Abstract
Estrogen receptors are ligand-activated transcription factors that regulate gene expression by binding to specific DNA sequences. To date, the effect of ligands on the conformation of estrogen receptor α (ERα)-DNA complex remains a poorly understood issue. In our study, we are introducing the quartz crystal microbalance with dissipation monitoring (QCM-D) as a new alternative to study the conformational differences in protein-DNA complexes. Specifically, we have used QCM-D, in combination with surface plasmon resonance (SPR) spectroscopy, to monitor the binding of ERα to a specific DNA (estrogen response element, ERE) and a nonspecific DNA in the presence of either the agonist ligand, 17b-estradiol, the partial antagonist ligand, 4-hydroxytamoxifen, or vehicle alone. Both with presence and absence of ligand, the specific ERα-ERE complexes are observed to adopt a more compact conformation compared to nonspecific complexes. This observation is well correlated to the biophysical changes occurring during protein-DNA interaction shown by past structural and mechanism studies. Notably, pretreatment of ERα with E2 and 4OHT affects not only the viscoelasticity and conformation of the protein-DNA complex but also ERα binding capacity to immobilized ERE. These results affirm that ligands have remarkable effects on ERα-DNA complexes. Understanding these effects will provide insight into how ligand binding promotes subsequent events required for gene transcription.
INTRODUCTION
Estrogen receptors (ERs), α- and β-subtypes, are ligand-activated transcription factors that regulate genes responsible for development and maintenance of reproductive tissues and are also involved in the maintenance of other physiological functions (1–3). Upon binding to cognate ligands such as estrogen, ERs undergo conformational changes (4) and subsequently dissociate from chaperone proteins (5), dimerize (6), and bind to specific DNA sequences known as estrogen response elements (EREs) (7,8) to change gene transcription levels. The exact mechanism(s) of how ERs differentially regulate genes is unclear, but it is known that recruitment of cell-specific factors such as coactivators to the ER-ERE complex through protein-protein interactions connects the regulatory effect of ERs to the transcription initiation complex (9,10).
Although the structural basis of how ligands affect the recruitment of coactivators or corepressors is understood through x-ray crystallography (11,12) and NMR studies (13) on the ligand binding domain (LBD) of human estrogen receptor α (hERα), it is controversial and debatable if ligands have effects on hERα interaction with DNA (8). Unfortunately, there is no structural information available on the atomic level of the full-length ERs (made up of six different domains) (8) after ligand and/or ERE binding to propel us nearer to understanding the regulation mechanism. On one hand, electrophoretic mobility shift assay (EMSA) experiments have demonstrated that antagonistic ligands, e.g., 4-hydroxytamoxifen (4-OHT)-bound hERα and ERE complexes display retarded mobility compared to those with nonliganded and 17β-estradiol-bound hERα-ERE (14,15), thus suggesting differences in charge, shape, or size of the 4OHT-bound hERα complex; on the other hand, protease digestion of hERα complexed with DNA have shown that binding of ligands does not lead to different digestion patterns of estrogen receptor α (ERα), i.e., no conformational difference of ERα after various ligand binding in the presence of ERE (14). Other studies have shown that ERE sequences are also important modulators of ER's conformation (particularly the conformation of the DNA binding domain, DBD) (16,17). These findings increase the level of complexity involved in controlling ERα's ability to recruit cofactors when bound to the complex (18).
To affirm if ligands have effects on the conformation of the overall ERα-DNA complex and to detect the conformational changes in ERE and ERα when forming specific complex, we employ an alternative technique, namely quartz crystal microbalance with dissipation monitoring (QCM-D), in combination with surface plasmon resonance (SPR) spectroscopy to monitor the complexes formed. QCM-D, as a surface analytical technique, has been increasingly used successfully to probe conformational changes during biointerface processes involving various biomolecules. The past studies on protein adsorption on various substrates (19–20), changes in their viscoelastic properties (21,22), self-assembly of supported lipid bilayers (23,24), DNA assembly and hybridization (25,26) etc. have demonstrated that the simultaneously measured frequency and dissipation changes reflect the viscoelastic behavior of the adsorbed biomolecules and can be related to the conformation of the adsorbed biomolecular films. We believe that the viscoelasticity-related conformation parameters such as flexibility and amount of water coupled are important biophysical parameters in seeking understanding of protein-DNA recognition processes.
To facilitate the study, we first examine the viscoelasticity behavior of an ERα-ERE complex that is formed based on specific sequence recognition and a nonspecific ERα-DNA complex formed based on mainly loose electrostatic contacts, without any ligand. After this, we investigate the effect of two ligands (an agonist 17β-estradiol, E2, and a partial antagonist, 4OHT) on the viscoelasticity behavior of ERα-ERE complexes. Both E2 and 4OHT are known to bind to the LBD of ERα and induce different conformational changes in LBD alone (14), but whether they affect the overall conformation of an ER-DNA complex seems unclear. The assessment of the ligand-free ER-DNA complexes provides a basis for understanding how QCM-D is sensitive to the conformation of ER-DNA complexes formed under different mechanisms, which then facilitates the understanding of conformational changes induced by ligand binding.
Since the QCM cannot provide direct quantification of the binding amount of protein and DNA (26–28), we use a complementary technique, SPR spectroscopy, to quantify the binding amounts of the proteins and elucidate ligand-dependent ERα binding capacity. By modeling the combined SPR and QCM-D data, the layer thickness, water content, and relative changes in viscoelastic properties of the differently liganded ERα can be found. Through this analysis, differences in viscoelasticity between the various ERα-ERE-complex biolayers and altered ability of liganded-ERα to interact with immobilized DNA were observed.
MATERIALS AND METHODS
Estrogen receptor
Purified recombinant hERα was purchased from PanVera (Madison, WI). The protein (2088 nM in HEPES buffer containing 10% glycerol) was stored in aliquots of 10 μl at −80°C for long-term storage. Before use, the aliquots were thawed in a room temperature water bath and diluted using HEPES buffer (40 mM HEPES-KOH binding buffer, pH 7.4, containing 10 mM MgCl2, 0.2% Triton X-100, 1 mM DTT (dithiothreitol), and 100 mM KCl) to form working solutions of 125 nM.
Oligodeoxyribonucleotides
Thirty-four basepair (bp) oligos, synthesized by Proligo Primers & Probes (Boulder, CO), were tagged with biotin at the 5′ end. The specific ERE (5′-biotin-GTCCAAAGTCAGGTCACAGTGACCTGATCAAAGT-3′), denoted ERE, contains core consensus sequence (underlined) from chicken vitellogenin A2 gene (29). A sequence-scrambled DNA (5′-biotin-GTCCAAAGTCAATCGCCAGCACGATGATCAAAGT-3′), denoted as non-ERE, was used as a negative control. The biotinylated strands and the antistrands were annealed in phosphate buffered saline (PBS, pH 7.4) containing 10 mM EDTA, pH 7.5. The double-stranded DNA solutions were stored at −27°C.
