we used an assay system developed by Stahl et al. (1995). This system consists of a set of dual reporter plasmids. Each plasmid codes for lacZ, a linker, and luciferase (Fig. 4A). The linker contains a readthrough sequence, one of the three stop codons, or a +1 or −1 frameshift. In the readthrough construct the luciferase sequence is translated as well as the lacZ sequence. In contrast, in each of the constructs with a modification (stop codon or frameshift) in the linker, the luciferase gene is only expressed if miscoding or a frameshift event occurs. For each strain to be tested, the luciferase activity is divided by the lacZ activity, which normalizes for the amount of mRNA available for translation, and thus eliminates any effects that might be due to transcription, export, or degradation of the message. In addition, this ratio is normalized by the luciferase/lacZ ratio of the construct with the readthrough linker, which excludes any effects that only affect the 3′ end of the message; for example, a decrease in transcription elongation efficiency. Thus, with this reporter set, only the frequency of miscoding or frameshifts in the linker region can be assessed. β-Galactosidase and luciferase activity of GAL1∷CTK1-TAP and wild-type cells grown in galactose (YPG, Ctk1 expressed) or shifted to glucose containing medium for 18 h (YPD, Ctk1 depleted) were measured for each of the reporter plasmids. Depletion of Ctk1 (Fig. 4B, YPD) leads to an approximately twofold increase in miscoding with each of the three stop codon constructs as compared with the same strain grown in galactose; that is, expressing Ctk1 (Fig. 4B, YPG, columns UAA, UAG, and UGA). In contrast, the frequency of a +1 or −1 frameshift is not changed when Ctk1 is depleted (Fig. 4B, columns −1 FS and +1 FS).
Figure 4.
Ctk1 plays a role in decoding fidelity in vivo. (A) Schematic showing the dual reporter system used for determination of miscoding frequency in B. (B) Loss of Ctk1 function leads to an increase in the frequency of miscoding. Cells depleted for Ctk1 by growth in glucose-containing medium (YPD) show an increase in the frequency of miscoding with all three stop codons (UAA, UAG, and UGA) compared with cells expressing Ctk1 (YPG), whereas an increased rate of frameshift events does not occur (−1 FS and +1 FS). (C) Depletion of Ctk1 leads to a slightly faster polysome run-off after inhibition of translation initiation. Wild-type (WT) and GAL1∷CTK1-TAP cells grown in YPD were shifted to glucose-lacking medium (YP) to inhibit translation initiation for the indicated time points and polysome profiles analyzed.
Furthermore, the lower translational accuracy in Ctk1-depleted cells might affect the translation elongation rate, which was assessed by polysome run-off experiments. After inhibition of general translation initiation by glucose starvation, polysome profiles were analyzed. Compared with wild-type cells, Ctk1-depleted cells show a slightly faster run-off of polysomes and a concomitant faster increase of the 80S peak, indicating that elongation is faster in Ctk1-depleted cells (Fig. 4C). This is consistent with earlier observations that hyperaccurate ribosomes have a decreased elongation rate, whereas a decrease in accuracy leads to faster elongation or has no effect on the elongation rate (Zimmermann et al. 1971; Andersson et al. 1982; Bjorkman et al. 1999; Carr-Schmid et al. 1999). Taken together, Ctk1 is needed for correct decoding in vivo.
Phosphorylation of Rps2 by Ctk1 is needed for translational accuracy
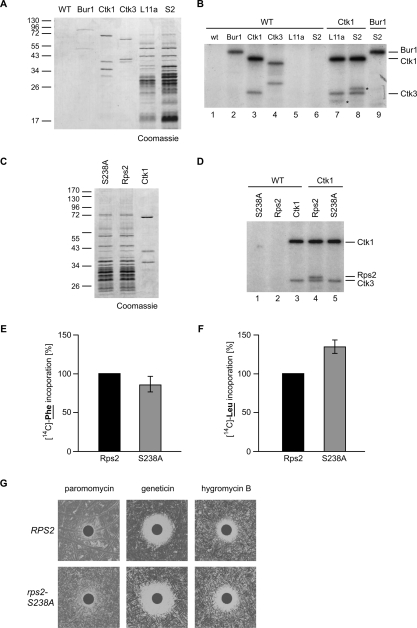
Since Ctk1 is a kinase, we set out to identify the substrate phosphorylated by Ctk1 in translation, which could be a translation factor or a ribosomal protein. Among other candidates we purified ribosomes using strains expressing TAP-tagged versions of a protein of the large ribosomal subunit (Rpl11a-TAP) or of the small ribosomal subunit (Rps2-TAP) and used them as substrates in in vitro kinase assays. The Ctk1–Ctk2–Ctk3 complex was purified using a CTK1-TAP or CTK3-TAP strain and buffer containing 1 M NaCl to avoid copurification of ribosomal proteins and potential contaminants. As negative control, the Bur1–Bur2 complex, which has a role similar to Ctk1-2-3 in transcription elongation (Keogh et al. 2003 and references therein), was purified. Figure 5A shows a Coomassie stain of the purified complexes. When Ctk1 was incubated with a mock eluate in the in vitro kinase assay, it (auto)phosphorylated itself and Ctk3 (Fig. 5B, lanes 3,4; Lee and Greenleaf 1991). Interestingly, an additional band appeared on the autoradiograph when ribosomes purified by TAP-tagged Rpl11a were added as a substrate (Fig. 5B, lane 7, the substrate of Ctk1 is marked with an asterisk). This band shifted up when ribosomes purified via Rps2-TAP were used as a substrate due to the remaining CBP (Calmodulin-binding protein) on Rps2 instead of Rpl11a (Fig. 5B, lane 8, marked with an asterisk), showing that Rps2 is phosphorylated by Ctk1. In contrast, Bur1–Bur2 phosphorylated itself (Fig. 5B, lane 2) but did not phosphorylate a component of the ribosome (Fig. 5B, lanes 2,9). Thus, a substrate of Ctk1 is Rps2, a protein of the small ribosomal subunit. Interestingly, Rps2 is known to be essential for translational accuracy (Eustice et al. 1986; Synetos et al. 1996; Stansfield et al. 1998).
