Abstract
Plant species have evolved a wide variety of flowering habits, each adapted to maximize reproductive success in their local environment. Even within a species, accessions from different environments can exhibit markedly different flowering behavior. In Arabidopsis, some accessions are rapid-cycling summer annuals, whereas others accessions are late flowering and vernalization responsive and thus behave as winter annuals. Two genes, FLOWERING LOCUS C (FLC) and FRIGIDA (FRI), interact synergistically to confer the winter-annual habit. Previous work has shown that many summer-annual accessions contain null mutations in the FRI gene; thus it appears that these summer-annual accessions have arisen from winter-annual ancestors by losing FRI function. In this work we demonstrate that naturally occurring allelic variation in FLC has provided another route to the evolution of summer-annual flowering behavior in Arabidopsis. We have identified two summer-annual accessions, Da (1)-12 and Shakhdara, that contain functional alleles of FRI, but are early flowering because of weak alleles of FLC. We have also determined that the weak allele of FLC found in Landsberg erecta is naturally occurring. Unlike accessions that have arisen because of loss-of-function mutations in FRI, the FLC alleles from Da (1)-12, Shakhdara, and Landsberg erecta are not nulls; however, they exhibit lower steady-state mRNA levels than strong alleles of FLC. Sequence analysis indicates that these weak alleles of FLC have arisen independently at least twice during the course of evolution.
Keywords: FRIGIDA, vernalization, winter annual, natural variation
Identification of the genetic mechanisms that underlie differences in life history is key to understanding the evolution of adaptive traits. Many plants species occur over a wide range of latitudes, and within a species, accessions from different regions can differ substantially in flowering behavior (1). As an adaptation to seasonal fluctuations in temperature experienced in temperate climates, flowering in many species is promoted by prolonged exposure to cold temperatures (vernalization) (2). Many accessions of Arabidopsis from temperate climates are relatively late flowering unless vernalized (3). This vernalization requirement prevents flowering before winter and promotes rapid flowering in the spring, and thus these accessions behave as winter annuals. Summer-annual Arabidopsis accessions, in contrast, flower rapidly in the absence of vernalization, and these accessions predominate in warmer climates that do not experience the cold temperatures required for vernalization (3).
The late-flowering phenotype of winter-annual accessions of Arabidopsis is created by the interaction of two genes, FLOWERING LOCUS C (FLC) and FRIGIDA (FRI) (2). FLC encodes a MADS domain-containing transcription factor whose expression is sufficient to inhibit flowering (4, 5); however, the native FLC gene is only expressed to high levels in the presence of FRI, which encodes a plant-specific gene of unknown biochemical activity (3). Thus, loss-of-function mutations in either gene eliminate the late-flowering phenotype. Vernalization promotes flowering in winter-annual strains of Arabidopsis by causing an epigenetic down-regulation of FLC (4, 5).
Genetic analysis of winter- and summer-annual accessions has revealed that variation at the FRI locus was a common determinate of flowering habit (6–9). Subsequent sequence analysis of the FRI gene from summer-annual accessions of Arabidopsis revealed that many contained lesions that create null alleles of FRI (3, 10). Thus, it appears that these summer-annual accessions were derived from winter-annual ancestors by losing FRI activity, and that FRI activity was lost multiple times during the course of evolution.
Here we describe a second mechanism by which the vernalization requirement of winter-annual Arabidopsis has been lost to produce a summer-annual flowering habit. We have identified summer-annual accessions of Arabidopsis that contain active alleles of FRI, but are early flowering because of novel weak alleles of FLC. Unlike the null alleles of FRI present in most summer-annual accessions, these alleles of FLC are not nulls; they are expressed and contain no nonsense or missense mutations. These alleles, however, cannot be effectively up-regulated by FRI.
Materials and Methods
Plant Materials. Arabidopsis accessions were obtained from the Arabidopsis Biological Resource Center: Abd-0 (CS932), Bla-2 (CS6194), Cnt-1 (CS1635), Co (CS3180), Da (1)-12(CS917) Kondara (CS916), Di-G (CS910), Di-M (CS919), Ema-1 (CS1637), ENF (CS8141), En-D (CS920), En-T (CS921), Est (CS911), Gr3 (CS3179), H55 (CS923), Je54 (CS924), Li5 (CS3178), Limeport (CS8070), LIN (CS8144), Litva (CS925), M3385S (CS3111), Nd-1 (CS1636), Oy-1 (CS1643), Petergof (CS926), Santa Clara (CS8069), Shakhdara (CS929), and Wei-0 (CS3110). FRI-SF2 in the Columbia (Col) background, FRI-SF2 in the Landsberg erecta (Ler) background, and FLC-Col in the Ler background have been described (11). FRI-SF2/FLC-Col in the Ler background was obtained from an F2 population generated by crossing FRI-SF2 in Ler and FLC-Col in Ler. flc-2, flc-3, FRI-SF2/flc-3 (4), fpa-7/flc-3, and ld-1/flc-3 (12) are all in the Col background and have also been described. LER was kindly provided by Maarten Koornneef.
