Abstract
Polygalacturonase-inhibiting proteins (PGIPs) are plant cell wall proteins that protect plants from fungal invasion. They interact with endopolygalacturonases secreted by phytopathogenic fungi, inhibit their enzymatic activity, and favor the accumulation of oligogalacturonides, which activate plant defense responses. PGIPs are members of the leucine-rich repeat (LRR) protein family that in plants play crucial roles in development, defense against pathogens, and recognition of beneficial microbes. Here we report the crystal structure at 1.7-Å resolution of a PGIP from Phaseolus vulgaris. The structure is characterized by the presence of two β-sheets instead of the single one originally predicted by modeling studies. The structure also reveals a negatively charged surface on the LRR concave face, likely involved in binding polygalacturonases. The structural information on PGIP provides a basis for designing more efficient inhibitors for plant protection.
Plants successfully defend themselves from the attack of a wide range of pathogenic microorganisms. Because they lack a system of circulating antibodies, their defense relies on the capability of each cell to recognize the presence of pathogens and subsequently activate defense responses. Many of the recognition events between plants and pathogens occur in the plant cell wall, which is the first barrier to come into contact with the invading organisms. The majority of microorganisms need to breach this barrier to gain access to the plant tissue and produce enzymes that degrade the wall polymers (1). Among the cell wall-degrading enzymes produced by phytopathogenic fungi, an important role is played by the endopolygalacturonases (EC 3.2.1.15) that cleave the linkages between d-galacturonic acid residues in nonmethylated homogalacturonan, a major component of pectin. Polygalacturonases (PGs) are among the first enzymes secreted by phytopathogenic fungi, and their action on the outer component of the cell wall is a prerequisite for further wall degradation by other degrading enzymes (2). PG-inhibiting proteins (PGIPs) are located in the plant cell wall and limit the fungal invasion by interacting with PGs (2). Their inhibitory activity modulates the PG activity, favoring the accumulation of cell wall fragments, the oligogalacturonides, which act as elicitors of plant defense responses (3). Plants have evolved many PGIPs with different and specific recognition capabilities against the many PGs secreted by pathogenic fungi. For example, in Phaseolus vulgaris the occurrence of four pgip genes is associated with a diversification of their biochemical function including the acquisition of new recognition specificities. The four mature products of these genes differ by between 8 and 81 aa and exhibit different inhibitory capabilities against the PGs of Botrytis cinerea, Colletotrichum gloeosporioides, Stenocarpella maydis, Fusarium moniliforme, and Aspergillus niger (A. Raiola, R. D'Ovidio, and G.D.L., unpublished work). In addition, the expression of the individual members of a PGIP family is differentially regulated by different signal molecules through separate transduction pathways (4).
PGIPs belong to the superfamily of leucine-rich repeat (LRR) proteins, a class of proteins specialized for protein–protein interactions (5). Plants use the LRR fold for their “immune” functions and recognition of non-self-molecules (6). Several plant resistance gene products or defense-related receptors display LRR motifs of the extracytoplasmic type homologous to that of PGIP (6, 7). These motifs are also found in proteins with important functions other than resistance or defense against microorganisms, such as a carrot antifreeze protein (8), several receptor kinases involved in steroid and peptide hormone perception (9, 10), development (11), defense responses against insects (12), or bacterial and fungal symbiosis (13, 14). Biochemical information on the mode of action of plant LRR proteins is still poor, and no structural information is available. Attempts to model the LRR proteins of the extracytoplasmic type on the basis of the structure of known LRR proteins of animal or microbial origin have been unsatisfactory thus far (5).
In this article we report the crystal structure of the isoform 2 of PGIP (PGIP2) from P. vulgaris at 1.7-Å resolution. This structure represents a model for studying the structural organization and the mode of interaction of plant LRR proteins. The structure also provides a molecular basis for understanding how PGIP inhibits PGs.