Ligands
17-β estradiol (E2) and 4OHT were purchased from Sigma-Aldrich (St. Louis, MO; E2257 and H7904, respectively). Both ligands were dissolved in ethanol and stored at 4°C.
Sensor surface preparation
QCM-D and SPR gold disks were first cleaned (10 min under ultraviolet/ozone followed by 2 min with hot piranha solution (caution!) and then treated overnight with a binary biotin-containing thiol mixture (30) of 10% biotin-PEG (polyethylene glycol) disulfide (LCC Engineering & Trading, Egerkingen, Switzerland) and 90% 11-mercaptol-1-undecanol (Sigma-Aldrich) at a net concentration of 1 mM in ethanol. The disks are ready for measurements after rinsing with ethanol followed by a drying step using nitrogen.
To prepare streptavidin (SA)-modified surfaces for DNA immobilization, the biotin-containing thiol-treated sensor disks were exposed to 0.1 mg/ml SA (Sigma-Aldrich) for 15 min. PBS buffer (10 mM phosphate buffer, pH 7.4, 150 mM NaCl) was used as a carrier buffer for SA and the successive DNA assembly. HEPES buffer was used for monitoring various ERα-DNA interactions.
Binding assay procedures
In the study of unliganded ERα-DNA complex, non-ERE or ERE at 200 nM was immobilized to SA-modified surfaces. Unliganded ERα (125 nM) was applied to the DNA-immobilized surfaces for 30 min in HEPES buffer at room temperature.
In the study of ligand effect, ERE immobilization was carried out using working concentrations of either 200 nM or 20 nM to produce immobilized DNA of different packing densities. Before application to ERE-immobilized surfaces, ERα (125 nM) was incubated with either 10 μM E2 or 4OHT (total ethanol content 0.1%) for 30 min in 4°C. All control experiments were performed with ERα, which was similarly pretreated with ethanol vehicle (Ctrl ERα).
To regenerate the immobilized DNA surface, 0.1% SDS (sodium dodecyl sulfate) was added to liquid cell and incubated for 2–3 min. HEPES buffer was then applied to replace the SDS and to reset the baseline for new cycles of receptor binding.
QCM-D measurement
The QCM-D measurements were conducted using Q-Sense electronics and 5-MHz AT-cut quartz crystals (Q-Sense, Göteborg, Sweden), which have a mass sensitivity factor of 1 Hz = 17.7 ng/cm2, valid for thin, rigid films. The QCM-D electronics allow for simultaneous measurements of frequency change (Δf) and energy dissipation change (ΔD) by periodically switching off the driving power over the crystal and recording the decay of the damped oscillation. The QCM-D setup allows for subsequent measurements of up to four harmonics (fundamental frequency and 15, 25, and 35 MHz, corresponding to the overtones n = 3, 5, and 7, respectively) of the 5-MHz crystal. For clarity, the normalized frequency shift (Δfnormalized = Δf5/5) and dissipation shift for the fifth overtone are presented. During the measurements, the crystal was mounted in a liquid chamber, designed to provide a rapid, nonperturbing exchange of the liquid over one side of the sensor. The measurements were conducted at controlled room temperature, and the short-term noise level in f and D with liquid load was 0.3 Hz and 0.2 × 10−6, respectively.
SPR measurement
The SPR measurements were conducted using the AutoLab ESPR (Eco Chemie, Utrecht, The Netherlands). A gold-coated glass disk mounted on a prism forms the base of a two-channel cuvette. In this study, different DNA samples are immobilized into the two independent channels for protein to bind. In kinetic measurement mode, the incoupling angle of the plasmon resonance (θ) is recorded over time for molecular adsorption at room temperature, and the noise level was 0.5 mDeg. In the Autolab ESPR system, the conversion factor from angle shift to absorbed mass is 833 ng/(cm2/deg) for proteins (31). Using the de Feijter formula with a conversion factor of 833 ng/(cm2/deg) for proteins (corresponding to dn/dc = 0.18) and 789 ng/(cm2/deg) for DNA (corresponding to dn/dc = 0.19), the mass of the adsorbed biomolecules, ΔmSPR, could be determined from the SPR angle shifts (32).
Data modeling
All QCM-D data were modeled using a Voigt-type viscoelastic model using the QTools 2 software (Q-Sense) (33). Two overtones were used for the modeling, and the third was used to verify the robustness of the results. Several approaches were tried assuming different layer structures. Based on the closeness of fit of the modeled to the measured data, it was determined that a one-layer model for simulating the ERE and ERα adsorption best represented the data. The SA layer was separately modeled and subtracted because of its low dissipation, and all effects of buffer changes on the data were removed by subtraction before modeling commenced.
First, the QCM-D data were modeled using an arbitrary density of 1100 g/dm3 to find the mass of the adsorbed biofilm, including coupled water, corrected for the viscoelastic response of the film (21). It has been shown that changing the density between 1000–1700 g/dm3 affects only the modeled thickness while essentially conserving the product, i.e., the mass (24,26). The water content of the film (total amount of water coupled in the film and not limited to water entrapped within the biomolecules) could then be calculated from 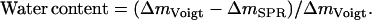 Knowing the water content and the respective mass of DNA and protein, the density of the film (DNA including protein) was calculated using
Knowing the water content and the respective mass of DNA and protein, the density of the film (DNA including protein) was calculated using
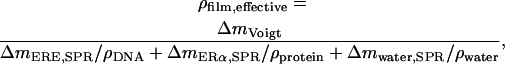 |
where ΔmERE,SPR and ΔmERα,SPR are the respective mass increases measured by SPR after adsorption of the hybridized DNA and the ligand or nonliganded ERα, respectively, 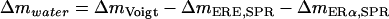 and ρDNA, ρprotein, and ρwater are the densities of DNA, protein, and water, respectively.
and ρDNA, ρprotein, and ρwater are the densities of DNA, protein, and water, respectively.
Using 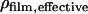 as input for iterated modeling, the proper acoustic thickness, viscosity, and shear modulus of the film could be obtained (27).
as input for iterated modeling, the proper acoustic thickness, viscosity, and shear modulus of the film could be obtained (27).