Plant Growth Conditions. All plants were grown at 22°C under long-days consisting of 16 h of cool-white fluorescent light followed by 8 h of darkness. For experiments involving vernalization, seeds plated on agar-solidified medium containing 0.65 g/liter Peters Excel 15-5-15 fertilizer (Grace Sierra, Milpitas, CA) and were kept at room temperature overnight to allow seeds to become metabolically active before being transferred to 2°C for 50 days. During cold treatment, samples were kept under short-day conditions (8 h light/16 h dark).
RNA-Blot Analysis. Total RNA was isolated by using RNA Isolator (Genosys Biotechnologies, The Woodlands, TX) according to the manufacturer's instructions. For RNA blots, 15 μg of RNA was separated by denaturing-formaldehyde-agarose gel electrophoresis as described (13). RNA blots were probed with a deoxyadenosine 5′-[α-32P]triphosphate-labeled cDNA fragment that did not contain the conserved MADS-box domains of FLC. Blots were also probed with an 18S rRNA probe as a control for the quantity of RNA loaded.
FLC Constructs. The full-length FLC clone consisted of an 8.1-kb Col genomic fragment containing 1.7 kb 5′ of the start site of translation and 0.5 kb 3′ of the translational stop in the binary vector pPZP211 (14). Promoter/coding region chimerics were created by using an NcoI site that occurs at the start site of translation of FLC. Sequences 5′ or 3′ of the NcoI site were PCR amplified from Ler, Da (1)-12, and Shakhdara and exchanged with the corresponding fragment in the full-length FLC clone from Col. Chimeric full-length Col FLC clones containing the 30-bp deletion or the 1.2-kb insertion from Ler were created by exchanging either an EcoRI/EcoRV fragment containing the 30-bp deletion or a AfeI/SphI fragment containing the 1.2-kb insertion with the corresponding Col fragments. Constructs were verified by sequencing.
Results and Discussion
Identification of Candidate Accessions with Weak Alleles of FLC. The late-flowering vernalization-responsive habit of winter-annual Arabidopsis requires active alleles of both FRI and FLC. Recent work with naturally occurring early-flowering accessions has shown that most contain null alleles of FRI, suggesting that summer-annual accessions arose from winter annuals by loss of FRI function (3). In the laboratory, however, we have shown that loss-of-function mutations in either FRI or FLC results in an equivalent early-flowering phenotype (4). This led us to investigate whether any naturally occurring summer-annual accessions have arisen because of natural allelic variation at FLC. FRI and FLC function was evaluated in 27 randomly chosen accessions by crossing to early-flowering tester lines containing an SF-2 allele of FRI with a null allele of flc (FRI/FRI;flc-3/flc-3 in the Col background) or FLC with a null allele of fri (fri/fri;FLC/FLC, which is wild-type Col). (The FRI allele from the winter-annual accession SF-2 and the FLC allele from Col are considered to be active alleles and are used as reference alleles in the evaluation of FRI and FLC alleles from other accessions.) Accessions containing functional FLC alleles will give rise to late-flowering F1 plants when crossed to the FRI-containing tester; likewise, accessions containing functional FRI alleles will give rise to late-flowering F1 plants when crossed to the FLC-containing tester.