Materials and Methods
Purification of PGIP2. PGIP2 from P. vulgaris was overexpressed in Nicotiana benthamiana plants infected with a modified potato virus X as described (15). Plants were harvested 3 weeks after the infection; leaves were homogenized in 1 M NaCl, and homogenates were incubated under shaking for 1 h at 4°C and centrifuged for 30 min at 18,000 × g. Ammonium sulfate was added to the filtered supernatant to achieve 30% saturation, and the solution was then shaken overnight at 4°C. The cloudy solution then was centrifuged for 30 min at 18,000 × g, and the supernatant was dialyzed against 20 mM sodium acetate, pH 4.7, and loaded on an SP-Sepharose column (Pharmacia). Adsorbed proteins were eluted with a linear gradient of 0–0.5 M NaCl in 20 mM sodium acetate, pH 4.7. Fractions containing PGIP2, detected by SDS/PAGE analysis, were dialyzed against 10 mM sodium acetate and purified by isoelectrofocusing in an IEF Rotofor system (Bio-Rad) in a pH 8.0–10.5 gradient. Fractions containing PGIP2 were collected and dialyzed against 20 mM sodium acetate, pH 4.7. PGIP2 was finally concentrated to 4.0 mg/ml by using VIVASPIN-4 concentrators with a molecular mass cutoff of 5,000 kDa.
Crystallography. Crystals were grown by the vapor-diffusion technique at 4°C under the following conditions: Drops were prepared by mixing 1.0 μl of protein solution and 0.8 μl of a reservoir solution consisting of 20–30% polyethylene glycol (PEG) 4000 (wt/vol), 0.18–0.2 M ammonium acetate, 0.1 M sodium acetate, pH 4.6, and allowed to equilibrate against 0.5 ml of the same reservoir solution. Crystals appeared after 5 days and reached final dimensions of ≈0.4 × 0.2 × 0.1 mm in 2 weeks. Crystals were cryoprotected by using a solution containing 35% PEG 4000 (wt/vol), 0.1 M sodium acetate, pH 4.6, 0.2 M ammonium acetate, and 15% PEG 200 (wt/vol) and flash-frozen in a nitrogen stream at 100 K (Oxford Cryosystems, Oxford). PGIP2 crystallized in two crystal forms (A and B), both belonging to the P21212 space group but with different unit cell dimensions. Form A (Native2 in Table 1) diffracted to 2.5-Å resolution and had cell dimensions of a = 139.59 Å, b = 65.64 Å, and c = 37.23 Å. Form B (Native1 in Table 1) diffracted better (up to 1.7-Å resolution) and showed the cell dimensions a = 134.84 Å, b = 65.45 Å, and c = 34.64 Å. Data for crystal form B to 1.7-Å resolution were collected at the XRD Beamline of the ELETTRA Synchrotron (Trieste, Italy) equipped with a Mar345 imaging-plate detector (MAR-Research, Hamburg, Germany). Data for crystal form A to 2.5-Å resolution were collected at BW7A Beamline of the Deutsches Elektronen Synchrotron (Hamburg, Germany), equipped with a MAR charge-coupled device detector. Reflection intensities were integrated and scaled by using denzo/scalepack (16). Phases were determined by single isomorphous replacement and anomalous scattering methods. Crystals were soaked in a solution containing 35% PEG 4000 (wt/vol), 0.2 M ammonium acetate, 0.1 M sodium acetate, and 1 mM K2OsO4 for 6 h. Heavy metal Patterson search with the derivatized and native crystals of form A was performed by using the program solve (17), and three peaks were found leading to an overall figure of merit of 0.45 in the 25- to 2.5-Å shell. Density modification was carried out with resolve (18), yielding a figure of merit of 0.65 at 2.5-Å resolution. The resulting electron density map was sufficiently connected to allow us to build a partial model out of two-thirds of the residues and to fit most of side chains by using the program quanta (Molecular Structure, The Woodlands, TX). The structure of the LRR domain of Internalin B (PDB code 1DOB) was a useful guide to locate sheet B1 inside the density. This partial model then was used to solve the structure at 1.7-Å resolution with native data from form B by molecular replacement with amore (19). Refinement was carried out with refmac (20), and water residues were added into the Fo – Fc density map, contoured at 4σ, with the X-SOLVATE tool of quanta. Several cycles of refinement, manual rebuilding, and addition of solvent molecules led to a crystallographic R factor of 0.194 and a free R factor of 0.244 for a complete model of PGIP2 consisting of 313 residues (1–313), 320 water molecules, four N-acetyl glucosamine residues (one linked at the Asn-35, two at Asn-112, and another one to Asn-274) (Table 2). Secondary structure assignment was performed by using the Kabsch and Sander algorithm (40) as implemented in the program ribbon (21). The model was monitored for geometrical quality by using procheck (22). Superposition of the LRR repeats of different proteins with the repeat of PGIP2 and rms deviation between related Cα atoms were performed and calculated by using the program insigthii (Molecular Structure)
Table 1. Data collection and phasing.