RESULTS AND DISCUSSION
Specific ERα-DNA binding leads to formation of a less dissipative complex
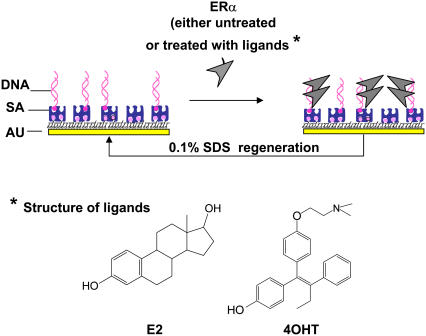
Fig. 1 is a schematic illustration of the assay procedures. Biotinylated-DNA, either specific or nonspecific sequence (ERE or non-ERE), was immobilized on the SA-modified surface. ERα (either liganded or unliganded) at a fixed concentration is then added to bind to the immobilized DNA. The DNA surface is regenerated for multiple subsequent binding events by using 0.1% SDS to remove the bound proteins.
FIGURE 1.
Schematic illustration of the assay procedures used in this study. Drawing is not to scale. The DNA can be specific ERE or non-ERE. The ERα can be unliganded or liganded with E2 or 4OHT.
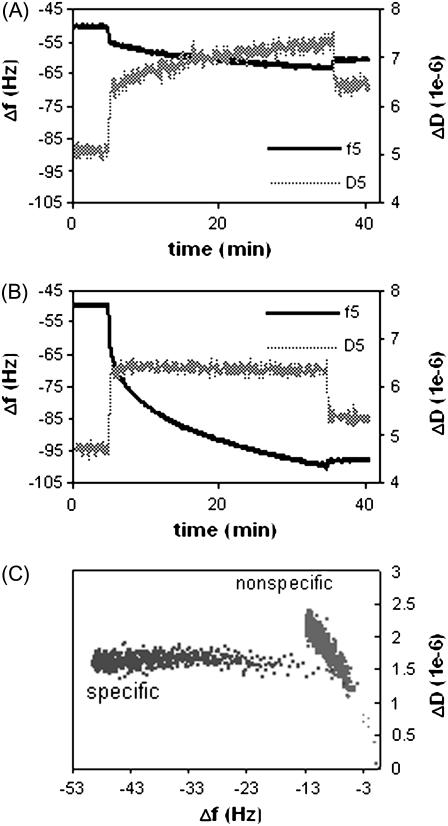
To understand how specific ERE binding modulates the conformation of ERα-ERE complex (with no ligand involved), real time frequency (Δf) and dissipation (ΔD) changes were recorded for the binding of ERα to the non-ERE (Fig. 2 A) and the specific ERE (Fig. 2 B) immobilized surfaces (immobilization of ERE and non-ERE has identical Δf and ΔD, (relative standard deviation) RSD < 1.5%, curves not shown). The initial change in both f and D observed upon addition of ERα samples includes not just protein adsorption but mainly a buffer change effect (the ERα working solution contains some glycerol absent in the baseline HEPES buffer; see Materials and Methods). Upon rinsing the surface at the end of the protein binding, this buffer effect is removed and the endpoint Δf and ΔD are recorded. Control experiments were performed to apply the ERα solution onto the SA-modified surface that carries no DNA. Only the response of the entirely reversible buffer effect is recorded, showing that there is no detectable nonspecific ERα adsorption on the surface.
FIGURE 2.
QCM-D measurements of normalized Δf and ΔD signals versus time at overtone n = 5 for unliganded ERα (125 nM) to bind to a (A) non-ERE and (B) specific ERE immobilized on the surface, forming nonspecific and specific complexes, respectively. (C) ΔD-Δf plots for the formation of nonspecific and specific complex.
In Fig. 2, a much larger frequency drop is observed for the specific binding (Fig. 2 B) compared to nonspecific binding (Fig. 2 A), showing that the ERα preferentially binds to the specific ERE sequence (14). The dissipation response also displays different trends for the specific and nonspecific binding. To observe how dissipation changes occur as more ERα binds to surface-immobilized DNA, ΔD induced by ERα binding is plotted against Δf (Fig. 2 C, buffer jump effect is removed according to the control experiments). This roughly corresponds to plotting the energy loss induced by the viscosity of the film versus the mass. Although the endpoint frequency changes are different, it is clear that for each unit frequency change down to Δf > −8, there were observable larger dissipation changes for nonspecific ERα binding than specific ERα binding. It should also be kept in mind that the first data points where binding looks similar still contain effects from the solution exchange and are not reflective of only the binding conformation.
Previous studies have shown that the QCM-D dissipation factor is a measure of internal energy lost in the biolayer due to periodic shear stress (34,35). Large dissipation changes or ΔD/Δf ratios are commonly associated with extended, flexible conformations of the individual biomolecules with a high water content (25) or loose bindings between interacting biomolecules (20,34), as loosely attached films tend to deform during the shear oscillation and dissipate more mechanical energy. On the other hand, a low dissipation is usually interpreted to be reflective of dehydrated, structured biomolecules packed to form a rigid biolayer (21,34).
The distinct dissipative behaviors we observed for the nonspecific and specific ERα-DNA complex can be explained based on 1), the above understandings of QCM-D capability, 2), available conformational analysis of ERα and DNA binding, and 3), current understandings of the mechanism of specific protein-DNA recognition.
In specific ERα-ERE interaction, structural analysis (e.g., x-ray crystallography and circular dichroism spectroscopy) shows that DNA binding causes the flexible and disordered region of the DBD of ERα to become ordered (36) and large-scale α-helical change is induced in ERα (37). During the specific ERα-ERE binding, DNA was also observed to bend toward its major groove (38,39) to induce significant conformation changes in both the protein and DNA. These changes are akin to that of the induced fit model (40), where initially the protein and DNA each have some degree of flexibility which allows them to interact and, upon specific binding, change conformation to “fit” to an energetically stable complex (40,41).
Current understanding of protein-DNA recognition using the Lac repressor model shows that nonspecific protein-DNA binding results in a more flexible complex, maintained by mainly electrostatic attraction (42). Similarly, ERα also interacts nonspecifically with DNA mainly through electrostatic interactions as demonstrated by a fluorescence anisotropy study (43). Molecular dynamics simulation further shows that low affinity of ERα to nonconsensus sequences can be attributed to a weak hydrogen-bonding network and failure to expel excess water molecules from the DBD-DNA interface as it does in the specific binding (44).
Using the combinational Δf and ΔD analysis on QCM-D measurement (ΔD-Δf plots), we successfully captured the distinct viscoelastic properties of the specific and nonspecific complexes, which are very well correlated to the biophysical properties of the complexes determined by the above mechanisms. The low dissipation per mass unit measured for the specific binding could be related to the formation of a well-structured complex with a high binding strength and less water entrapment due to dehydration. On the other hand, the higher dissipation per mass measured for the nonspecific complex could be attributable to the loose binding of the protein, the higher disorder of the DBD, and a high degree of hydration.