Figure 5.
Phosphorylation of Rps2 on Ser 238 by Ctk1 is needed for translational accuracy. (A) Complexes purified for the in vitro kinase assay shown in B. The CTDK-I complex was purified by a TAP tag on Ctk1 or Ctk3, the Bur1–Bur2 complex via TAP-tagged Bur1, and ribosomes were purified via TAP-tagged Rpl11a (L11a) or Rps2 (S2). A nontagged wild-type (WT) strain served as negative control. Proteins were separated by SDS-PAGE and stained with Coomassie. The left lane shows a molecular weight marker. (B) Ctk1 phosphorylates Rps2 in vitro. The protein complexes indicated by the component that was TAP-tagged for the purification were used in in vitro kinase assays. After incubation with radioactively labeled ATP, proteins were separated by SDS-PAGE, and phosphorylated proteins were visualized by autoradiography. An eluate of a nontagged wild-type strain served as negative control. Incubation of the control eluate alone (WT; lane 1) or of Rpl11a (L11) or Rps2 (S2) containing ribosomes with control eluate (lanes 5,6) did not give a radioactive signal. Incubation of Bur1, Ctk1, and Ctk3 with the control eluate (WT; lanes 2–4) gave signals for (auto)phosphorylated Bur1, Ctk1, and Ctk3. (Lane 7) Incubation of Ckt1 with Rpl11a-purified ribosomes yields an additional phosphorylated protein (indicated by a star). (Lane 8) This band shifts up when CBP-tagged Rps2-containing ribosomes (S2) are incubated with Ctk1, identifying Rps2 as the phosphorylated product (Rps2-TAP indicated by a star). (Lane 9) Bur1, used as a control kinase, does not phosphorylate Rps2. (C) Coomassie staining of purified ribosomes containing Rps2 or Rps2–S238A and CTDK-I complex used for the in vitro kinase assays shown in D. (D) Ctk1 phosphorylates Rps2 on S238. (Lane 4) Ctk1 phosphorylates itself, Ctk3, and Rps2. (Lane 5) In contrast, when ribosomes containing Rps2–S238A–CBP are used as substrate, no phosphorylation of Rps2 can be observed. (E,F) rps2-S238A extracts show a minor decrease in translation elongation and an increase in miscoding events. Experiments were performed as described in Figure 3B. (G) rps2-S238A cells are sensitive to drugs that impair translation elongation.
In order to identify the residue of Rps2 phosphorylated by Ctk1, we mutated all minimal CDK consensus motifs in Rps2 from S-P to A-P. Purification of TAP-tagged Rps2 containing a Ser 238 to alanine (S238A) mutation yields ribosomes comparable to purification of Rps2-TAP (Fig. 5C). However, Rps2-S238A is not phosphorylated by Ctk1-2-3 in the in vitro kinase assay (Fig. 5D, cf. lanes 4,5), showing that Ctk1 phosphorylates Rps2 on S238. In order to assess whether this S238A mutation leads to the expected increase in miscoding, the translation activity of extracts prepared from rps2-S238A cells and from wild-type cells was measured in the poly(U) assay (Fig. 3B). rps2-S238A extracts show a minor decrease in translation elongation efficiency (Fig. 5E) and an increase in the rate of miscoding (Fig. 5F). Consistent with these in vitro results, the rps2-S238A mutant is more sensitive to drugs that impair translational fidelity than an isogenic wild-type strain (Fig. 5G). These results are consistent with the model that phosphorylation of S238 on Rps2 by Ctk1 enhances translational accuracy.
The function of Ctk1 in translation may be conserved
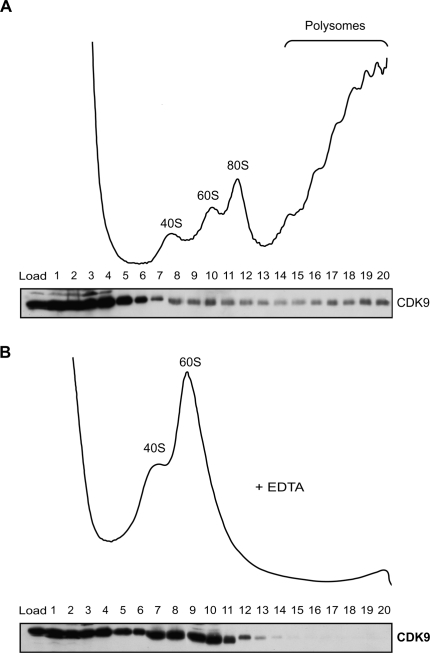
We were curious to know whether the role of Ctk1 in translation is conserved in higher eukaryotes. CDK9, the potential metazoan homolog of Ctk1, constitutes together with one of the cyclins T1, T2, or K the positive elongation factor P-TEFb. Like Ctk1, P-TEFb is needed for efficient transcription elongation and phosphorylates S2 of the CTD (Price 2000). To gain a first insight into the potential conservation of Ctk1 function, extracts of the human embryonic kidney cell line 293T were loaded onto sucrose density gradients to separate soluble proteins from (poly)ribosomes. The presence of CDK9 in each fraction was assessed by Western blotting using an antibody directed against CDK9. CDK9 was found mainly in the soluble pool, but a subpopulation of CDK9 clearly comigrated with ribosomal subunits, monosomes, and polysomes (Fig. 6A). This reflects an association of CDK9 with polysomes since CDK9 shifted to the fractions containing ribosomal subunits, when monosomes and polysomes were disrupted by EDTA (Fig. 6B). The association of CDK9 with translating ribosomes is an indication that the novel function of Ctk1 in translation described here is conserved in evolution.