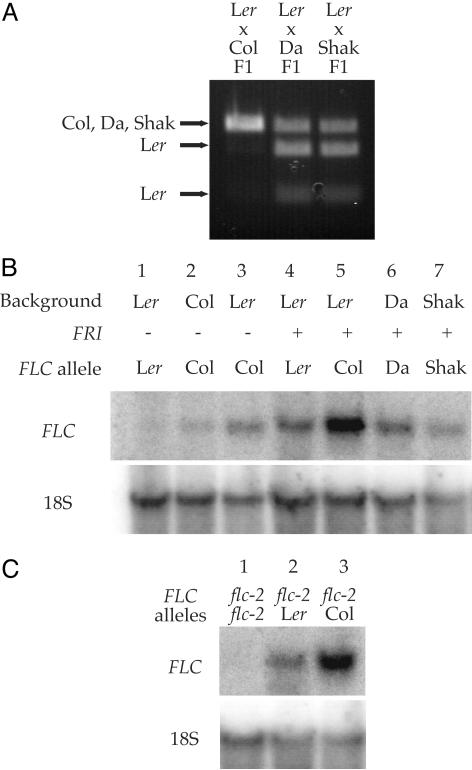
The majority of accessions (Abd-0, Bla-2, Cnt-1, Co, Di-G, Di-M, Ema-1, ENF, En-D, En-T, Est, Gr3, H55, Je54, Li5, Limeport, LIN, Litva, M3385S, Nd-1, Oy-1, Petergof, Santa Clara, and Wei-0) gave rise to late-flowering F1 progeny only when crossed to the FRI-containing tester, indicating that these accessions likely contain functional alleles of FLC and nonfunctional alleles of FRI. Kondara, Da (1)-12, and Shakhdara, however, only gave rise to late-flowering progeny in crosses to the FLC-containing tester (Fig. 1A), indicating that these lines may contain weak or nonfunctional alleles of FLC and functional alleles of FRI. (Independent work using recombinant inbred lines has also suggested that Shakhdara contains a weak allele of FLC and a functional allele of FRI; ref. 15) Consistent with these accessions containing functional alleles of FRI, none of the lesions previously described in FRI genes from naturally occurring summer-annual accessions are present in Kondara, Shakhdara (3) or Da (1)-12 (data not shown). To confirm that the late-flowering phenotype observed in crosses to the FLC-containing tester was caused by the restoration of FLC activity, Kondara, Da (1)-12 and Shakhdara were crossed to a tester line with null alleles of both flc and fri (fri/fri;flc-3/flc-3 in Col). F1 progeny of this cross were early flowering (Fig. 1 A), indicating that the late flowering observed in crosses to the FLC-containing tester is indeed caused by the restoration of FLC activity in the F1.
Fig. 1.
Determination of FRI and FLC activity in early-flowering accessions. (A) Da (1)-12 (open bars), Shakhdara (gray bars), and Kondara (cross-hatched bars) were crossed to tester lines homozygous for the indicated genotypes, and the rosette leaf numbers of the resulting F1 plants are presented. A line homozygous for FRI and flc-3 was used as a control (black bars). (B)Ler (black bars) and LER (open bars) were crossed to tester lines homozygous for the indicated genotypes, and the rosette leaf numbers of the resulting F1 plants are presented.
Among the accessions containing weaker alleles of FLC, Kondara formed approximately twice as many rosette leaves before flowering as Da (1)-12 and Shakhdara. One possible explanation for the later flowering of Kondara is that it contains an FLC allele that is intermediate in strength relative to the strong Col allele and the weak Da (1)-12 and Shakhdara alleles. Because Da (1)-12 and Shakhdara were likely to have most severely reduced FLC function, these accessions were chosen for further analysis. To determine whether Da (1)-12 and Shakhdara are closely related accessions, PCR was performed by using 12 simple sequence length polymorphism markers that detect polymorphisms between Col and Ler (16). Da (1)-12 and Shakhdara showed polymorphisms for 8 of the 12 markers tested (data not shown), indicating that these two accessions are not closely related.
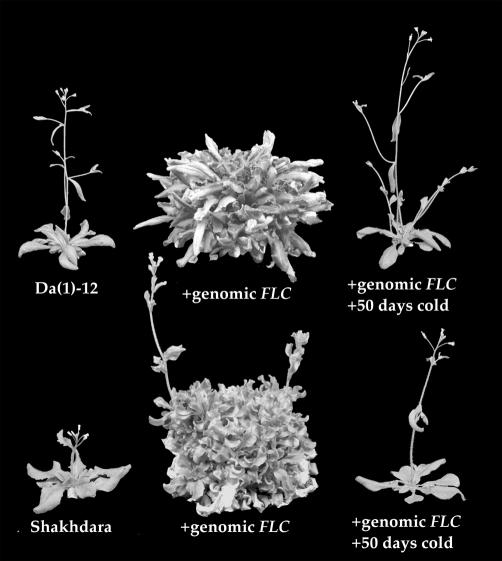
Restoration of FLC Activity Is Sufficient to Confer Winter-Annual Behavior in Da (1)-12 and Shakhdara. If the summer-annual strains Da (1)-12 and Shakhdara arose from winter-annual ancestors by an attenuation of FLC function, then introducing a functional allele of FLC into these accession should restore the winter-annual habit. To test this hypothesis, Da (1)-12 and Shakhdara were transformed with a construct containing the FLC genomic region from Col (4). The majority of the resulting T1 plants were late flowering and vernalization responsive (Fig. 2). Thus, transformation with an active allele of FLC is able to convert Da (1)-12 and Shakhdara from summer to winter annuals.
Fig. 2.
Transformation with a genomic FLC construct is sufficient to transform Da (1)-12 and Shakhdara from summer annuals to winter annuals.
