Abstract
The primary photochemical process of the visual function has been investigated using the three crystallographic models, 11-cis-rhodopsin, all-trans-bathorhodopsin, and the artificial isomeric 9-cis-rhodopsin. Detailed examination of the atomic displacements and dihedral angle changes of the retinal chromophore involved in the interconversion among these isomers suggests the mechanism of isomerization efficiency.
Ultrafast, highly efficient photoisomerization of the chromophore is an outstanding feature of retinal proteins such as bacteriorhodopsin and rhodopsin (1). It is well known that ocular visual rhodopsins use the 11-cis form of retinal (vitamin A aldehyde) exclusively as the chromophore, and the strict selection of this isomer seems to have occurred early in the evolution of visual function. On the other hand, both the 9-cis and 11-cis isomers can bind to the apoprotein (opsin) and form pigments with similar but distinct properties, e.g., the speed of isomerization and quantum efficiency (2). To understand the molecular mechanism of the isomerization reaction, extensive spectroscopic studies have been performed on 9-cis and other rhodopsin analogs whereas high-resolution structural data of such artificial pigments is lacking.
The detailed atomic environment of 11-cis-retinal in the dim-light receptor rhodopsin has been revealed recently by x-ray crystallography (3,4). Subsequent advances in the analysis of the primary photo-intermediate, bathorhodopsin (5), and in the preparation of artificial rhodopsin crystals containing 9-cis-retinal (6) also provide an opportunity to carry out comparative structural studies of the primary photoreaction in rhodopsin and 9-cis-rhodopsin.
Crystals of 9-cis-rhodopsin belong to the same space group as rhodopsin (P41). Details of crystallization and data collection are described elsewhere (6). Structure refinement of 9-cis-rhodopsin has been done using data to 2.95 Å resolution for the two molecules in the asymmetric unit (Table 1). Refinement of the retinal chromophore was carried out using the starting parameters (bond lengths, bond angles, and dihedral angles) derived from the high-resolution structures of microbial retinal proteins as applied recently to native rhodopsin (3) except for some of the dihedral angles. The resolution of the diffraction data is currently limited to 2.95 Å; however, the conformation of the crystallographic 9-cis-retinal chromophore model has been supported by a recent quantum mechanical study (7).
TABLE 1.
Refinement statistics
| Resolution range, Å | 50.0–2.95 |
|---|---|
| Twin fraction | 0.001 |
| Completeness, % (outer shell) | 94.3 (68.1) |
| Protein atoms | 5990 |
| Retinal atoms | 40 |
| Rcryst*, % (outer shell) | 24.3 (36.1) |
| Rfree†, % (outer shell) | 28.9 (41.0) |
| RMSD of bonds, Å | 0.012 |
| RMSD of angles, ° | 1.40 |
Rcryst = Σhkl | Fo − Fc | / Σhkl | Fo |.
Rfree was calculated from a set of 5% randomly selected reflections that were omitted from refinement.
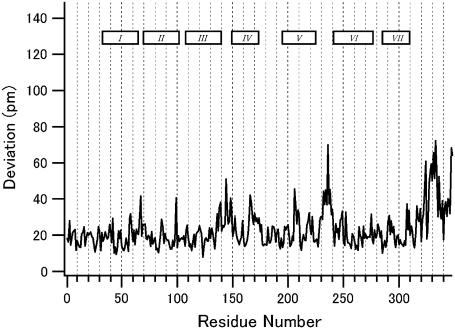
First we compare the backbone structures of rhodopsin and 9-cis-rhodopsin to see whether incorporation of the artificial retinal isomer has caused any substantial perturbations. After least-squares fitting of the individual molecules in the asymmetric unit, root mean square deviations (RMSD) are calculated for each of the two molecules, and the averaged values are plotted against the residue number (Fig. 1). Large deviations are found only in the interhelical loops and the carboxyl-terminal tail region where the crystallographic temperature factors are high. An exception is the second extracellular loop that connects helices IV and V and makes close contact with the retinal chromophore. In addition to this loop, the side chains of the retinal binding pocket keep nearly the same orientation as in rhodopsin. In conclusion, opsin accommodates 9-cis-retinal without suffering any significant structural changes in the transmembrane region.