Figure 6.
The function of Ctk1 in translation might be conserved. (A) CDK9, the human homolog of Ctk1, associates with polysomes. Extracts of 293T cells were separated by sucrose density centrifugation. (Top panel) The ribosomal fractions were visualized by OD254 nm measurement of the gradient fractions. (Bottom panel) The presence of CDK9 in each fraction was analyzed by Western blotting with an antibody recognizing CDK9. (B) The association of CDK9 with polysomes is specific. OD254 nm measurement (top panel) and CDK9 Western blot (bottom panel) of a sucrose gradient containing EDTA, which dissociates mono- and polysomes.
Discussion
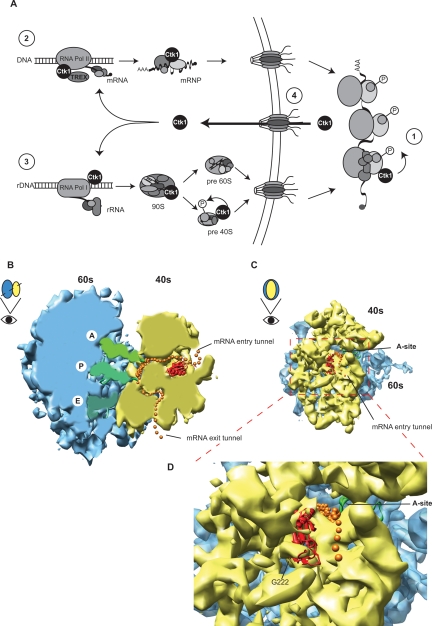
A plethora of cellular processes is regulated by kinases. A function of the conserved yeast CTDK-I complex in the regulation of transcription has been well established (Cho et al. 2001; Bouchoux et al. 2004). Here, we describe a novel function of the catalytic subunit of this complex, Ctk1, in translation. Ctk1 associates with translating ribosomes in vivo, and loss of Ctk1 function leads to a reduction of translational activity. Importantly, this translation defect is evident in Ctk1-depleted cells before their growth is impaired, indicating that this is a direct effect. A direct role of Ctk1 in translation is further corroborated by our reconstitution experiments, where we could show that addition of purified CTDK-I complex to extracts of Ctk1-depleted cells rescues their translation defects. More specifically, Ctk1 is needed to maintain translation accuracy. A molecular explanation of Ctk1’s action in translation fidelity is suggested by our identification of Rps2 as a substrate of Ctk1. Mutation of Rps2 is known to lead to an increase in the frequency of miscoding events (Eustice et al. 1986; Synetos et al. 1996; Stansfield et al. 1998). We show that Ctk1 phosphorylates Rps2 on Ser 238 (S238) in vitro and that mutation of S238 to alanine causes a decrease in translational fidelity. Thus, phosphorylation of Rps2 on S238 is most likely the mechanism by which Ctk1 increases the fidelity during decoding of the message (Fig. 7A, 1).
Figure 7.
(A, 1) Model of Ctk1’s function in translation. Phosphorylation of Rps2 on S238 by Ctk1 is needed for a higher accuracy of mRNA decoding. (2) Since Ctk1 is implicated in transcription of RNA Pol II and interacts with the TREX complex, it could exit the nucleus bound to correctly processed mRNPs. (3) Alternatively, Ctk1 could travel to the cytoplasm with preribosomes, as Ctk1 has been implicated in RNA Pol I transcription. After phosphorylation of Rps2 (1), Ctk1 most likely rapidly reenters the nucleus (4). See Discussion for details. (B–D) Cryo-EM model of an elongating 80S ribosome with messenger RNA, A, P, and E site tRNA, and Rps2 modeled in according to Spahn et al. (2001). (B) Cartoon of a cross-section allowing a view of the elongation process. Rps2 is positioned at the beginning of the mRNA entry tunnel. (C) View of the 40S subunit from the solvent site. The transparency was reduced to visualize the path of the mRNA, the tRNAs, and Rps2. Yeast Rps2 is present with reduced electron density in the 80S EM structure and as crystal structure of E. coli S5. (D) Closeup of the localization of Rps2 on the small ribosomal subunit at the entry tunnel of the mRNA. The large ribosomal subunit is shown in blue, the small ribosomal subunit is in yellow, mRNA is in orange (single spheres show the likely path of the mRNA, which is not seen in the electron density), A, P, and E site tRNAs are in green, and Rps2 is in red.
In order to explain how phosphorylation of Rps2 by Ctk1 might influence translation fidelity we looked at the position of Rps2 within the ribosome. Figure 7,B–D, shows the structure of the S. cerevisiae 80S ribosome (Spahn et al. 2001) with the large ribosomal subunit shown in blue, the small ribosomal subunit shown in yellow, the path of the mRNA (as seen in the electron density as well as modeled) shown in orange, and the tRNAs in the A, P, and E sites shown in green. The position of Rps2 is based on the position of its E. coli homolog S5 in the crystal structure of the 70S ribosome (Spahn et al. 2001) and was modeled into the electron density of the yeast ribosome in red. Rps2 is positioned in the middle of the small ribosomal subunit right next to the mRNA entry tunnel (Fig. 7B [side view of the 80S ribosome], C [view of the 40S subunit from the solvent side]). Unfortunately, the last 30 amino acids of Rps2, including S238 (the residue of Rps2 phosphorylated by Ctk1), are not present in the crystal structure, indicating that the C terminus of Rps2 is flexible. However, the last amino acid present, Gly 222 (G222), points toward the solvent side (Fig. 7D, G222). Thus, even though it is not known how the C terminus of Rps2 is positioned within the structure, S238 is probably accessible for phosphorylation by Ctk1.