| Native1 | Native2 | K2OsO4 | |
|---|---|---|---|
| Space group | P21212 | P21212 | P21212 |
| Unit cell dimensions, Å | a = 134.84, b = 65.45, c = 34.64 | a = 139.59, b = 65.64, c = 37.23 | a = 139.72, b = 65.81, c = 37.47 |
| Wavelength, Å | 0.95 | 0.9537 | 0.8431 |
| Resolution limits, Å | 30.0-1.7 | 25.0-2.5 | 25.0-2.5 |
| Reflections | |||
| Total (N) | 330,460 | 656,453 | 367,444 |
| Unique (N) | 34,715 | 25,439 | 12,694 |
| Completeness (last shell) | 94.1 (65.5) | 87.1 (86.0) | 93.1 (86.7) |
| Average l/σ | 11.4 | 15.3 | 13.4 |
| Rmerge,* % | 0.064 (0.45) | 0.080 (0.312) | 0.125 (0.317) |
| Derivatives | |||
| Concentration, mM | 1 | ||
| Soaking time, h | 6 | ||
| FOM (before DM) | 0.45 | ||
| FOM (after DM) | 0.65 |
Rsym = Σhkl|〈l〉 — l/(Σhkl|Fo(hkl) — Fc(hkl)|/Σhkl Fo(hkl). Rfree is Rcryst for 5% of the reflections excluded from the refinement.
Table 2. Refinement parameters.
| Refinement statistic | |
| Resolution, Å | 25.0-1.7 |
| Rcryst, % | 0.194 |
| Rfree, % | 0.244 |
| Ramachandran statistics | |
| % of residues in allowed regions | 77.4 |
| % of residues in generously allowed | 22.6 |
| % of residues in not allowed | 0.0 |
| Rms deviation | |
| Bond lengths, Å | 0.015 |
| Bond angles, ° | 2.6 |
| Model | |
| 313 | Amino acid residues |
| 4 | GlcNAc |
| 320 | Water residues |
Results and Discussion
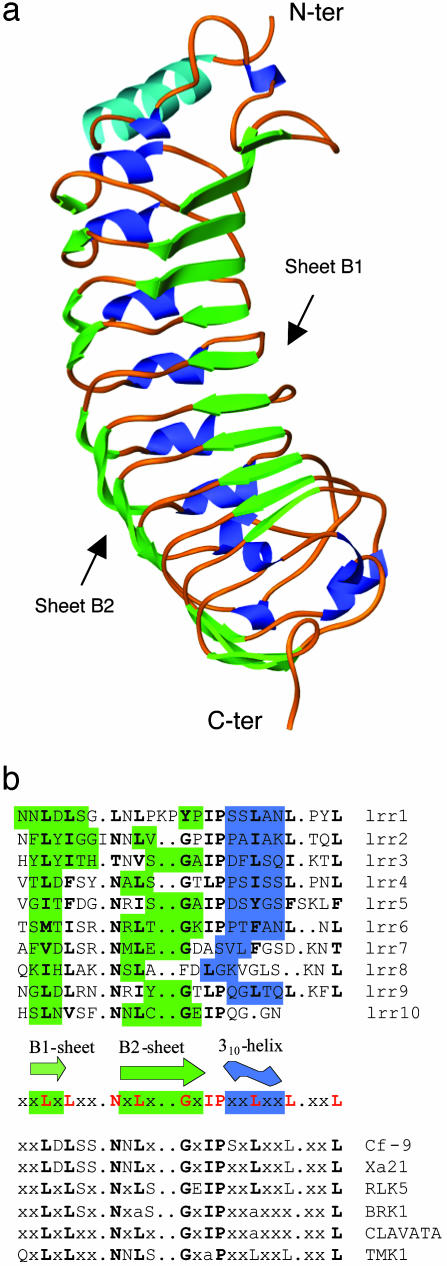
Overall Structure Description. The structure of PGIP2 from P. vulgaris was determined at 1.7-Å resolution by using the single isomorphous replacement and anomalous scattering method. PGIP2 is the first LRR protein belonging to the plant-specific subfamily, according to the classification proposed by Kajava (23), the structure of which has been solved. The overall structure of PGIP2 shows a typical curved and elongated shape; however, its scaffold appears more twisted than other LRR proteins (24–28) (Fig. 1a). The central LRR domain, folded in a right-handed superhelix (residues 53–289), consists of a set of 10 tandemly repeating units, each made up of 24 residues. A long parallel β-sheet (B1), consisting of three strands formed by six residues on the N-terminal part and of seven shorter strands on the C-terminal portion, occupies the concave inner side of the structure. The β-sheet B1 corresponds to the predicted β-sheet where the residues determining the affinity and specificity of PGIP2 are known to reside (15). On the opposite side of the protein, nine 310-helices are almost parallel to sheet B1. The first six helices have a comparable length and are regularly spaced, whereas the three helices in the C-terminal portion are variable in length and position.
Fig. 1.