Ler Contains a Naturally Occurring Weak Allele of FLC. FLC was first identified in crosses of late-flowering lines containing FRI or mutations in the autonomous-pathway gene LUMENIDEPENDENS to the Ler strain of Arabidopsis (11, 17). Ler contains a weak allele of FLC that is able to partially suppress the late-flowering phenotype of FRI and autonomous-pathway mutants (18). Thus, the Ler allele of FLC behaves similarly to the FLC alleles of Da (1)-12 and Shakhdara in that it is able to suppress the late-flowering phenotype of FRI. Ler, however, was isolated from a mutagenized population of heterogeneous plants (19) and it was therefore unclear whether the weak allele of FLC present in Ler is naturally occurring or the result of mutagenesis.
To address the origin of the FLC allele present in Ler, we crossed Ler and a line thought to be the wild-type, unmutagenized parent of Ler (LER) to the same set of tester lines used above for the analysis of Da (1)-12 and Shakhdara. If the weak allele of FLC found in Ler is naturally occurring, then both lines should behave similarly in their interaction with FRI; however, if it is the result of mutagenesis, then the wild-type parent of Ler should contain a stronger allele of FLC. In all crosses the LER parent behaved similarly to Ler (Fig. 1B), demonstrating that both lines contain weak alleles of FLC. Thus, the weak allele of FLC present in Ler is naturally occurring and not the result of mutagenesis. To confirm that LER was in fact the parent of Ler, both lines were tested with 12 highly polymorphic sequence length polymorphism markers. If LER is the Ler parent, the markers from both lines should be identical in size. Indeed, for all 12 markers, LER and Ler gave identical products, indicating that these strains are closely related. This result is consistent with previous amplified fragment length polymorphism analysis performed on LER and Ler (20).
The Da (1)-12, Shakhdara, and Ler Alleles of FLC Do Not Contain Missense or Nonsense Mutations. As discussed above, summer-annual Arabidopsis accessions with null alleles of FRI have been previously described. To determine whether the weak alleles of FLC present in naturally occurring accessions are likewise the result of loss-of-function mutations, the FLC cDNA sequence was determined for Da (1)-12, Shakhdara, and Ler. As has been previously reported, Ler contains no missense or nonsense mutations in FLC (21) when compared with the Col sequence. We found that Ler contains only a single nucleotide difference located 369 bp 3′ of the start of translation (Col:ACACCTT; Ler:ACATCTT). The cDNA sequences from Da (1)-12 and Shakhdara were identical to Col throughout the coding region. Thus, the weak alleles of FLC present in Da (1)-12, Shakhdara, and Ler are not caused by alterations in the coding regions of FLC.
FLC Alleles from Da (1)-12, Shakhdara, and Ler Result in Lower Steady-State Levels of FLC RNA. FLC steady-state mRNA levels are closely correlated with flowering time (4, 5). FLC expression is low in early-flowering summer-annual accessions and is upregulated by FRI in winter annuals. Vernalization of winter-annual accessions causes in an epigenetic reduction of FLC levels and results in early flowering. Because the regulation of FLC levels plays a critical role in the regulation of flowering time, we investigated whether the weak FLC alleles of Da (1)-12, Shakhdara and Ler might exhibit reduced steady-state mRNA levels.
The expression levels of the Ler allele of FLC were initially compared with the alleles from Da (1)-12, Shakhdara, and Col by crossing Ler to Da (1)-12, Shakhdara, and Col and examining FLC expression in the resulting F1 plants by allele-specific RT-PCR. The coding regions of the four alleles are identical except for a single base change in Ler that creates a unique Hpy188III site. By performing RT-PCR with primers that flank the base change and digesting the product with Hpy188III, expression of the Ler allele can be distinguished from the Da (1)-12, Shakhdara, and Col alleles in the F1 plants. This approach has the advantage that both FLC alleles are in an identical F1 genetic background; thus, differences in expression must be caused by locus-specific effects. In the F1 generated from a Ler/Col cross the Col allele is expressed at much higher levels than the Ler allele (Fig. 3A). In contrast, the Ler, Da (1)-12, Shakhdara alleles are expressed to similar levels in the F1 progeny. Thus, the strong allele of Col is expressed to higher levels than the weak alleles from Ler, Da (1)-12, and Shakhdara.
Fig. 3.
Analysis of FLC mRNA levels. (A) Allele-specific RT-PCR was used to examine the relative expression levels of different FLC alleles in F1 plants resulting from the indicated crosses. RT-PCR products were digested with Hpy188III, resolved on a 1.5% agarose gel, and visualized by staining with ethidium bromide. A unique Hpy188III site in the Ler RT-PCR product allows the Ler product to be separated from the Col, Da (1)-12, and Shakhdara products. (B) RNA blot analysis of FLC expression in Ler, Col, Da (1)-12, and Shakhdara, and in near isogenic lines containing FRI, FLC-Col, or FRI and FLC-Col in the Ler genetic background. (C) RNA blot analysis of FLC expression in F1 plants resulting from crosses between a line containing FRI and flc-2 in the Col background and Ler or a near isogenic line containing FLC-Col in the Ler genetic background. The flc-2 allele of FLC is a large deletion that completely removes the FLC gene.