Interesting in this context, the E. coli homolog of Rps2, S5, is one of three proteins that constitute the so-called decoding accuracy center. It has been shown in E. coli that mutations in these proteins—namely, S4 and S5—decrease the accuracy of decoding and were therefore called ram (ribosomal ambiguity) mutations (Rosset and Gorini 1969; Cabezon et al. 1976). These two proteins, together with the third one, S12, are localized in a cluster on the opposite side of the site of codon–anti-codon interaction (Capel et al. 1987). They interact with a highly conserved 530-base-pair (bp) stem–loop region of the 16S rRNA (Powers and Noller 1991), and mutation of these proteins may therefore perturb the 16S rRNA structure, leading to an increased frequency of miscoding (Allen and Noller 1989; Ogle and Ramakrishnan 2005). Importantly, mutation of yeast RPS2 was shown to cause a decrease in translation fidelity (Eustice et al. 1986; Synetos et al. 1996; Stansfield et al. 1998). These findings are consistent with an increased sensitivity of RPS2 mutants (Fig. 5G; Eustice et al. 1986; Synetos et al. 1996; Stansfield et al. 1998) as well as Ctk1-depleted cells (Fig. 3A) to paromomycin. Paromomycin binds to the 16S rRNA and induces a conformational change (Fourmy et al. 1998; Jerinic and Joseph 2000) that stabilizes the binding of the aminoacyl-tRNA in the A site not only when the anti-codon is cognate, but also when it is near cognate (Rodnina and Wintermeyer 2001). Thus, phosphorylation of Rps2 on S238 by Ctk1 could induce a structural change in Rps2 and thereby contribute to an accurate decoding process. Without Ctk1, Rps2 remains nonphosphorylated, and thus the ribosome might be more prone to misreading.
Ctk1 could either act constitutively or regulate translation, for example, in response to cellular stress. Translation is the final step of gene expression and a prime target for regulation as it allows a rapid response (Gebauer and Hentze 2004; Holcik and Sonenberg 2005). On the other hand, translational control can be global or mRNA specific. Since deletion of Ctk1 leads to a major decrease in translational activity in the in vitro translation assays with endogenous, exogenous, and luciferase mRNA, Ctk1 most likely regulates translation at a global level. This notion is also supported by our finding that Rps2, a protein of the small ribosomal subunit, is a substrate of Ctk1. Even though we cannot exclude that Ctk1 function is regulated, we favor a model according to which Ctk1 enhances the translation of correctly processed, assembled, and exported mRNPs (also see below) in a constitutive manner.
Ctk1 localizes mainly to the nucleus (Lee and Greenleaf 1991), whereas ribosomes preside in the cytoplasm. Thus, the main pools of Ctk1 and ribosomes are physically separated. However, these steady-state distributions do not permit conclusions about the in vivo dynamics. The observed interaction between Ctk1 and ribosomes most likely takes place in the cytoplasm as indicated by Ctk1’s association with translating ribosomes (Fig. 1). Moreover, that only a small amount of Ctk1 associates with cytoplasmic ribosomes at any given time is consistent with our cross-linking experiment (Fig. 1E) and an enzymatic function of Ctk1. Furthermore, CDK9, the human homolog of Ctk1, has been shown to shuttle between the nucleus and the cytoplasm (Napolitano et al. 2002). Thus, Ctk1 probably travels to the cytoplasm, enhances translation by phosphorylating Rps2 early in translation (Fig. 7A, 1), and rapidly returns to the nucleus (Fig. 7A, 4).
Two possible routes could be taken by Ctk1 to travel from the nucleus to the cytoplasm. Since Ctk1 is necessary for efficient transcription by RNA Pol I, it could remain associated with preribosomal particles as they mature into 60S and 40S subunits and be exported to the cytoplasm bound to them (Fig. 7A, 3). However, we do not favor this possibility as the majority of Ctk1 does not cosediment with preribosomal particles (data not shown), and we could not detect any Ctk1 copurifying with preribosomal particles (S. Röther, unpubl.). A second possible route for Ctk1 to reach the cytoplasm is via the exported mRNP (Fig. 7A, 2). This possibility seems likely, as Ctk1 regulates RNA Pol II transcription (Cho et al. 2001) and interacts genetically as well as biochemically with TREX, a complex that couples transcription to early mRNA export (Strasser et al. 2002; Hurt et al. 2004). Also, as Rps2 is located in close proximity of the mRNA entry channel (see above), it is in a prime position to be phosphorylated by Ctk1 arriving bound to an mRNP. The question of whether Ctk1 remains associated with mRNPs at these later stages of the mRNA export pathway or with preribosomes will be the subject of future investigations.
The function of Ctk1 in translation is most likely conserved in evolution as indicated by our finding that its human homolog, CDK9, also associates with polysomes. Recently, CDK955, a longer isoform of CDK9 with an N-terminal extension, has been described (Shore et al. 2003). Ctk1 also contains an extension N-terminal of its kinase domain. The N termini of CDK955 and Ctk1, however, do not share any striking sequence similarity. It will be interesting to assess the common and different functions of the CDK942 and CDK955 isoforms in transcription as well as translation. Rps2, on the other hand, is also highly conserved over all kingdoms. The residue of Rps2 phosphorylated by Ckt1, Ser 238, is conserved in higher eukaryotes (with the exception of, e.g., Paramecium tetraurelia, Dictyostelium discoideum, Anopheles gambiae, Drosophila melanogaster, and Schizosaccharomyces pombe, where serine is exchanged for a threonine), but not in eubacteria or archaea. Thus, the phosphorylation of Rps2 by Ctk1 described here might have evolved to fine-tune the intricate regulation of gene expression of compartmentalized cells and is most likely conserved among eukaryotes.