Ligand binding results in different viscoelastic behavior of ERα-ERE complexes
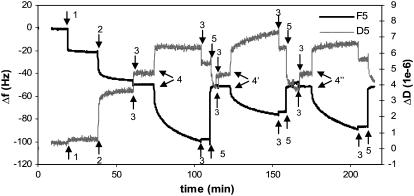
To investigate the effect of ligands on the viscoelastic behavior of ERα-ERE complexes, various ERα, unliganded (Ctrl ERα) or liganded by 17β-estradiol (E2-ERα) or 4OHT-ERα, are added separately to a surface functionalized with immobilized ERE as outlined in Fig. 1. Fig. 3 shows a typical QCM-D plot, with real time changes in frequency (Δf) and dissipation (ΔD) recorded for SA immobilization followed by ERE assembly in PBS buffer (t = 20 min) and finally ERα bindings in HEPES buffer (t = 30 min). Successful regeneration of the ERE-immobilized surface is evidenced by the retainable baseline after regeneration using 0.1% SDS (t = 3 min). This ensures the surface can be reused for binding of ERα with different ligands.
FIGURE 3.
QCM-D measurements of the binding reactions outlined in Fig. 1. Immobilization of SA (0.1 mg/ml) and ERE (200 nM) was done in PBS buffer, and binding of proteins was carried out in HEPES buffer containing 100 mM KCl. The initial change in both f and D observed upon addition of ERα samples includes not just protein adsorption but also a buffer change effect (the ERα working solution contains some glycerol absent in the baseline HEPES buffer). Upon rinsing the surface at the end of the protein binding, this buffer effect is removed and the endpoint Δf and ΔD are recorded. The surface with immobilized ERE is regenerated using 0.1% SDS to remove bound proteins and the baseline is reset with HEPES buffer.
Table 1 lists the normalized endpoint Δf and ΔD values averaged over all measurements recorded in the HEPES buffer for the binding of Ctrl and liganded-ERα to immobilized-ERE (specific bindings) and Ctrl ERα to immobilized-non-ERE (nonspecific binding). The ratio of ΔD/Δf, the amount of energy dissipated within the layer per unit mass coupled, is calculated. Addition of 0.1% ethanol or 10 μM ligands alone to immobilized ERE have no significant irreversible effect on frequency and dissipation response, showing that the f and D changes obtained are caused by the event of various ERα binding to the ERE.
TABLE 1.
Summary of QCM-D results
| Sample | Δf* (Hz) | ΔD* (× 10−6) | ΔD/Δf† (× 10−9 Hz−1) |
|---|---|---|---|
| Ctrl ERα-ERE | 39.9 | 0.56 | 11.7 |
| E2-ERα-ERE | 33.0 | 0.93 | 26.5 |
| 4OHT-ERα-ERE | 19.6 | 1.63 | 71.0 |
| Ctrl ERα-non-ERE | 9.3 | 1.36 | 146.2 |
Δf and ΔD are averaged from normalized signals from overtones 3, 5, and 7 measured from experiments repeated 3–4 times. The experimental variation for Δf and ΔD within each binding step is ∼±15% but similar ΔD/Δf (RSD < 5.0%) were obtained.
ΔD/Δf value gives an indication of the dissipation induced per coupled unit mass (31).
Among the three specific interactions, the highest ΔD/Δf value obtained for binding of 4OHT-ERα indicates that this complex assumes a water-rich, and less well-structured conformation; whereas the E2- and unliganded ERα-ERE complexes are relatively more dehydrated, compact, and rigid. The distinct conformation of the 4OHT-ERα-ERE complex compared to that of the E2-ERα- and Ctrl ERα-ERE complexes observed here correlates with its retarded mobility observed during gel electrophoresis (14,15).
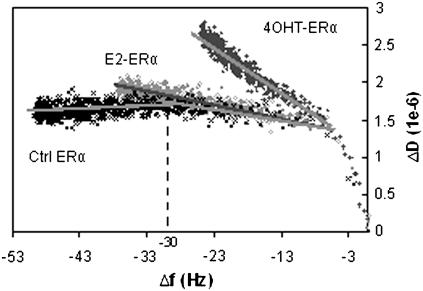
To understand if there are coverage-induced conformational changes in the complexes (23,32), ΔD is plotted against Δf for the three specific ERα bindings (Fig. 4). For the Ctrl ERα, after Δf of −30, the slope gradient of the ΔD-Δf curves goes downward, indicating an increase in rigidity of the biolayer. The trend of E2-ERα binding is similar, except that there is no obvious downward gradient of the ΔD/Δf slope after Δf of −30 Hz. This could mean that E2-treated ERα forms complexes having a similar conformation with the complex formed with Ctrl ERα, but due to some reason, the surface does not allow tighter packing, leading to condensation of E2-ERα-ERE complexes at a higher capacity.
FIGURE 4.
ΔD-Δf plots for the binding of Ctrl ERα, E2-ERα, and 4OHT-ERα to an ERE-immobilized surface (derived from data shown in Fig. 3). Slope gradients identified for all three curves are shown as dark shaded lines.
Taken together, the viscoelasticity differences of the ERα-DNA complexes detectable through the endpoint ΔD/Δf values and ΔD-Δf slopes affirm that the ligands have an effect on overall conformation of the ERα-DNA complex. Although it is hard to deduce the exact structural origins of these differences in dissipation behavior, it is clear that E2 and 4OHT have different effects: E2-ERα-ERE shows dissipative behavior very similar to Ctrl ERα, except that the surface density is higher; 4OHT-ERα-ERE complexes form a much more dissipative protein-DNA complex, but it is still relatively more rigid than the nonspecific complexes where nonspecific interaction occurs, indicating stronger binding of a similarly disordered conformation.
Ligands affect ERα binding capacity to immobilized ERE
To investigate if the amount of ERα bound on the DNA is related to the conformation and to determine whether the measured differences in ΔD/Δf were due to mainly a difference in water content (thickness of the film) or conformational differences between the molecules (mainly density and viscosity increase), SPR—a complementary surface analytical technique—is utilized to monitor the same binding reactions and to provide an independent measurement of ERα binding amounts, ΔmSPR, i.e., molecular mass that does not include coupled water (27,45).