Structure of PGIP2 from P. vulgaris. (a) ribbon (21) representation of the structure of PGIP2. Sheets B1 and B2 are colored green, and helices are colored blue (light blue for the N-terminal α-helix and dark blue for 310-helices in the LRR central portion of the molecule). (b) Secondary structure organization of the LRR motif (residues 53–289) in PGIP2. The consensus sequence of PGIP2 and other plant-derived extracytoplasmic LRR (6) are shown together with the secondary structure elements found in PGIP2. Residues contributing to form the secondary structure elements are colored green (sheets B1 and B2) and blue (310-helix).
An additional extended parallel β-sheet (B2) characterizes the fold of PGIP2. This is absent in the majority of other LRR proteins, in which helices and β-sheet B1 are connected simply by loops or β-turns. The only exception is represented by dynein LC1 (29) belonging to SDS22-like subfamily, for which the LRR domain is formed by six β-β-α motifs. Due to the twisted shape of the molecule, sheet B2 is distorted and the length of its β-strands is fairly variable. By careful inspection of the H-bonding pattern, B2 may be considered a single sheet formed by the clustering of four shorter sheets that interact together. The presence of the additional sheet B2 places the fold of PGIP2 between the typical LRR structure and the β-helical architecture found in pectate lyases and PGs that contain three to four β-sheets (30–33).
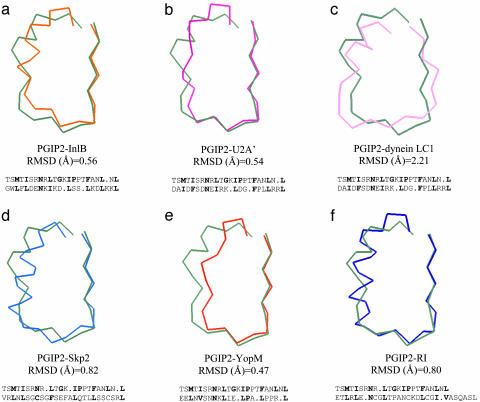
PGIP2 belongs to the plant-specific LRR subfamily characterized by the consensus sequence Lt/sGxIP in the region following the conserved “β-structure + Asn-ladder” (23). The superimposition of the backbone of the β-structure + Asn-ladder region in the sixth LRR repeat of PGIP2 to the corresponding region of proteins representative of other LRR subfamilies is shown in Fig. 2. For most of the proteins used these regions are well superimposed while the following regions do not overlap. Only the repeat of dynein LC1 exhibits significant differences from PGIP2 also in the typically conserved region. In PGIP2 the characteristic sequence Lt/sGxIP is partly involved in the formation of sheet B2, where the conserved glycine residues invariably show a stereochemical conformation that is strictly forbidden to any other residue and make the bending of sheet B2 possible. We speculate that this second β-sheet contributes to the formation of an additional surface for interactions. Other plant-derived extracytoplasmic LRR proteins exhibit the same repeating motif Lt/sGxIP and are therefore likely to form a second β-sheet (Fig. 1b), which possibly allows the interaction among different components of a receptor signal transduction complex after binding of the ligands (34, 35).