FIGURE 1.
The RMSD between the main chain atoms of 9-cis and 11-cis rhodopsin versus the residue number. The plotted values are obtained by averaging the two molecules in the asymmetric unit. The seven rectangles in the panel represent the positions of the transmembrane helices.
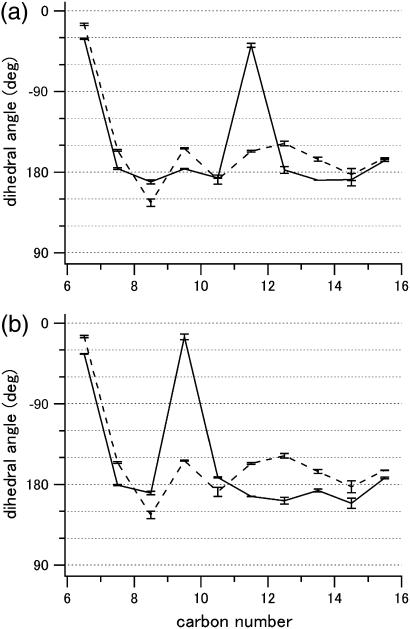
Both the different speed of isomerization and the quantum efficiency of rhodopsin and its 9-cis analog have been studied experimentally (2,8,9). To understand the molecular mechanism of these differences, two issues should be considered: the intrinsic properties of the chromophore itself and the effect of the protein on the chromophore conformation. With respect to the former, retinal Schiff bases and their protonated forms exhibit varying quantum yields depending on the isomeric state. Experiments have demonstrated that 9-cis-retinal protonated Schiff base is inherently less efficient in photoisomerization than the 11-cis isomer (10,11). However, it is doubtful whether the reported quantum yields in the isolated systems can explain the large differences observed for the protein-bound isomers. With regard to details of the chromophore conformation in the protein binding pocket, the pattern of bond lengths and angles is not much different from that seen in the chromophore of rhodopsin (supplemental Fig. 1, Supplementary Material) (3). The most striking feature found in the latter is the large negative dihedral angle (−38°, Fig. 3 a; calculated −18° (3)) about the C11=C12 bond. That this pretwist should arise in rhodopsin is not really surprising considering the steric repulsion between the C13 methyl and the C10 hydrogen expected for the 11-cis isomer in the 12s-trans conformation. Such an internal atomic contact does not occur in the 9-cis form. However, the crystallographic model indicates that the C9=C10 bond is also twisted, though significantly less (−15°, Fig. 3 b; calculated −9° (7)). This confirms a previous theoretical study of 13-demethyl-11-cis-rhodopsin where it was shown that the protein itself, by fixing the β-ionone ring and the Schiff base nitrogen within the binding pocket, causes a substantial negative twist of the chromophore chain that accumulates in the cis-bond (12).
FIGURE 3.
Change of the dihedral angles before (solid lines) and after (dashed lines) photoisomerization of (a) rhodopsin and (b) 9-cis-rhodopsin. Error bars are based on the values from the two molecules in the asymmetric unit. Carbon number 16 corresponds to the Schiff base nitrogen. Dashed lines in panels a and b are identical and derived from the coordinates of bathorhodopsin (2G87).
In contrast to rhodopsin the quantum yield of the 9-cis-rhodopsin photoreaction is wavelength dependent (8,9). Also, the yield decreases with decreasing temperature suggesting that there is a small but not negligible barrier to the 9-cis to 9-trans photoisomerization in the excited state (9). We propose that this barrier is caused by the small value of the pretwist angle. According to ab initio semiclassical trajectory calculations of retinal model chromophores (13,14) the conversion of the initially excited bond stretching coordinate into bond torsion and eventual isomerization takes place on a relatively flat bifurcating region of the potential energy surface. With a small pretwist angle the system is likely to spend more time in the stretching region with its high-energy turning points, whereas a larger angle will shift it more readily out of the Franck-Condon region and accelerate it toward the point of isomerization.