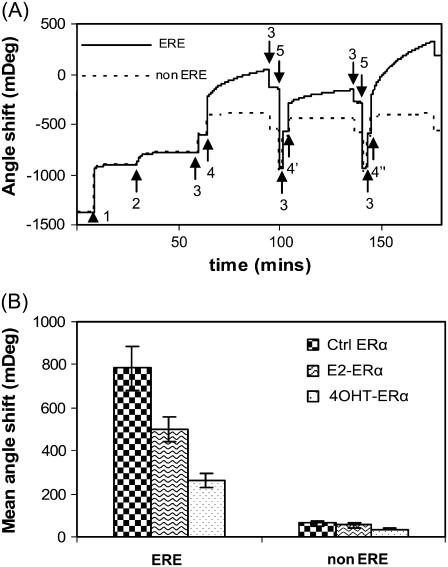
Fig. 5 A shows the SPR measurement of the binding reactions outlined in Fig. 1 (corresponding QCM-D measurement is shown in Fig. 3). The double-channel feature of the SPR equipment, ERα binding to the ERE and non-ERE preimmobilized in different channels to be monitored simultaneously. For the ERα binding, we observed the reversible buffer jump effects as we did in the QCM-D measurements. Rinsing the surface with HEPES buffer corrects the buffer jump signals. Different angle shifts (Δθ) obtained for the binding of various ERα to ERE (data are summarized in Fig. 5 B) confirmed that there is indeed a significant difference in protein binding amounts after ligand binding. Within each ERα case, the binding to non-ERE is always obviously lower than binding to a consenus ERE, as expected.
FIGURE 5.
(A) SPR sensorgram showing the binding reactions outlined in Fig. 1. Legend: Step 1: SA; 2: DNA immobilization (ERE, solid line, and scrambled non-ERE, dashed line, each in one channel); 3: HEPES buffer; 4: E2-ERα, 4′: 4OHT-ERα, 4″: Ctrl ERα; 5: 0.1% SDS. Immobilization of SA and DNA are carried out in PBS buffer. Identical SA (464 ± 5 mDeg) and DNA binding amounts (129 ± 6 mDeg) are achieved for the ERE and non-ERE channels. ERα (125 nM) was either preincubated with 10 μM 17β-estradiol (E2-ERα), 10 μM 4OHT-ERα, or ethanol (Ctrl ERα). Similar to the QCM-D experiment, addition of ERα samples to DNA layer introduced a buffer change effect (the ERα working solution contains some glycerol absent in the baseline HEPES buffer). Upon rinsing the surface at the end of the protein binding, this buffer effect is removed and the endpoint Δθ are recorded. (B) The SPR angle shifts caused by the binding of various ERα (liganded or unliganded) are averaged from three to five experiments.
The results we have presented up to now are based on a 129 ± 6 mDeg of DNA assembled from 200 nM DNA solution on a standard SA layer of 464 ± 5 mDeg. This results in a DNA packing density of 0.78 molar ERE per molar SA (molecular mass of SA and ERE are 60 kDa and 21.4 kDa, respectively) or 5.0 pmol DNA/cm2. In a next series of SPR experiment, we repeated the ERα bindings but used a lower ERE concentration (20 nM), with which the packing density of immobilized DNA is decreased to 0.23 ERE/SA or 1.5 pmol/cm2. The binding capacity of various ERα (liganded and unliganded) at two ERE densities is calculated (Table 2).
TABLE 2.
Summary of binding capacity of various ERα (liganded or unliganded) measured at two different ERE packing densities
| DNA packing density
|
||||
|---|---|---|---|---|
| Sample | 0.78 ERE/SA ratio (mol/mol) or 5.0 (pmol/cm2) | 0.23 ERE/SA ratio (mol/mol) or 1.5 (pmol/cm2) | ||
| ERα binding capacity (pmol/cm2) | ERα/ERE (mol/mol) | ERα binding capacity (pmol/cm2) | ERα/ERE (mol/mol) | |
| Ctrl ERα | 9.8 ± 1.3 | 2.0 ± 0.3 | 5.8 ± 0.6 | 3.9 ± 0.4 |
| E2-ERα | 6.2 ± 0.7 | 1.2 ± 0.1 | 5.3 ± 1.0 | 3.5 ± 0.7 |
| 4OHT-ERα | 3.2 ± 0.4 | 0.6 ± 0.1 | 2.4 ± 0.7 | 1.6 ± 0.5 |
*ERα binding capacity (pmol/cm2) or ERα per DNA ratio (mol/mol) are calculated from the angle shift values using an AutoLab SPR sensitivity of 833 ng/(cm2/deg) and the molecular mass of 21.4, 66.4, 66.7, and 66.8 kDa for the 34-bp ERE, Ctrl ERα, E2-ERα, and 4OHT-ERα, respectively. Values are mean ± SD obtained from three to five experiments.
Results in Table 2 show that at the lower DNA packing density condition, 125 nM Ctrl ERα saturated the immobilized DNA to give a binding ratio (or stoichiometry) of ∼4 ERα per ERE. This is consistent with previous reports that ERα binds ERE as a tetramer (46–48). At this DNA density E2-ERα is found to bind to ERE at a similar amount (or capacity) as Ctrl ERα, on the surface, which confirms the previous reports that E2-treated ERα has a similar affinity as untreated ERα in liquid phase measurements (14,47,48). In contrast, the 4OHT-ERα displayed a significantly lower binding capacity compared to Ctrl and E2-treated samples.
At the higher DNA density condition, 125 nM ERα is not sufficient to saturate the immobilized ERE, thus for liganded and Ctrl ERα, the binding ratio is lower than that at the lower DNA density condition, as expected. Although Ctrl and E2-ERα are known to have similar affinity to ERE in liquid phase, when ERE is closely packed on the SPR surface the E2-ERα is found to bind at a significantly lower capacity (1.2 molar protein per molar ERE) or a lower apparent affinity than Ctrl ERα (2.0 molar protein per molar ERE). This lower apparent affinity of E2-ERα may be attributable to the formation of bigger complexes (as indicated by QCM-D) to which steric effect plays a role to prevent the proteins from binding in a high capacity.
At both high and low ERE packing densities, the 4OHT-ERα binding amount is always significantly lower than E2-ERα and Ctrl ERα. The significantly lower binding capacity of 4OHT-ERα at both the high and low DNA density conditions (SPR results) and the high dissipation (QCM-D results) may reflect not only altered conformation of the complex but also significantly altered binding behavior or affinity. This proposition is supported by a fluorescent anisotropy study which shows that 4OHT-ERα has a lower affinity toward ERE (48).
Modeling of SPR and QCM-D data
The mass of the adsorbed biomolecules, ΔmSPR, determined from the SPR angle shifts is listed in Table 3. By comparing ΔmSPR to ΔmVoigt, which includes the contribution from water trapped in the biofilm, the water content could be determined and then used to give a quantitative estimate of the conformational differences of the overall biofilms and provide further verification of the trends observed in the corresponding ΔD/Δf plot (Fig. 4). Furthermore, by discriminating the response of the different biomolecules and water, the density of the biofilm could be calculated as outlined in Materials and Methods, and the thickness, viscosity, and shear modulus of the film estimated (25,27). The obtained values for the different ERα-ERE films can be found in Table 2.
TABLE 3.