Fig. 2.
Backbone superposition of a single LRR motif of proteins representative of the LRR subfamilies defined by Kajava (23) with LRR6 of PGIP2 repeat (green). (a) Internalin B (25) (orange). (b) U2A′ (36) (dark pink). (c) Dynein LC1 (29) (light pink) belonging to the SDS22-like subfamily. (d) Skp2 (27) (light blue) belonging to the cysteine-containing subfamily. (e) YopM (26) (red) belonging to the bacterial subfamily. (f) RI (24) (dark blue) belonging to the RI-like subfamily. rms deviation (RMSD) values are calculated for the superposition of the first eight residues of each repeat forming the so-called β-structure + Asn-ladder region.
The N-terminal region of PGIP2 (residue 1–52) consists of a 13-residue-long α-helix and a short β-strand forming H bonds with sheet B1 and resembling the β-hairpin conformation observed in the N-terminal domains of the U2A′ spliceosomal protein (36) and GPIbα (28). The C-terminal region consists of the last two 310-helices, the last strand of sheet B2, and a short loop. Four disulfide bridges flank the LRR domain: two bridges are located in the N-terminal region (Cys-3–Cys-33 and Cys-34–Cys-43), and the other two are in the C-terminal region (Cys-281–Cys-303 and Cys-305–Cys-312). Both N- and C-terminal regions contribute to cap the hydrophobic core of the protein solenoid.
The LRR conserved residues (represented by Leu, Ile, and Val and the bulkier Phe and Tyr) are involved in stacking interactions that are crucial for the formation of the hydrophobic core in the LRR domain. Interestingly, several aromatic residues (Phe-133, Phe-156, Tyr-169, Phe-172, Phe-176, and Phe-194) are positioned almost in the middle of the protein, exactly where the protein bends. The conserved LRR asparagine ladder is typically located in the interior part of the repeat and forms H bonds with the main-chain carbonyl and amide groups, thus contributing to the creation of the bend of the protein.
The only secondary structure elements present in the convex face of the LRR region are the short 310-helices. As in Internalin B (25) and YopM (26), this part of the LRR fold is stabilized by the formation of H bonds between the protein backbone carbonyls and water molecules. These are organized in spines, and their low-temperature factors suggest that their presence has a structural relevance. The ensuing structural flexibility may be necessary to facilitate the adaptation of the PGIP2 scaffold to the surface of its interacting proteins.
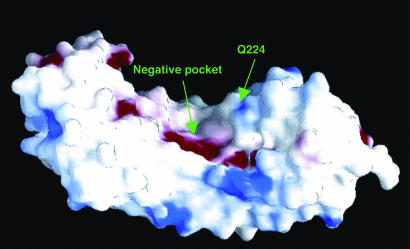
PGIP–PG Interaction. PGIP2 inhibits fungal PGs through the formation of bimolecular complexes, and the residues of PGIP2 critical for its affinity and recognition capability are located in sheet B1 (15). In a recent study we observed that the interaction between the PG of F. moniliforme (FmPG) and PGIP2 is mediated by at least two positively charged residues of the enzyme (Arg-267 and Lys-269), which are located at the edge of its active site and are involved in substrate binding. The involvement of these two residues in the interaction with PGIP2 provides an explanation for the competitive inhibition observed (33). Examination of the electrostatic potential surface of PGIP2 reveals a negative pocket formed by the charged residues Asp-131, Asp-157, and Asp-203 and the polar residues Ser-133, Thr-155, and Thr-180, located approximately in the center of sheet B1 (Fig. 3). Interestingly, the three aspartic residues are highly conserved in all PGIPs (2). The pocket is sufficiently large and deep to accommodate the positively charged residues Arg-267 and Lys-269 on the surface of the enzyme and may completely cover its active site, thus preventing access to the substrate. The residue Gln-224 of PGIP2, which is crucial for the specificity of the inhibitor toward FmPG, is located in sheet B1 immediately above the negative pocket putatively involved in PG binding (Fig. 3). We hypothesize that this residue may interact with an unidentified partner residue of FmPG to correctly lock Arg-267 and Lys-269 into the negative pocket. In PGIP1, which is unable to interact with FmPG (15), this role may not be fulfilled by the corresponding Lys-224.