In the past couple of years, it has become increasingly clear that the different steps in gene expression are intimately linked (Maniatis and Reed 2002; Proudfoot et al. 2002). Whereas a lot of data documenting the interconnection between different processes such as transcription elongation and termination, processing of the mRNA, mRNA export, and cytoplasmic nonsensemediated decay (NMD) exist, examples connecting nuclear events with translation are still scarce. One example for such a connection is the finding that the human mRNA exporter Tap-p15, which localizes to the nucleus at steady state but shuttles between the nucleus and the cytoplasm, promotes the translation of unspliced CTE mRNA, a viral RNA it exports (Jin et al. 2003). In addition, shuttling SR proteins, which function primarily in splicing, were recently shown to play a role in mRNA translation (Sanford et al. 2004). In yeast, the SR-like TREX components Gbp2 and Hrb1 as well as Npl3, another SR-like protein, seem to accompany the mRNA to the ribosome, but need to be dissociated for efficient translation to take place (Windgassen et al. 2004). In contrast, our findings suggest a positive function of Ctk1 in translation and might thus provide a connection between translation and a process as distant as transcription. Ctk1 may function to mark correctly processed mRNAs, accompany them to the cytoplasm, and ensure their efficient translation (Fig. 7A, 2 and 1). Alternatively, Ctk1 might have a function in the cytoplasm that is independent of its nuclear function. In conclusion, we have shown that one further role of Ctk1 in addition to its function in transcription is to contribute to the fidelity of decoding the message. This finding might provide another thread in the network connecting different steps in gene expression.
Materials and methods
Sucrose density gradients
For sucrose density fractionation, yeast cells were grown in YPD to an optical density of 0.5. After addition of 0.1 mg/mL cycloheximide, cells were further incubated for 10 min at 30°C. Cells were resuspended in 1 mL of polysome buffer (20 mM HEPES-KOH at pH 7.6, 75 mM KCl, 2.5 mM MgCl2, 1 mM EGTA, 1 mM DTT, plus or minus 0.1 mg/mL cycloheximide or 15 mM EDTA). Cells were lysed by vortexing the cell suspension with glass beads for 5 min in the cold. When cycloheximide was omitted, cell lysates were incubated for 15 min at 22°C prior to removal of cell debris. Puromycin treatment was carried out by incubating the culture with 1 mM puromycin/2 mM GTP for 15 min prior to harvesting. Lysis was carried out in polysome buffer without cycloheximide plus puromycin/GTP. After removal of cell debris by centrifugation (5 min, 16,000g), extract corresponding to 250 μg of RNA was loaded onto a 10-mL 10%–50% or 15%–35% (EDTA) linear sucrose gradient in polysome buffer ± 15 mM EDTA and was centrifuged for 2 h at 257,000g. Fractions were obtained by connecting a peristaltic pump to the UV flow cell of the Äkta Basic System (Amersham Biosciences). Absorbance was measured at 254 nm. Fractions were TCA-precipitated for Western analysis.
HCOH cross-linking was performed as described (Nielsen et al. 2004), except that the salt concentration of the buffer was raised to 500 mM KCl. Analysis of polysome run-off after glucose starvation was done as described (Ashe et al. 2000). Extracts from wild-type and GAL1∷CTK1-TAP cells after 18 h of growth in YPD were prepared in polysome buffer after 0-, 0.5-, 1-, and 2-min shift to YP medium.
For sucrose density gradients with extracts of mammalian cells, 2 × 107 adherent 293T cells were treated with 50 μg/mL cycloheximide for 5 min at 37°C. Cells were harvested and washed twice in ice-cold PBS plus cycloheximide. Lysis was performed in 600 μL of mammalian polysome buffer (20 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 50 mM KCl, 1 mM DTT, 50 μg/mL cycloheximide) plus 0.3% NP-40 with or without 15 mM EDTA. After 5 min of incubation on ice, cell debris was removed by 5 min of centrifugation at 10,000g. The supernatant was loaded onto a 10-mL 10%–50% linear sucrose gradient in mammalian polysome buffer and centrifuged for 2 h at 257,000g. Fractionation and visualization were done as described above.
In vitro translation assay and preparation of RNA
In vitro translation extracts were performed according to Altmann and Trachsel (1997). Polysomal RNA was extracted from the polysomes generated during preparation of the extract by the technique of Gallis et al. (1975). Capped luciferase mRNA was in vitro transcribed from pSP6P (Verge et al. 2004) using the AmpliCap high-yield message maker kit (Epicentre).
Translation assays with endogenous mRNA as template were performed as described in Altmann and Trachsel (1997). For translation assays with exogenous total RNA, extracts were treated with 30 U/mL micrococcal nuclease for 15 min at 22°C before addition of 80 μg/mL RNA. For elongation/miscoding assays, extracts were treated with 30 U/mL micrococcal nuclease for 15 min at 22°C and incubated with either [14C]phenylalanine or [14C]leucine using 15 mg/mL poly(U) as template. In the miscoding experiments, the Mg concentration was 10 mM. To analyze the effect of addition of purified CTDK-I complex on the activity of the depleted extracts, the CTK1-TAP and a nontagged wild-type control strain were purified using TAP, and increasing amounts were incubated with translation extracts from wild-type and Ctk1-depleted cells (18 h) for 30 min at 22°C prior to the in vitro translation assays. Incorporated [35S]methionine, [14C]phenylalanine, or [14C]leucine was analyzed after a 30-min translation reaction at 22°C by spotting 3 μL of the reaction onto a filter paper and preparation for liquid scintillation counting as described. Counting was performed using a Tri Carb Liquid Scintillation Analyzer (Packard). Luciferase activity was measured 30 min after addition of 40 μg/mL Luc transcript to the in vitro translation reaction by analyzing 20 μL of the reaction in 100 μL of Luc assay buffer (25 mM glycylglycine at pH 7.8, 15 mM potassium phosphate at pH 7.8, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, 0.2 mM Luciferin) in a Lumat LB Luminometer. Translation activity was calculated from the average counts per minute of incorporated radioactivity or relative Luciferase units, respectively, of two independent experiments of three independent wild-type, Ctk1-depleted, or rps2-S238A extracts. Since the efficiency of nuclease treatment varied, the translation activity of experiments with exogenously added total or poly(U) RNA was determined as fold over translation activity after treatment with micrococcal nuclease without addition of RNA. To determine the percentage of miscoding events, the counts per minute of incorporated leucine was divided by the counts per minute of phenylalanine, and the ratio between the analyzed strains and the wild type was calculated. For all experiments, wild-type activity was set to 100%. The activity of the reconstitution experiment was calculated as the activity of the CTDK-I-treated extract over the activity of the mock-treated extract. Due to variation in the CTDK-I purifications, one representative experiment of three independent experiments is shown for the reconstitution experiment.