Mass adsorption and viscoelastic properties calculated from QCM-D and SPR results
| Sample | ΔmVoigt* (ng/cm2) | ΔmSPR† (ng/cm2) | Water content (mass %) | ρVoigt‡ (g/dm3) | dVoigt‡ (nm) | ηVoigt‡ (mPa/s) | μVoigt‡ (MPa) |
|---|---|---|---|---|---|---|---|
| ERE | 655 | 102 | 84 | 1069 | 6.2 | 2.0 | 0.19 |
| Ctrl ERα-ERE | 1470 | 755 | 49 | 1168 | 12.6 | 4.3 | 0.55 |
| E2-ERα-ERE | 1270 | 516 | 59 | 1133 | 11.2 | 3.4 | 0.42 |
| 4OHT-ERα-ERE | 1155 | 319 | 72 | 1093 | 10.6 | 2.4 | 0.23 |
ΔmVoigt value is obtained through viscoelastic modeling using the Voigt model for a one-layer film (see Materials and Methods).
ΔmSPR value calculated from their corresponding angle shift using the de Feijter formula with a conversion factor of 833 ng/(cm2/deg) for proteins (corresponding to dn/dc = 0.18) and 789 ng/(cm2/deg) for DNA (corresponding to dn/dc = 0.19).
The effective density of the film was calculated by weighting from the modeled and measured masses ΔmVoigt and ΔmSPR, using a density of 1000, 1350, and 1700 g/dm3 for buffer, protein, and DNA, respectively (see Materials and Methods). This density was used in a subsequent iteration of modeling to find dVoigt, ηVoigt, and μVoigt.
The water content obtained in this way confirms the qualitative analysis based on the ΔD/Δf ratios. The original 34-bp duplex ERE film consists mostly of water (similar to most other DNA films as previously demonstrated (25–27)). Upon binding of the ERα, the water content decreases strongly. The decrease in water content is even more pronounced for the low dissipation binding. For the Ctrl ERα the water content is almost halved, which means that the density of the layer has strongly increased through the binding of the protein. Although the water content has decreased and the density strongly increased for binding of the E2-ERα as well, the 4OHT-ERα shows only a small decrease in water content, from 84% to 72%.
A more detailed analysis of the change in layer mechanical properties is possible by calculating the density of the layer from the QCM-D and SPR data for the different adsorbed species and then reiterate the modeling at near-equilibrium adsorption (30-min adsorption time). Thus, the acoustic thickness of the biolayer is obtained and can be compared, although surface roughness and the natural errors associated with measuring and modeling requires us to not take the values we get as exact layer thickness for the multi-layer film. A fully extended 34-bp duplex DNA strand has a length of ∼11.56 nm (∼0.34 nm rise per bp); however, the ERE film thickness is about half of this value, pointing toward a tilted, random conformation of the DNA strands or at least incomplete coupling of the water within the film. However, after binding of the Ctrl ERα the total film thickness corresponds to and even exceeds the expected ERE film thickness. As the density of the film simultaneously strongly increases, it can be interpreted to mean that tight rigid binding of the Ctrl ERα allows packing at a high density and strongly reduces the conformational and orientational freedom of the ERE-ERα biolayer. This is corroborated by the change in viscoelastic parameters for the film. A similar, but not as large, thickness increase of 4–5 nm takes place when the E2-ERα and 4OHT-ERα binds to the ERE. The lower decrease in water content as well as the lower viscosities and shear moduli for these films suggest the less conformationally constrained and less close-packed nature of these films. In particular, 4OHT-ERα stands out, with the ligand strongly influencing both the density of packing of the protein into the layer and the rigidity of the formed complex between ERE and ERα. The water content and viscoelastic parameters of the E2-ERα is always in between the Ctrl-ERα and 4OHT-ERα, although the simulated density and thickness of the film can vary between batches to approach values closer to one or the other.
Since the modeling data were obtained based on protein-DNA films formed in 30 min, which leads to different protein coverage for the liganded and Ctrl ERα, the results are protein coverage dependent. To remove the protein coverage as a variable when comparing the viscoelastic values for Ctrl and liganded ERα and to provide an unambiguous correlation of viscoelastic property with different protein binding modes, we modeled the various data at time points when the SPR mass of all three proteins are similar (RSD ∼2%, 5, 7, and 26 min for Ctrl ERα, E2-ERα, and 4OHT-ERα, respectively). Results show that, upon binding of the ERα, the water content decreased from 84% to 73%, 72%, and 74% for Ctrl ERα, E2-ERα, and 4OHT-ERα, respectively. Similar water contents are obtained for all three ERα-ERE films probably because at a low protein coverage, the water content is largely contributed by entrapped water in the entire biofilm comprising mostly ERE. Similar thickness increases to 10.8, 11.5, and 11.4 nm, respectively, and similar effective density of the films (1085–1094 g/dm3) are also obtained compared to their increments from 6.2 nm and 1069 g/dm3 of the ERE film. However, different viscosity was still observed for the 4OHT-ERα−ERE complex (2.15 mPa/S) compared to the other two ERα-ERE complexes (2.43 mPa/S). At this fixed, low protein coverage, both E2-ERα-ERE and Ctrl ERα-ERE seemingly displayed similar viscoelastic properties, evidenced by the identical viscosity and dissipative behavior as shown in the ΔD/Δf plot (Fig. 4). With this modeling the protein coverage is removed as a variable, the difference in the viscosity values can then be a true reflection of the conformation of the ERα-ERE complexes. The smaller viscosity of the 4OHT-ERα−ERE complex can be readily related to its loose conformation (loose binding and bigger complex) that may tender deformation easily during the shear oscillation.
The large difference in water content and viscosity between the differently liganded ERα shown in the modeling at near-equilibrium coverage (Table 3) is mainly due to the higher packing demonstrated for the Ctrl ERα and E2-ERα. That significantly different packing and viscoelastic properties between both liganded and nonliganded ERα observed when the ERE density is increased indicates a difference in binding conformation of both liganded ERα complexes that is not captured by previous measurements of liquid phase binding affinities. It is also clear from the modeling results at the same coverage of ERα that 4OHT has the most significant effect on the conformation of the ERα-ERE complex, showing up as quantitative differences already at low coverage. Furthermore, it is demonstrated that higher packing is only possible—and thus correlates—with a tight, rigid, binding conformation between ERE and ERα.