Fig. 3.
grasp (39) electrostatic potential surface of PGIP2. Regions of negative and positive potential are shown in red and blue, respectively. A wide negative pocket, putatively involved in PG recognition, is located in the middle of the inner concave surface of the protein. The residue Gln-224, crucial for PGIP2 specificity, is also indicated.
In conclusion, the 1.7-Å resolution crystal structure of PGIP2 from P. vulgaris presented in this work provides insight into the architecture of the plant-specific LRR subfamily. With respect to the vast majority of previously known LRR proteins, a characteristic feature of this structure is the presence of two extended β-sheets that are likely to be conserved in other plant LRR proteins. A peculiarity of the PGIP2 structure is the presence of a cluster of negatively charged residues in the concave surface of the protein likely involved in the interaction with PGs. We expect that this structure will be useful for understanding the mode of interaction of PGIPs as well as for designing better inhibitors with tailor-made specificities. This structure may also be useful for modeling other plant LRR proteins, which are known to perform important functions in defense and development, and pave the way to elucidate their multiple interactive properties and their mechanism of recognition (37, 38).
Acknowledgments
This paper is dedicated to the memory of Noel Keen. We thank Maurizio Brunori for encouragement and valuable advice. We also thank Veronica Morea for the careful reading of the manuscript. This research was supported by European Community Grant QLK3-CT99-089, the Institute Pasteur Fondazione Cenci Bolognetti, and the Armenise–Harvard Foundation.
Abbreviations: PG, polygalacturonase; PGIP, PG-inhibiting protein; LRR, leucine-rich repeat.
Data deposition: The atomic coordinates and structure factors for PGIP2 from Phaseolus vulgaris have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1OGQ).
References
- 1.De Lorenzo, G. & Ferrari, S. (2002) Curr. Opin. Plant Biol. 5, 295–299. [DOI] [PubMed] [Google Scholar]
- 2.De Lorenzo, G., D'Ovidio, R. & Cervone, F. (2001) Annu. Rev. Phytopathol. 39, 313–335. [DOI] [PubMed] [Google Scholar]
- 3.Cervone, F., Hahn, M. G., De Lorenzo, G., Darvill, A. & Albersheim, P. (1989) Plant Physiol. 90, 542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrari, S., Vairo, D., Ausubel, F. M., Cervone, F. & De Lorenzo, G. (2003) Plant Cell 15, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobe, B. & Kajava, A. V. (2001) Curr. Opin. Struct. Biol. 11, 725–732. [DOI] [PubMed] [Google Scholar]
- 6.Jones, J. D. (2001) Curr. Opin. Plant Biol. 4, 281–287. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Gomez, L., Bauer, Z. & Boller, T. (2001) Plant Cell 13, 1155–1163. [PMC free article] [PubMed] [Google Scholar]
- 8.Worrall, D., Elias, L., Ashford, D., Smallwood, M., Sidebottom, C., Lillford, P., Telford, J., Holt, C. & Bowles, D. (1998) Science 282, 115–117. [DOI] [PubMed] [Google Scholar]
- 9.Li, J. & Chory, J. (1997) Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- 10.Montoya, T., Nomura, T., Farrar, K., Kaneta, T., Yokota, T. & Bishop, G. J. (2002) Plant Cell 14, 3163–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, S. E., Williams, R. W. & Meyerowitz, E. M. (1997) Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- 12.Scheer, J. M. & Ryan, C. A., Jr. (2002) Proc. Natl. Acad. Sci. USA 99, 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., et al. (2002) Nature 417, 959–962. [DOI] [PubMed] [Google Scholar]
- 14.Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kalo, P. & Kiss, G. B. (2002) Nature 417, 962–966. [DOI] [PubMed] [Google Scholar]
- 15.Leckie, F., Mattei, B., Capodicasa, C., Hemmings, A., Nuss, L., Aracri, B., De Lorenzo, G. & Cervone, F. (1999) EMBO J. 18, 2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski, Z. & Minor, W. (1997) in Methods in Enzymology, eds. Carter, C. W. & Sweet, R. M. (Academic, New York), pp. 307–326.