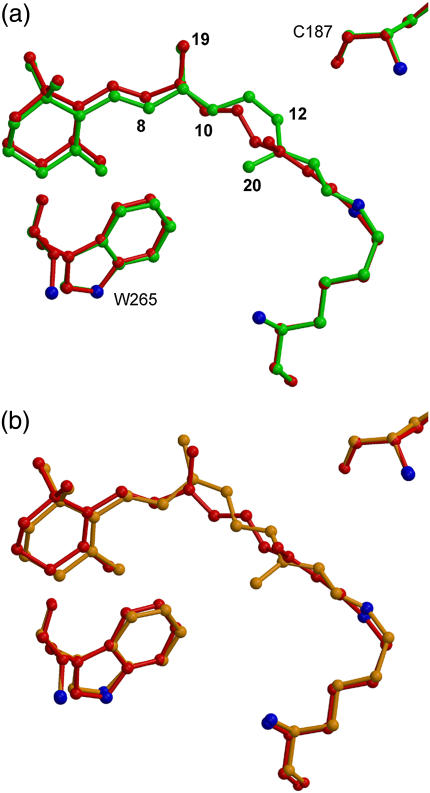
At low temperature (e.g., 100 K), continuous illumination of rhodopsin results in the formation of a photosteady-state mixture of rhodopsin, bathorhodopsin, and 9-cis-rhodopsin, indicating reversible interconversion among these species (1). It is also well accepted that photoisomerization from both rhodopsin and 9-cis-rhodopsin results in the formation of the same photointermediate (bathorhodopsin) (15). Taking the reasonable assumption that these features are independent of the molecular environment of the protein (membrane, detergent, or crystal) our experimental models of the three states constructed so far should contain valuable information. In Fig. 2, the two conversion processes modeled by x-ray crystallography are compared to see the overall change of the chromophores and how the isomerization reactions might proceed.
FIGURE 2.
Structural change of the chromophore before and after the photoisomerization; (a) rhodopsin and (b) 9-cis-rhodopsin. The common intermediate bathorhodopsin (PDB code, 2G87) is shown in red. Movies for these conversion processes are available as Supplementary Material.
As reported recently, 11-cis to all-trans conversion of retinal in rhodopsin involves large atomic displacement of C11 and C12 (Fig. 2 a) (5). In contrast, overlay of the crystallographic models of 9-cis-rhodopsin and bathorhodopsin suggests rather complicated movement during isomerization (Fig. 2 b). In both rhodopsin and 9-cis-rhodopsin, the large rotation of the main double bond (C11=C12 and C9=C10, respectively) is accompanied by counterrotations of the other double bonds in the polyene chain.
In Fig. 3, crossings of the two (solid and dashed) plots indicate the change of the direction of dihedral rotation. Detailed analysis of the dihedral angle changes demonstrates that, in 9-cis-rhodopsin, two adjacent single bonds are rotating in the same direction as the C9=C10 double bond. As a result, opposite twists of the two blocks in the polyene chain (C8-C11, C11-C15) characterize the isomerization process of 9-cis-rhodopsin, whereas rather alternating directional rotation along the entire polyene can be seen in the conversion from rhodopsin to bathorhodopsin. These differences in the dihedral changes during isomerization of the two cis forms might contribute to the distinct features of the two pigments.
In summary, the crystallographic models of rhodopsin, bathorhodopsin, and 9-cis-rhodopsin provide a detailed view of chromophore isomerization within a confined binding pocket of dim-light opsin and give insights to understanding the highly developed primary visual function.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
We are grateful to H. Sakai and M. Kawamoto for excellent support at BL41XU of SPring-8. The coordinate file of 9-cis-rhodopsin has been deposited with the Protein Data Bank (code, 2PED).
This research was supported in part by grants from Japanese Ministry of Education Culture, Sports, Science and Technology, New Energy and Industrial Technology Development Organization, and the Deutsche Forschungsgemeinschaft (FOR480).