To further investigate the relationship between FLC allele strength and steady-state mRNA levels, RNA blot analysis was also performed on Ler, Da (1)-12, Shakhdara and Col, and near isogenic lines in the Ler background (see Methods) containing either FRI-SF2, FLC-Col, or both FRI-SF2 and FLC-Col (Fig. 3B). As with the allele-specific RT-PCR, RNA blot analysis indicates that the weak alleles of FLC are expressed to lower levels than the strong Col allele in similar genetic backgrounds. As shown previously, in the near isogenic lines, the Ler allele of FLC is expressed to lower levels than the Col allele both in the absence (Fig. 3B lanes 1 and 3) or presence of FRI (Fig. 3b lanes 4 and 5) (5, 22). Consistent with Da (1)-12 and Shakhdara having strong alleles of FRI and weak alleles of FLC, Da (1)-12 and Shakhdara show levels of FLC expression similar to the near isogenic line containing FRI-SF2 in the Ler background (Fig. 3B lanes 4, 6, and 7). Ler and the near isogenic line containing FLC-Col were also crossed to a line containing FRI-SF2 and flc-2 in the Col background and FLC expression was examined by RNA blot analysis (Fig. 3C). flc-2 is a deletion allele that completely eliminates the FLC gene. Thus, F1 plants are hemizygous for either the Ler or Col allele of FLC. In this case, the Col allele of FLC is also expressed to higher levels than the Ler allele.
Sequences Downstream of the Start Site of Translation Are Responsible for the Weak Nature of the Ler, Da, and Shakhdara FLC Alleles. Because the weak nature of the Ler, Da (1)-12, and Shakhdara alleles of FLC is caused by reduced expression rather than changes in the coding region of the gene, it was of interest to localize the region of the gene responsible for the relatively low level of expression. The translational start site of FLC is located in an NcoI restriction enzyme site. This NcoI site was used to create chimeric genomic constructs containing the Col 5′ upstream region fused to the coding regions from Ler, Da (1)-12, and Shakhdara and, conversely, constructs containing the 5′ upstream regions from Ler, Da (1)-12, and Shakhdara fused to the Col coding region. The constructs were used to transform a line homozygous for FRI-SF2 and flc-3 in the Col background, and the flowering time of the resulting T1 plants was determined (Table 1). All three of the weak alleles of FLC behaved similarly in the chimeric constructs. Constructs containing the Col 5′ upstream region fused to the Ler, Da (1)-12, and Shakhdara coding regions were early flowering, whereas constructs containing the 5′ upstream regions from Ler, Da (1)-12, and Shakhdara fused to the Col coding region were late flowering. Thus, the weak nature of the Ler, Da (1)-12, and Shakhdara FLC allele is caused by sequences downstream of the start site of translation.
Table 1. Analysis of chimeric FLC constructs.
| Promoter | Coding | T1 flowering time* |
|---|---|---|
| Col | Col | 55.5 (13.2) |
| Col | Ler | 11.0 (0.8) |
| Col | Da | 11.0 (1.1) |
| Col | Shahkdara | 12.4 (1.7) |
| Ler | Col | 50.9 (12.2) |
| Shahkdara | Col | 51.7 (15.8) |
| Da | Col | 48.1 (19.8) |
Rosette leaves formed before flowering (SD).
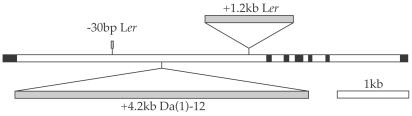
The Weak Behavior of the Ler allele of FLC Is Caused By an Insertion in the First Intron. The ability of the Ler allele of FLC to suppress the late-flowering vernalization-responsive phenotype of FRI and autonomous-pathway mutations has been extensively characterized (11, 17, 18). Thus, it was of interest to identify the region downstream of the start codon that was responsible for the weak nature of the Ler allele. The first intron of FLC was chosen as a candidate region and was sequenced from Ler (GenBank accession AY303833). Like many MADS-box transcription factors in Arabidopsis, the Col allele of FLC has a small first exon (180 bp) and a relatively large first intron (3.5 kb). Two major differences were observed between the Ler and Col intron I sequences (Fig. 4). First, Col contains a direct repeat of the 30-bp sequence 5′-CACAACCTTTGTATCTCGTGTCTTTTGTCA-3′, whereas Ler contains only a single copy of this element. Second, Ler contains an insertion near the 3′ end of the intron, 5,236 bp from the translational start site. The 1,224-bp insertion is defined by imperfect inverted repeats and is flanked by an apparent 9-bp target site duplication. These structural features suggest that this element represents a miniature inverted repeat transposable element (MITE) (23, 24), which are nonautonomous Class II transposable elements (24, 25). Elements related to the Ler insertion are found on all five chromosomes of the Col genome. Related elements on bacterial artificial chromosomes T32N4, MBK23, and F24H14 are >90% identical to the Ler insertion, and ≈15 other sequences exhibit a clear degree of relatedness.