Readthrough frequency analysis
The analysis of in vivo readthrough frequencies was carried out as described in Kwapisz et al. (2005) using dual reporter plasmids (Stahl et al. 1995; Bidou et al. 2000; also see Fig. 4A). The frequency of miscoding, expressed as percentages compared with the wild type, was calculated by dividing the luciferase/β-galactosidase ratio for the GAL1∷CTK1-TAP strain after growth in YPG or YPD for 18 h for each reporter plasmid by the ratio for the readthrough control, followed by normalization by values for the wild-type strain. Results are the mean of three independent experiments.
Acknowledgments
We are grateful to G. Dieci for providing polyclonal rabbit antibodies against ribosomal proteins Rpl6 and Rps8; M. Altmann for plasmid pSP6P; and J.P. Rousset for plasmids pACTy, pAC1789, pACTMV, pACTGA, pACTAA, and pACTQ to test in vivo miscoding. We thank R.-P. Jansen, R. Beckmann, and H. Tschochner for suggestions on this work; P. Cramer, R.-P. Jansen, and D. Wilson for critical reading of the manuscript; and T. Becker for help with preparation of the ribosome structures. This work is supported by research grants of the Deutsche Forschungsgemeinschaft (STR 697/1-1 and SFB646), the EMBO Young Investigator Programme, and the “Fonds der Chemischen Industrie.”
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.428407
References
- Allen P.N., Noller H.F., Noller H.F. Mutations in ribosomal proteins S4 and S12 influence the higher order structure of 16 S ribosomal RNA. J. Mol. Biol. 1989;208:457–468. doi: 10.1016/0022-2836(89)90509-3. [DOI] [PubMed] [Google Scholar]
- Altmann M., Trachsel H., Trachsel H. Translation initiation factor-dependent extracts from yeast Saccharomyces cerevisiae. Methods. 1997;11:343–352. doi: 10.1006/meth.1996.0432. [DOI] [PubMed] [Google Scholar]
- Andersson D.I., Bohman K., Isaksson L.A., Kurland C.G., Bohman K., Isaksson L.A., Kurland C.G., Isaksson L.A., Kurland C.G., Kurland C.G. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol. Gen. Genet. 1982;187:467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- Ashe M.P., De Long S.K., Sachs A.B., De Long S.K., Sachs A.B., Sachs A.B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidou L., Stahl G., Hatin I., Namy O., Rousset J.P., Farabaugh P.J., Stahl G., Hatin I., Namy O., Rousset J.P., Farabaugh P.J., Hatin I., Namy O., Rousset J.P., Farabaugh P.J., Namy O., Rousset J.P., Farabaugh P.J., Rousset J.P., Farabaugh P.J., Farabaugh P.J. Nonsense-mediated decay mutants do not affect programmed −1 frameshifting. RNA. 2000;6:952–961. doi: 10.1017/s1355838200000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman J., Samuelsson P., Andersson D.I., Hughes D., Samuelsson P., Andersson D.I., Hughes D., Andersson D.I., Hughes D., Hughes D. Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol. Microbiol. 1999;31:53–58. doi: 10.1046/j.1365-2958.1999.01142.x. [DOI] [PubMed] [Google Scholar]
- Bouchoux C., Hautbergue G., Grenetier S., Carles C., Riva M., Goguel V., Hautbergue G., Grenetier S., Carles C., Riva M., Goguel V., Grenetier S., Carles C., Riva M., Goguel V., Carles C., Riva M., Goguel V., Riva M., Goguel V., Goguel V. CTD kinase I is involved in RNA polymerase I transcription. Nucleic Acids Res. 2004;32:5851–5860. doi: 10.1093/nar/gkh927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezon T., Herzog A., De Wilde M., Villarroel R., Bollen A., Herzog A., De Wilde M., Villarroel R., Bollen A., De Wilde M., Villarroel R., Bollen A., Villarroel R., Bollen A., Bollen A. Cooperative control of translational fidelity by ribosomal proteins in Escherichia coli. III. A ram mutation in the structural gene for protein S5 (rpx E) Mol. Gen. Genet. 1976;144:59–62. doi: 10.1007/BF00277305. [DOI] [PubMed] [Google Scholar]
- Capel M.S., Engelman D.M., Freeborn B.R., Kjeldgaard M., Langer J.A., Ramakrishnan V., Schindler D.G., Schneider D.K., Schoenborn B.P., Sillers I.Y., Engelman D.M., Freeborn B.R., Kjeldgaard M., Langer J.A., Ramakrishnan V., Schindler D.G., Schneider D.K., Schoenborn B.P., Sillers I.Y., Freeborn B.R., Kjeldgaard M., Langer J.A., Ramakrishnan V., Schindler D.G., Schneider D.K., Schoenborn B.P., Sillers I.Y., Kjeldgaard M., Langer J.A., Ramakrishnan V., Schindler D.G., Schneider D.K., Schoenborn B.P., Sillers I.Y., Langer J.A., Ramakrishnan V., Schindler D.G., Schneider D.K., Schoenborn B.P., Sillers I.Y., Ramakrishnan V., Schindler D.G., Schneider D.K., Schoenborn B.P., Sillers I.Y., Schindler D.G., Schneider D.K., Schoenborn B.P., Sillers I.Y., Schneider D.K., Schoenborn B.P., Sillers I.Y., Schoenborn B.P., Sillers I.Y., Sillers I.Y., et al. A complete mapping of the proteins in the small ribosomal subunit of Escherichia coli. Science. 1987;238:1403–1406. doi: 10.1126/science.3317832. [DOI] [PubMed] [Google Scholar]