References
- 1.Birge, R. R. 1990. Nature of the primary photochemical events in rhodopsin and bacteriorhodopsin. Biochim. Biophys. Acta. 1016:293–327. [DOI] [PubMed] [Google Scholar]
- 2.Schoenlein, R. W., L. A. Peteanu, Q. Wang, R. A. Mathies, and C. V. Shank. 1993. Femtosecond dynamics of cis-trans isomerization in a visual pigment analog: isorhodopsin. J. Phys. Chem. 97:12087–12092. [Google Scholar]
- 3.Okada, T., M. Sugihara, A. N. Bondar, M. Elstner, P. Entel, and V. Buss. 2004. The retinal conformation and its environment in the light of new 2.2 Å crystal structure. J. Mol. Biol. 342:571–581. [DOI] [PubMed] [Google Scholar]
- 4.Li, J., P. C. Edwards, M. Burghammer, C. Villa, and G. F. X. Schertler. 2004. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 343:1409–1438. [DOI] [PubMed] [Google Scholar]
- 5.Nakamichi, H., and T. Okada. 2006. Crystallographic analysis of primary visual photochemistry. Angew. Chem. Int. Ed. Engl. 45:4270–4273. [DOI] [PubMed] [Google Scholar]
- 6.Nakamichi, H., and T. Okada. 2007. X-ray crystallographic analysis of 9-cis-rhodopsin, a model analogue visual pigment. Photochem. Photobiol. 83:232–235. [DOI] [PubMed] [Google Scholar]
- 7.Sekharan, S., M. Sugihara, O. Weingart, T. Okada, and V. Buss. 2007. Protein assistance in the photoisomerization of rhodopsin and 9-cis-rhodopsin—insights from experiment and theory. J. Am. Chem. Soc. 129:1052–1054. [DOI] [PubMed] [Google Scholar]
- 8.Hurley, J. B., T. G. Ebrey, B. Honig, and M. Ottolenghi. 1977. Temperature and wavelength effects on the photochemistry of rhodopsin, isorhodopsin, bacteriorhodopsin and their photoproducts. Nature. 270:540–542. [DOI] [PubMed] [Google Scholar]
- 9.Birge, R. R., C. M. Einterz, H. M. Knapp, and L. P. Murray. 1988. The nature of the primary photochemical events in rhodopsin and isorhodopsin. Biophys. J. 53:367–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman, K. A., and R. S. Becker. 1986. Comparative investigation of the photoisomerization of the protonated and unprotonated n-butylamine Schiff base of 9-cis-, 11-cis-, 13-cis, and all-trans-retinals. J. Am. Chem. Soc. 108:1245–1251. [Google Scholar]
- 11.Koyama, Y., K. Kubo, M. Komori, H. Yasuda, and Y. Mukai. 1991. Effect of protonation on the isomerization properties of n-butylamine Schiff base of isomeric retinal as revealed by direct HPLC analyses: selection of isomerization pathways by retinal proteins. Photochem. Photobiol. 54:433–443. [DOI] [PubMed] [Google Scholar]
- 12.Sugihara, M., J. Hufen, and V. Buss. 2006. Origin and consequences of steric strain in the rhodopsin binding pocket. Biochemistry. 45:801–810. [DOI] [PubMed] [Google Scholar]
- 13.Vreven, T., F. Bernardi, M. Garavelli, M. Olivucci, M. A. Robb, and H. B. Schlegel. 1997. Ab initio photoisomerization dynamics of a simple retinal chromophore model. J. Am. Chem. Soc. 119:12687–12688. [Google Scholar]
- 14.Weingart, O., A. Migani, M. Olivucci, M. A. Robb, V. Buss, and P. Hunt. 2004. Probing the photochemical funnel of a retinal chromophore model via zero-point energy sampling semiclassical dynamics. J. Phys. Chem. A. 108:4685–4693. [Google Scholar]
- 15.Hug, S. J., J. W. Lewis, and D. S. Kliger. 1988. Evidence for a common BATHO-intermediate of rhodopsin and isorhodopsin. J. Am. Chem. Soc. 110:1998–1999. [DOI] [PubMed] [Google Scholar]