Fig. 4.
Summary of the polymorphisms detected in the first intron of FLC in Ler and Da (1)-12. The structure of the Col allele of FLC is shown with exons as black boxes and introns as open boxes. Ler and Da (1)-12 insertions and the Ler 30-bp deletion are shown as gray boxes.
To determine whether either of these polymorphisms is responsible for the weak nature of the Ler FLC allele, a Col genomic FLC clone was engineered to contain either the 30-bp deletion or the 1.2-kb insertion from Ler. To assay FLC activity of the constructs, they were transformed into a line homozygous for FRI and flc-3 (FRI/FRI;flc-3/flc-3) and the flowering time of the resulting T1 plants was determined (Fig. 5). The untransformed FRI/FRI;flc-3/flc-3 line is early flowering because of the lack of FLC activity (Fig. 5A). Transformation with the wild-type Col genomic FLC clone or the chimeric clone containing the 30-bp deletion from Ler (Fig. 5 B and C) gave rise to a similar range of late-flowering plants. Thus, the 30-bp deletion does not have a significant effect on FLC activity. Transformed plants containing the 1.2-kb insertion, however, were uniformly early flowering indicating that the 1.2-kb insertion is sufficient to dramatically reduce FLC function (Fig. 5D).
Fig. 5.
Effect of Ler polymorphisms on FLC activity in a FRI-containing or autonomous-pathway-mutant background. The number of rosette leaves formed by the primary meristem was determined for a line homozygous for FRI and flc-3 in the Col background alone (A) or after transformation with a wild-type Col FLC genomic construct (B), a Col FLC genomic construct containing the 30-bp deletion found in Ler (C), or a Col FLC genomic construct containing the 1.2-kb insertion found in Ler (D). In addition, rosette leaves formed before flowering were determined for a line homozygous for fpa-7 and flc-3 either alone (E) or after transformation with a wild-type Col FLC genomic construct (F) or a Col FLC genomic construct containing the 1.2-kb insertion found in Ler (G).
Because the Ler allele of FLC has been shown to suppress the late-flowering phenotype of both FRI and autonomous-pathway mutations, we also tested the affect of the 1.2-kb insertion in the autonomous-pathway-mutant fpa-7. A line homozygous for fpa-7 and flc-3 was transformed with the wild-type Col genomic FLC clone or the chimeric clone containing the 1.2-kb insertion from Ler, and the flowering time of the resulting T1 plants was determined. Similar to the FRI/flc-3 line, the fpa-7/fpa-7;flc-3/flc-3 line is early flowering because of the lack of FLC activity (Fig. 5E) (12). Late flowering is restored by the wild-type Col genomic clone but not by the chimeric clone containing the 1.2-kb insertion from Ler. Similar results were also obtained in the ld-1 autonomous-pathway mutant background (data not shown). Thus, the 1.2-kb insertion present in Ler is sufficient to dramatically reduce the activity of the Col allele of FLC in either a FRI-containing line or an autonomous-pathway mutant background.
It is interesting to note that although FRI and autonomouspathway mutations are modestly late flowering in the Ler background, the chimeric Col clone containing the 1.2-kb insertion from Ler has little effect on flowering time in the FRI, fpa, or ld backgrounds. Thus, the chimeric clone appears to be weaker than the Ler allele of FLC. One possible explanation for this observation is that, in addition to the 1.2-kb insertion, which has a negative impact on FLC expression, the Ler allele of FLC may also contain other sequence differences relative to Col that enhance FLC expression. Such polymorphisms may allow partial activity in the Ler FLC allele despite the 1.2-kb insertion event.