- 17.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terwilliger, T. C. (1999) Acta Crystallogr. D 55, 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navaza, J. (1994) Acta Crystallogr. D 50, 157–163. [Google Scholar]
- 20.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 21.Carson, M. (1997) Methods Enzymol. 277, 493–505. [PubMed] [Google Scholar]
- 22.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 23.Kajava, A. V. (1998) J. Mol. Biol. 277, 519–527. [DOI] [PubMed] [Google Scholar]
- 24.Kobe, B. & Deisenhofer, J. (1996) J. Mol. Biol. 264, 1028–1043. [DOI] [PubMed] [Google Scholar]
- 25.Marino, M., Braun, L., Cossart, P. & Ghosh, P. (1999) Mol. Cell 4, 1063–1072. [DOI] [PubMed] [Google Scholar]
- 26.Evdokimov, A. G., Anderson, D. E., Routzahn, K. M. & Waugh, D. S. (2001) J. Mol. Biol. 312, 807–821. [DOI] [PubMed] [Google Scholar]
- 27.Schulman, B. A., Carrano, A. C., Jeffrey, P. D., Bowen, Z., Kinnucan, E. R., Finnin, M. S., Elledge, S. J., Harper, J. W., Pagano, M. & Pavletich, N. P. (2000) Nature 408, 381–386. [DOI] [PubMed] [Google Scholar]
- 28.Uff, S., Clemetson, J. M., Harrison, T., Clemetson, K. J. & Emsley, J. (2002) J. Biol. Chem. 277, 35657–35663. [DOI] [PubMed] [Google Scholar]
- 29.Wu, H., Maciejewski, M. W., Marintchev, A., Benashski, S. E., Mullen, G. P. & King, S. M. (2000) Nat. Struct. Biol. 7, 575–579. [DOI] [PubMed] [Google Scholar]
- 30.Yoder, M. D., Keen, N. T. & Jurnak, F. (1993) Science 260, 1503–1507. [DOI] [PubMed] [Google Scholar]
- 31.Pickersgill, R., Smith, D., Worboys, K. & Jenkins, J. (1998) J. Biol. Chem. 273, 24660–24664. [DOI] [PubMed] [Google Scholar]
- 32.Herron, S. R., Benen, J. A., Scavetta, R. D., Visser, J. & Jurnak, F. (2000) Proc. Natl. Acad. Sci. USA 97, 8762–8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Federici, L., Caprari, C., Mattei, B., Savino, C., Di Matteo, A., De Lorenzo, G., Cervone, F. & Tsernoglou, D. (2001) Proc. Natl. Acad. Sci. USA 98, 13425–13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rojo, E., Sharma, V. K., Kovaleva, V., Raikhel, N. V. & Fletcher, J. C. (2002) Plant Cell 14, 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E. & Walker, J. C. (2002) Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- 36.Price, S. R., Evans, P. R. & Nagai, K. (1998) Nature 394, 645–650. [DOI] [PubMed] [Google Scholar]
- 37.Axtell, M. J. & Staskawicz, B. J. (2003) Cell 112, 369–377. [DOI] [PubMed] [Google Scholar]
- 38.Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R. & Dangl, J. L. (2003) Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- 39.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]
- 40.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577–2637. [DOI] [PubMed] [Google Scholar]