CONCLUSION
QCM-D and SPR were employed to study how ERα interacts differently with a specific ERE and nonspecific DNA and—more importantly—the ligand binding effects. Significant differences in viscoelastic behavior observed between nonspecific complexes and specific complexes correlate with previous structural studies. Using QCM-D analysis, 4OHT-ERα was observed to form distinctly less dense and more dissipative complexes with immobilized ERE compared to E2-ERα and unliganded ERα. Both ligands were affirmed to have effects on ERα-ERE conformation as well as binding capacity and water content of the formed biolayer at high ERE and ERα coverage. Without ligand, ERα-ERE forms a rigid, extended complex with a high packing density. Combined with SPR studies, we showed by modeling the biolayer viscoelastic properties that the water content, viscosity, and shear modulus correlate with the binding capacity to immobilized DNA. Importantly, this study shows that QCM-D can extend its usefulness and is sufficiently sensitive to offer an efficient alternative and new perspective to the study of the conformation differences of protein-DNA interactions.
Acknowledgments
This work was supported by the Institute of Materials Research and Engineering under the Agency for Science, Technology & Research (A-Star) of Singapore.
References
- 1.Deroo, B. J., and K. S. Korach. 2006. Estrogen receptors and human disease. J. Clin. Invest. 116:561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green, P. S., and J. W. Simpkins. 2000. Neuroprotective effects of estrogen: potential mechanisms of action. Int. J. Devl. Neurosci. 18:347–358. [DOI] [PubMed] [Google Scholar]
- 3.Jansson, L., and R. Holmdahl. 1998. Estrogen-mediated immunosupression in autoimmune diseases. Inflamm. Res. 47:290–301. [DOI] [PubMed] [Google Scholar]
- 4.Kraichely, D. M., J. Sun, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2000. Conformational changes and coactivator recruitment by novel ligands for estrogen receptor-α and estrogen receptor-β: correlations with biological character and distinct differences among SRC coactivator family members. Endocrinology. 141:3534–3545. [DOI] [PubMed] [Google Scholar]
- 5.Devin-Leclerc, J., X. Meng, F. Delahaye, P. Leclerc, E. Baulieu, and M. Catelli. 1998. Interaction and dissociation by ligands of estrogen receptor and Hsp90: the antiestrogen RU 58668 induces a protein synthesis-dependent clustering of the receptor in the cytoplasm. Mol. Endocrinol. 12:842–854. [DOI] [PubMed] [Google Scholar]
- 6.Wang, H., G. A. Peters, X. Zeng, M. Tang, W. Ip, and S. A. Khans. 1995. Yeast two-hybrid system demonstrates that estrogen receptor dimerization is ligand-dependent in vivo. J. Biol. Chem. 270:23322–23329. [DOI] [PubMed] [Google Scholar]
- 7.Zilliacus, J., A. P. H. Wright, J. Carlstedt-Duke, and J. Gustafsson. 1995. Structural determinants of DNA-binding specificity by steroid receptors. Mol. Endocrinol. 9:389–400. [DOI] [PubMed] [Google Scholar]
- 8.Klinge, C. M. 2001. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 29:2905–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muramatsu, M., and S. Inoue. 2000. Estrogen receptors: how do they control reproductive and nonreproductive functions? Biochem. Biophys. Res. Commun. 270:1–10. [DOI] [PubMed] [Google Scholar]
- 10.Klinge, C. M. 2000. Estrogen receptor interaction with co-activators and co-repressors. Steriods. 65:227–251. [DOI] [PubMed] [Google Scholar]
- 11.Brzozowski, A. M., A. C. W. Pike, Z. Dauter, R. E. Hubbard, T. Bonn, O. Engström, L. Öhman, G. L. Greene, J. Gustafsson, and M. Carlquist. 1997. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 389:753–758. [DOI] [PubMed] [Google Scholar]
- 12.Shiau, A. K., D. Barstad, P. M. Loria, L. Cheng, P. J. Kushner, D. A. Agard, and G. L. Greene. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 95:927–937. [DOI] [PubMed] [Google Scholar]
- 13.Luck, L. A., J. L. Barse, A. M. Luck, and C. H. Peck. 2000. Conformational changes in the human estrogen receptor observed by 19F NMR. Biochem. Biophys. Res. Commun. 270:988–991. [DOI] [PubMed] [Google Scholar]
- 14.Yi, P., M. D. Driscoll, J. Huang, S. Bhagat, R. Hilf, R. A. Bambara, and M. Muyan. 2002. The effects of estrogen-responsive element- and ligand-induced structural changes on the recruitment of cofactors and transcriptional responses by ERα and ERβ. Mol. Endocrinol. 16:674–693. [DOI] [PubMed] [Google Scholar]
- 15.Metzger, D., M. Berry, S. Ali, and P. Chambon. 1995. Effect of antagonists on DNA binding properties of the human estrogen receptor in vitro and in vivo. Mol. Endocrinol. 9:579–591. [DOI] [PubMed] [Google Scholar]
- 16.Wood, J. R., G. L. Greene, and A. M. Nardulli. 1998. Estrogen response elements function as allosteric modulators of estrogen receptor conformation. Mol. Cell. Biol. 18:1927–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood, J. R., V. S. Likhite, M. A. Loven, and A. M. Nardulli. 2001. Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol. Endocrinol. 15:1114–1126. [DOI] [PubMed] [Google Scholar]
- 18.Geserick, C., H. Meyer, and B. Haendler. 2005. The role of DNA response elements as allosteric modulators of steroid receptor function. Mol. Cell. Endocrinol. 236:1–7. [DOI] [PubMed] [Google Scholar]
- 19.Brewer, S. H., W. R. Glomm, M. C. Johnson, M. K. Knag, and S. Franzen. 2005. Probing BSA binding to citrate-coated gold nanoparticles and surfaces. Langmuir. 21:9303–9307. [DOI] [PubMed] [Google Scholar]
- 20.Höök, F., M. Rodahl, B. Kasemo, and P. Brzezinski. 1998. Structural changes in hemoglobin during adsorption to solid surfaces: effects of pH, ionic strength, and ligand binding. Proc. Natl. Acad. Sci. USA (Biophysics). 95:12271–12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Höök, F., and B. Kasemo. 2001. Variations in coupled water, viscoelastic properties, and film thickness of a Mefp-1 protein film during adsorption and cross-linking: a quartz crystal microbalance with dissipation monitoring, ellipsometry, and surface plasmon resonance study. Anal. Chem. 73:5796–5804. [DOI] [PubMed] [Google Scholar]
- 22.Stengel, G., F. Höök, and W. Knoll. 2005. Viscoelastic modeling of template-directed DNA synthesis. Anal. Chem. 77:3709–3714. [DOI] [PubMed] [Google Scholar]
- 23.Morigaki, K., and K. Tawa. 2006. Vesicle fusion studied by surface plasmon resonance and surface plasmon fluorescence spectroscopy. Biophys. J. 91:1380–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svedhem, S., D. Dahlborg, J. Ekeroth, J. Kelly, F. Höök, and J. Gold. 2003. In situ peptide-modified supported lipid bilayers for controlled cell attachment. Langmuir. 19:6730–6736. [Google Scholar]
- 25.Larsson, C., M. Rodahl, and F. Höök. 2003. Characterization of DNA immobilization and subsequent hybridization on a 2D arrangement of streptavidin on a biotin-modified lipid bilayer supported on SiO2. Anal. Chem. 75:5080–5087. [DOI] [PubMed] [Google Scholar]
- 26.Su, X. D., Y. J. Wu, and W. Knoll. 2005. Comparison of surface plasmon resonance spectroscopy and quartz crystal microbalance techniques for studying DNA assembly and hybridization. Biosens. Bioelectron. 21:719–726. [DOI] [PubMed] [Google Scholar]
- 27.Reimhult, E., C. Larsson, B. Kasemo, and F. Höök. 2004. Simultaneous surface plasmon resonance and quartz crystal microbalance with dissipation monitoring measurements of biomolecular adsorption events involving structural transformations and variations in coupled water. Anal. Chem. 76:7211–7220. [DOI] [PubMed] [Google Scholar]
- 28.Zelander, G. 2006. QCM-D real-time monitoring of structural changes in an adsorbed protein layer. Nat. Methods. Application Notes:an41–an42.