The 1.2-kb Insertion Responsible for the Weak Nature of the Ler Allele of FLC Is Not Found in Da (1)-12 or Shakhdara. Because Da (1)-12 and Shakhdara contain FLC alleles that behave similarly to the Ler allele and, like Ler, the sequences that confer the weak behavior are located downstream of the translational start site, it was of interest to determine whether Da (1)-12 or Shakhdara might contain Ler-like insertions in the first intron of FLC. Thus, the first intron of FLC from Da (1)-12 and Shakhdara was amplified, sequenced, and compared with the sequences from Ler and Col. Neither Da (1)-12 nor Shakhdara (GenBank accession nos. AY303834 and AY303835, respectively) contains an insertion event in the region where the 1.2-kb Ler insertion is found; in this region the Da (1)-12 and Shakhdara sequences are similar to Col. Thus, the sequences responsible for the weak behavior of the FLC alleles from Da (1)-12 and Shakhdara appear to be different from that of the Ler allele. Shakhdara did not contain any large insertions or deletions; however, Da (1)-12 contains a 4.2-kb insertion in approximately the middle of the first intron of FLC, 4,009 bp from the translational start site (Fig. 4). The sequence of the insertion in Da (1)-12 is unrelated to the 1.2-kb insertion in Ler and exhibits similarity to copia-type retrotransposons (26). Two similar sequences are present in the Col genome on BACs F9O13 and F8A12 that are >95% identical to the insertion in Da (1)-12. Given that the 1.2-kb insertion found in Ler is sufficient to strongly inhibit FLC function, perhaps this 4.2-kb insertion has a similar attenuating effect on the Da (1)-12 allele of FLC.
In addition to Da (1)-12 and Shakhdara, the C24 accession of Arabidopsis has also been reported to contain a functional FRI allele and a weak or nonfunctional FLC allele (27). The sequence and expression of the C24 allele of FLC has been determined (5), and it is worth noting that, like Shakhdara, it is expressed at relatively low levels and does not contain any large insertions or deletions relative to Col (the one exception is that the C24 allele contains a 30-bp deletion in the first intron of FLC that is identical to that found in the Ler). Whether C24 should be considered a naturally occurring accession, however, is unclear. Its origin is unknown and the glabrous phenotype of C24 suggests that it may have been isolated from a mutagenized population (27).
To date, four accessions that contain weak alleles of FLC have been identified. Ler and Da (1)-12 contain insertion events, whereas Shakhdara and C24 do not contain any large insertions or deletions relative to the Col allele. How these polymorphisms lead to FLC attenuation is not known. One possibility is that certain polymorphisms lead to general defects in RNA processing. For example the large insertions in Ler and Da (1)-12 may reduce splicing efficiency. Another possibility is that polymorphisms affect cis-acting regulatory elements that are required for high levels of FLC expression.
Evolution of Weak Alleles of FLC. Analysis of the intron I sequences of the Ler, Da (1)-12, and Shakhdara alleles of FLC provides evidence that attenuation of FLC activity has occurred at least twice during evolution. Because Da (1)-12 and Shakhdara do not contain the 1.2-kb insertion responsible for the weak nature of the Ler allele of FLC, they likely arose independently from the Ler allele. The Da (1)-12 and Shakhdara alleles of FLC may have also arisen independently from each other. The 4.2-kb insertion present in Da (1)-12 is not present in Shakhdara; further Da (1)-12 and Shakhdara do not have any sequence polymorphisms in common in intron I. Thus, it is possible that the Ler, Da (1)-12, and Shakhdara alleles of FLC have all arisen independently.
In is interesting to note that, in contrast to summer-annual accessions that have arisen by FRI loss-of-function mutations, the FLC alleles from Ler, Da (1)-12, and Shakhdara are not nulls; they contain no nonsense or missense codons and have detectable levels of transcription. The explanation of why the sequence changes responsible for the weak nature of these FLC alleles have arisen in the noncoding regions of the gene may be simply statistical; the six introns of FLC account for 89.5% of the genomic sequence of the gene. Thus, it is much more likely that a mutation would take place in an intron rather than an exon. Another possibility, however, is that a total loss of FLC activity is detrimental. FLC plays a central role in the regulation of flowering time in Arabidopsis and is positively regulated by FRI, negatively regulated by the autonomous floral promotion pathway, and is epigenetically down-regulated by vernalization (2). FLC may also have roles other than the regulation of flowering time. For example, it has been shown that allelic variation at the FLC locus is strongly associated with circadian period in quantitative trait locus mapping studies using recombinant inbred lines derived from a Ler/Col cross, and that flc loss-of-function mutations have shorter circadian periods (28).
The work presented here, along with previous work, indicates that the vernalization requirement of winter-annual accessions of Arabidopsis has been lost several times during evolution by either loss-of-function mutations in FRI or reduction of FLC function. It is interesting to note, however, that among the summer-annual accessions thus far examined, Ler is unique in that it contains both a weak allele of FLC and a null allele of FRI. Two other accessions, Dijon and Gr, contain Ler-like lesions in FRI (3), but contain strong alleles of FLC (this work). Thus, it appears that FRI function was lost in a common ancestor of Ler, Dijon, and Gr, and that FLC activity was subsequently attenuated in the lineage that gave rise to Ler. Presumably, during the course of evolution, there was a pressure for earlier flowering that selected for the loss of FRI activity. The same evolutionary pressure may have also selected for a reduction of FLC activity. A near isogenic line containing the Col allele of FLC in the Ler background flowers after forming approximately six more rosette leaves than Ler (11). Thus, if Ler originally contained a Col-like FLC allele, the attenuation of FLC activity in a fri background would have resulted in a significant additional acceleration of flowering time.