- Carr-Schmid A., Valente L., Loik V.I., Williams T., Starita L.M., Kinzy T.G., Valente L., Loik V.I., Williams T., Starita L.M., Kinzy T.G., Loik V.I., Williams T., Starita L.M., Kinzy T.G., Williams T., Starita L.M., Kinzy T.G., Starita L.M., Kinzy T.G., Kinzy T.G. Mutations in elongation factor 1β, a guanine nucleotide exchange factor, enhance translational fidelity. Mol. Cell. Biol. 1999;19:5257–5266. doi: 10.1128/mcb.19.8.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.J., Kobor M.S., Kim M., Greenblatt J., Buratowski S., Kobor M.S., Kim M., Greenblatt J., Buratowski S., Kim M., Greenblatt J., Buratowski S., Greenblatt J., Buratowski S., Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustice D.C., Wakem L.P., Wilhelm J.M., Sherman F., Wakem L.P., Wilhelm J.M., Sherman F., Wilhelm J.M., Sherman F., Sherman F. Altered 40 S ribosomal subunits in omnipotent suppressors of yeast. J. Mol. Biol. 1986;188:207–214. doi: 10.1016/0022-2836(86)90305-0. [DOI] [PubMed] [Google Scholar]
- Fourmy D., Yoshizawa S., Puglisi J.D., Yoshizawa S., Puglisi J.D., Puglisi J.D. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J. Mol. Biol. 1998;277:333–345. doi: 10.1006/jmbi.1997.1551. [DOI] [PubMed] [Google Scholar]
- Gallis B.M., McDonnell J.P., Hopper J.E., Young E.T., McDonnell J.P., Hopper J.E., Young E.T., Hopper J.E., Young E.T., Young E.T. Translation of poly(riboadenylic acid)-enriched messenger RNAs from the yeast, Saccharomyces cerevisiae, in heterologous cell-free systems. Biochemistry. 1975;14:1038–1046. doi: 10.1021/bi00676a024. [DOI] [PubMed] [Google Scholar]
- Gebauer F., Hentze M.W., Hentze M.W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M., Sonenberg N., Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Hurt E., Luo M.J., Rother S., Reed R., Strasser K., Luo M.J., Rother S., Reed R., Strasser K., Rother S., Reed R., Strasser K., Reed R., Strasser K., Strasser K. Cotranscriptional recruitment of the serine–arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl. Acad. Sci. 2004;101:1858–1862. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T.H., Dower K., Libri D., Rosbash M., Dower K., Libri D., Rosbash M., Libri D., Rosbash M., Rosbash M. Early formation of mRNP: License for export or quality control? Mol. Cell. 2003;11:1129–1138. doi: 10.1016/s1097-2765(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Jerinic O., Joseph S., Joseph S. Conformational changes in the ribosome induced by translational miscoding agents. J. Mol. Biol. 2000;304:707–713. doi: 10.1006/jmbi.2000.4269. [DOI] [PubMed] [Google Scholar]
- Jimeno S., Rondon A.G., Luna R., Aguilera A., Rondon A.G., Luna R., Aguilera A., Luna R., Aguilera A., Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Guzik B.W., Bor Y.C., Rekosh D., Hammarskjold M.L., Guzik B.W., Bor Y.C., Rekosh D., Hammarskjold M.L., Bor Y.C., Rekosh D., Hammarskjold M.L., Rekosh D., Hammarskjold M.L., Hammarskjold M.L. Tap and NXT promote translation of unspliced mRNA. Genes & Dev. 2003;17:3075–3086. doi: 10.1101/gad.1155703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M.C., Podolny V., Buratowski S., Podolny V., Buratowski S., Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:7005–7018. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwapisz M., Cholbinski P., Hopper A.K., Rousset J.P., Zoladek T., Cholbinski P., Hopper A.K., Rousset J.P., Zoladek T., Hopper A.K., Rousset J.P., Zoladek T., Rousset J.P., Zoladek T., Zoladek T. Rsp5 ubiquitin ligase modulates translation accuracy in yeast Saccharomyces cerevisiae. RNA. 2005;11:1710–1718. doi: 10.1261/rna.2131605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Greenleaf A.L., Greenleaf A.L. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Greenleaf A.L., Greenleaf A.L. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J. Biol. Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H.F., Noller H.F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Napolitano G., Licciardo P., Carbone R., Majello B., Lania L., Licciardo P., Carbone R., Majello B., Lania L., Carbone R., Majello B., Lania L., Majello B., Lania L., Lania L. CDK9 has the intrinsic property to shuttle between nucleus and cytoplasm, and enhanced expression of cyclin T1 promotes its nuclear localization. J. Cell. Physiol. 2002;192:209–215. doi: 10.1002/jcp.10130. [DOI] [PubMed] [Google Scholar]
- Nielsen K.H., Szamecz B., Valasek L., Jivotovskaya A., Shin B.S., Hinnebusch A.G., Szamecz B., Valasek L., Jivotovskaya A., Shin B.S., Hinnebusch A.G., Valasek L., Jivotovskaya A., Shin B.S., Hinnebusch A.G., Jivotovskaya A., Shin B.S., Hinnebusch A.G., Shin B.S., Hinnebusch A.G., Hinnebusch A.G. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle J.M., Ramakrishnan V., Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Palmer E., Wilhelm J.M., Sherman F., Wilhelm J.M., Sherman F., Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- Powers T., Noller H.F., Noller H.F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger A., Dye M.J., Furger A., Dye M.J., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Reed R., Hurt E., Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Wintermeyer W., Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: Kinetic and structural mechanisms. Annu. Rev. Biochem. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- Rosset R., Gorini L., Gorini L. A ribosomal ambiguity mutation. J. Mol. Biol. 1969;39:95–112. doi: 10.1016/0022-2836(69)90336-2. [DOI] [PubMed] [Google Scholar]