- 29.Klein-Hitpass, L., G. U. Ryffel, E. Heitlinger, and A. B. C. Cato. 1998. A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res. 16:647–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su, X. D., R. Robelek, Y. J. Wu, and W. Knoll. 2004. Detection of point mutation and insertion mutations in DNA using a quartz crystal microbalance and MutS, a mismatch binding protein. Anal. Chem. 76:489–494. [DOI] [PubMed] [Google Scholar]
- 31.User Manual for Autolab ESPRIT, Version 2. 2002. Chapter 7, p. 105.
- 32.de Feijter, J. A., J. Benjamins, and F. A. Veer. 1978. Ellipsometry as a tool to study the adsorption behavior of synthetic and biopolymers at the air-water interface. Biopolymers. 17:1759–1772. [Google Scholar]
- 33.Voinova, M. V., M. Rodahl, M. Jonson, and B. Kasemo. 1999. Viscoelastic acoustic response of layered polymer films at fluid-solid interfaces: continuum mechanics approach. Phys. Scr. 59:391–396. [Google Scholar]
- 34.Höök, F., M. Rodahl, P. Brzezinski, and B. Kasemo. 1998. Energy dissipation kinetics for protein and antibody-antigen adsorption under shear oscillation on a quartz crystal microbalance. Langmuir. 14:729–734. [Google Scholar]
- 35.Zhou, C., J. M. Friedt, A. Angelova, K. H. Choi, W. Laureyn, F. Frederix, L. A. Francis, A. Campitelli, Y. Engelborghs, and G. Borghs. 2004. Human immunoglobin adsorption investigated by means of quartz crystal microbalance dissipation, atomic force microscopy and surface acoustic wave, and surface plasmon resonance techniques. Langmuir. 20:5870–5878. [DOI] [PubMed] [Google Scholar]
- 36.Schwabe, J. W. R., L. C. Chapman, J. T. Finch, D. Rhodes, and D. Neuhaus. 1993. DNA recognition by the oestrogen receptor: from solution to the crystal. Structure. 15:187–204. [DOI] [PubMed] [Google Scholar]
- 37.Greenfield, N., V. Vijayanathan, T. J. Thomas, M. A. Gallo, and T. Thomas. 2001. Increase in the stability and helical content of estrogen receptor α in the presence of the estrogen response elements: analysis by circular dichroism spectroscopy. Biochemistry. 40:6646–6652. [DOI] [PubMed] [Google Scholar]
- 38.Nardulli, A. M., G. L. Greene, and D. J. Shapiro. 1993. Human estrogen receptor bound to an estrogen response element bends DNA. Mol. Endocrinol. 7:331–340. [DOI] [PubMed] [Google Scholar]
- 39.Sabbah, M., S. L. Ricousse, G. Redeuilh, and E. E. Baulieu. 1992. Estrogen receptor-induced bending of the Xenopus vitellogenin A2 gene hormone response element. Biochem. Biophys. Res. Commun. 185:944–952. [DOI] [PubMed] [Google Scholar]
- 40.Günther, S., K. Rother, and C. Frömmel. 2006. Molecular flexibility in protein-DNA interactions. Biosystems. 85:126–136. [DOI] [PubMed] [Google Scholar]
- 41.Spolar, R. S., and M. T. Jr. Record. 1994. Coupling of local folding to site-specific binding of proteins to DNA. Science. 263:777–784. [DOI] [PubMed] [Google Scholar]
- 42.Kalodimos, C. G., N. Byres, A. M. J. J. Bonvin, M. M. Levandoski, M. Guennuegues, R. Boelens, and R. Kaptein. 2004. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 305:386–398. [DOI] [PubMed] [Google Scholar]
- 43.Ozers, M. S., J. J. Hill, K. Ervin, J. R. Wood, A. M. Nardulli, C. A. Royer, and J. Gorski. 1997. Equilibrium binding of estrogen receptor with DNA using fluorescence anisotropy. J. Biol. Chem. 272:30405–30411. [DOI] [PubMed] [Google Scholar]
- 44.Kosztin, D., T. C. Bishop, and K. Schulten. 1997. Binding of the estrogen receptor to DNA. The role of waters. Biophys. J. 73:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Höök, M., J. Rodahl, R. Vörös, P. Kurrat, J. J. Böni, M. Ramsden, N. D. Textor, P. Spencer, J. Tengvall, B. Gold, and B. Kasemo. 2002. A comparative study of protein adsorption on titanium oxide surfaces using in situ ellipsometry, optical waveguide lightmode spectroscopy, and the quartz crystal microbalance/dissipation. Colloids Surf. B Biointerfaces. 24:155–170. [Google Scholar]
- 46.Teh, H. F., W. Y. X. Peh, X. D. Su, and J. S. Thomsen. 2006. Characterization of protein-DNA interactions using surface plasmon resonance spectroscopy with various assay schemes. Biochemistry. 46:2127–2135. [DOI] [PubMed] [Google Scholar]
- 47.Boyer, M., N. Poujol, E. Margeat, and C. A. Royer. 2000. Quantitative characterization of the interaction between purified human estrogen receptor α and DNA using fluorescence anisotropy. Nucleic Acids Res. 28:2494–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margeat, E., A. Bourdoncle, R. Margueron, N. Poujol, V. Cavaillès, and C. A. Royer. 2003. Ligands differentially modulate the protein interactions of the human estrogen receptors α and β. J. Mol. Biol. 326:77–92. [DOI] [PubMed] [Google Scholar]