Acknowledgments
We thank William R. Belknap for discussion of the nature of the transposable element in the FLC-Ler intron and Mark Doyle for comments on the manuscript. This work was supported by the College of Agricultural and Life Sciences and the Graduate School of the University of Wisconsin, the U.S. Department of Agriculture National Research Initiative Competitive Grants Program, and National Science Foundation Grant 0133663 (to R.M.A.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Col, Columbia; Ler, Landsberg erecta.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY303833–AY303835).
References
- 1.Cumming, B. G. (1969) Can. J. Bot. 41, 901–926. [Google Scholar]
- 2.Michaels, S. & Amasino, R. (2000) Plant Cell Environ. 23, 1145–1154. [Google Scholar]
- 3.Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R. & Dean, C. (2000) Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- 4.Michaels, S. & Amasino, R. (1999) Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J. & Dennis, E. S. (1999) Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Napp-Zinn, K. (1979) in La Physiologie de la Floraison, eds. Champagnat, P. & Jaques, R. (Centre National de la Recherche Scientifique, Paris), pp. 217–220.
- 7.Clarke, J. H. & Dean, C. (1994) Mol. Gen. Genet. 242, 81–89. [DOI] [PubMed] [Google Scholar]
- 8.Burn, J. E., Smyth, D. R., Peacock, W. J. & Dennis, E. S. (1993) Genetica 90, 147–155. [Google Scholar]
- 9.Lee, I., Bleecker, A. & Amasino, R. (1993) Mol. Gen. Genet. 237, 171–176. [DOI] [PubMed] [Google Scholar]
- 10.Corre, V. L., Roux, F. & Reboud, X. (2002) Mol. Biol. Evol. 19, 1261–1271. [DOI] [PubMed] [Google Scholar]
- 11.Lee, I., Michaels, S. D., Masshardt, A. S. & Amasino, R. M. (1994) Plant J. 6, 903–909. [Google Scholar]
- 12.Michaels, S. D. & Amasino, R. M. (2001) Plant Cell 13, 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 14.Hajdukiewicz, P., Svab, Z. & Maliga, P. (1994) Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- 15.Loudet, O., Chaillou, S., Camilleri, C., Bouchez, D. & Daniel-Vedele, F. (2002) Theor. Appl. Genet. 104, 1173–1184. [DOI] [PubMed] [Google Scholar]
- 16.Bell, C. J. & Ecker, J. R. (1994) Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- 17.Koornneef, M., Blankestijn-de Vries, H., Hanhart, C., Soppe, W. & Peeters, T. (1994) Plant J. 6, 911–919. [Google Scholar]
- 18.Sanda, S. L. & Amasino, R. M. (1996) Mol. Gen. Genet. 251, 69–74. [DOI] [PubMed] [Google Scholar]
- 19.Redei, G. P. (1992) in Methods in Arabidopsis Research, eds. Koncz, C., Chua, N. H. & Schell, J. (World Scientific, Singapore), pp. 3–15.
- 20.Fransz, P., Armstrong, S., Alonso-Blanco, C., Fischer, T., Torres-Ruiz, R. & Jones, G. (1998) Plant J. 13, 867–876. [DOI] [PubMed] [Google Scholar]
- 21.Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J. & Dennis, E. S. (2000) Proc. Natl. Acad. Sci. USA 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlappi, M. (2001) Int. J. Plant Sci. 162, 527–537. [Google Scholar]
- 23.Bureau, T. E., Ronald, P. C. & Wessler, S. R. (1996) Proc. Natl. Acad. Sci. USA 93, 8524–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang, G. & Hall, T. C. (2003) J. Mol. Evol. 56, 255–264. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, X., Feschotte, C., Zhang, Q., Jiang, N., Eggleston, W. B. & Wessler, S. R. (2001) Proc. Natl. Acad. Sci. USA 98, 12572–12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voytas, D. F., Cummings, M. P., Koniczny, A., Ausubel, F. M. & Rodermel, S. R. (1992) Proc. Natl. Acad. Sci. USA 89, 7124–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanda, S. L. & Amasino, R. M. (1995) Weeds World 2, 2–8. [Google Scholar]
- 28.Swarup, K., Alonso-Blanco, C., Lynn, J. R., Michaels, S. D., Amasino, R. M., Koornneef, M. & Millar, A. J. (1999) Plant J. 20, 67–77. [DOI] [PubMed] [Google Scholar]