- Sanford J.R., Gray N.K., Beckmann K., Caceres J.F., Gray N.K., Beckmann K., Caceres J.F., Beckmann K., Caceres J.F., Caceres J.F. A novel role for shuttling SR proteins in mRNA translation. Genes & Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore S.M., Byers S.A., Maury W., Price D.H., Byers S.A., Maury W., Price D.H., Maury W., Price D.H., Price D.H. Identification of a novel isoform of Cdk9. Gene. 2003;307:175–182. doi: 10.1016/s0378-1119(03)00466-9. [DOI] [PubMed] [Google Scholar]
- Singh A., Ursic D., Davies J., Ursic D., Davies J., Davies J. Phenotypic suppression and misreading Saccharomyces cerevisiae. Nature. 1979;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- Spahn C.M., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J., Penczek P.A., Sali A., Blobel G., Frank J., Sali A., Blobel G., Frank J., Blobel G., Frank J., Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Stahl G., Bidou L., Rousset J.P., Cassan M., Bidou L., Rousset J.P., Cassan M., Rousset J.P., Cassan M., Cassan M. Versatile vectors to study recoding: Conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I., Jones K.M., Herbert P., Lewendon A., Shaw W.V., Tuite M.F., Jones K.M., Herbert P., Lewendon A., Shaw W.V., Tuite M.F., Herbert P., Lewendon A., Shaw W.V., Tuite M.F., Lewendon A., Shaw W.V., Tuite M.F., Shaw W.V., Tuite M.F., Tuite M.F. Missense translation errors in Saccharomyces cerevisiae. J. Mol. Biol. 1998;282:13–24. doi: 10.1006/jmbi.1998.1976. [DOI] [PubMed] [Google Scholar]
- Sterner D.E., Lee J.M., Hardin S.E., Greenleaf A.L., Lee J.M., Hardin S.E., Greenleaf A.L., Hardin S.E., Greenleaf A.L., Greenleaf A.L. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin–cyclin-dependent kinase complex. Mol. Cell. Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R., Rondon A.G., Aguilera A., Struhl K., Reed R., Aguilera A., Struhl K., Reed R., Struhl K., Reed R., Reed R., et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Stutz F., Izaurralde E., Izaurralde E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 2003;13:319–327. doi: 10.1016/s0962-8924(03)00106-5. [DOI] [PubMed] [Google Scholar]
- Synetos D., Frantziou C.P., Alksne L.E., Frantziou C.P., Alksne L.E., Alksne L.E. Mutations in yeast ribosomal proteins S28 and S4 affect the accuracy of translation and alter the sensitivity of the ribosomes to paromomycin. Biochim. Biophys. Acta. 1996;1309:156–166. doi: 10.1016/s0167-4781(96)00128-5. [DOI] [PubMed] [Google Scholar]
- Verge V., Vonlanthen M., Masson J.M., Trachsel H., Altmann M., Vonlanthen M., Masson J.M., Trachsel H., Altmann M., Masson J.M., Trachsel H., Altmann M., Trachsel H., Altmann M., Altmann M. Localization of a promoter in the putative internal ribosome entry site of the Saccharomyces cerevisiae TIF4631 gene. RNA. 2004;10:277–286. doi: 10.1261/rna.5910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen M., Sturm D., Cajigas I.J., Gonzalez C.I., Seedorf M., Bastians H., Krebber H., Sturm D., Cajigas I.J., Gonzalez C.I., Seedorf M., Bastians H., Krebber H., Cajigas I.J., Gonzalez C.I., Seedorf M., Bastians H., Krebber H., Gonzalez C.I., Seedorf M., Bastians H., Krebber H., Seedorf M., Bastians H., Krebber H., Bastians H., Krebber H., Krebber H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol. Cell. Biol. 2004;24:10479–10491. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F., Minet M., Dufour M.E., Vo L.T., Lacroute F., Minet M., Dufour M.E., Vo L.T., Lacroute F., Dufour M.E., Vo L.T., Lacroute F., Vo L.T., Lacroute F., Lacroute F. Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast. Mol. Cell. Biol. 2000;20:3538–3549. doi: 10.1128/mcb.20.10.3538-3549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra P., Wyss J.C., Stutz F., Vinciguerra P., Wyss J.C., Stutz F., Wyss J.C., Stutz F., Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Pe’ery T., Peng J., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M.B., Price D.H., Pe’ery T., Peng J., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M.B., Price D.H., Peng J., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M.B., Price D.H., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M.B., Price D.H., Marshall N., Marshall T., Amendt B., Mathews M.B., Price D.H., Marshall T., Amendt B., Mathews M.B., Price D.H., Amendt B., Mathews M.B., Price D.H., Mathews M.B., Price D.H., Price D.H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes & Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R.A., Garvin R.T., Gorini L., Garvin R.T., Gorini L., Gorini L. Alteration of a 30S ribosomal protein accompanying the ram mutation in Escherichia coli. Proc. Natl. Acad. Sci. 1971;68:2263–2267. doi: 10.1073/pnas.68.9.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]