Abstract
Following genotoxic stress, transcriptional activation of target genes by p53 tumor suppressor is critical in cell fate determination. Here we report that the restoration of p53 function in human cancer cell lines that are deficient in p53 function upregulated the expression of Notch1. Interestingly, the expression of wild-type p53 in human prostate and breast cancer cell lines correlated well with increased expression of Notch1. Furthermore, knockdown of p53 expression in cancer cells that express wild-type p53 resulted in reduced expression of Notch1. Importantly, genotoxic stress to cancer cells that resulted in activation of p53 also upregulated the expression of Notch1. Moreover, p53-mediated induction of Notch1 expression was associated with stimulation of the activity of Notch-responsive reporters. Notably, p53 differentially regulated the expression of Notch family members: expression of Notch2 and Notch4 was not induced by p53. Significantly, treatment of cells with gamma secretase inhibitor, an inhibitor of Notch signaling, increased susceptibility to apoptosis in response to genotoxic stress. Together, our observations suggest that p53-mediated upregulation of Notch1 expression in human cancer cell lines contributes to cell fate determination after genotoxic stress.
Keywords: p53, Notch, transcription, DNA damage, apoptosis
Introduction
p53 is a key tumor suppressor that is mutated in > 50% of human cancers [1–3]. p53 is a transcription factor that activates the transcription of its target genes by binding to a p53 DNA-binding consensus sequence [2]. Importantly, proteins encoded by p53 target genes contribute to the determination of cell fate after genotoxic stress [4,5]. p53 mediates its tumor-suppressive functions by inducing cell growth arrest, apoptosis, or senescence [3,6].
A recent study [7] has identified a potential p53 DNA-binding site about 3.7 kb upstream in the promoter region of the human Notch1 gene. Notch1 gene encodes a protein that belongs to a family of large (∼ 300 kDa), single-pass, evolutionarily conserved membrane-associated receptors [8,9]. Other members of the mammalian Notch family include Notch2, Notch3, and Notch4. Proteins in the family primarily regulate cell fate and lineage specification during embryonic and postembryonic development. These proteins have conserved structural elements in their extracellular transmembrane and intracellular domains [8,9]. Mammalian cells are shown to express five ligands (i.e., Jagged-1, Jagged-2, Delta-1, Delta-3, and Delta-4) for Notch family receptors [10]. Notch signaling is initiated by receptor-ligand interactions between neighboring cells, resulting in two successive proteolytic cleavages by tumor necrosis factor-α-converting enzyme and gamma secretase/presenilin complex [11,12]. These two proteolytic cleavages of the Notch receptor protein result in the release of the intracellular domain [notch intracellular domain (NIC), the functionally active form of Notch], which translocates to the nucleus and modulates the transcription of target genes. Gamma secretase inhibitors (GSIs), such as the tripeptide inhibitor z-Leu-Leu-Nle-CHO, inhibit the generation of NIC [13,14]. Consequently, treatment of cells with GSI is shown to inhibit Notch signaling [13,15] and to decrease cell survival.
After nuclear translocation, NIC binds to CBF1 (also termed RBP-Jκ), a DNA-binding protein [16,17]. In the absence of NIC, CBF1 acts as a transcriptional repressor. However, the binding of NIC to CBF1 converts it to a transcriptional activator, which results in Notch1-dependent transcriptional activation of genes. Notch1 target genes include Hes (Hairy/Enhancer of Split), p21CIP1/WAF1, and cyclin D [16,17].
Similar to its role in development, the role of Notch family members in oncogenesis appears to be cell type/organ-specific. Aberrant Notch1 signaling has been cited as causative in T-cell lymphoblastic leukemias, certain lymphomas, breast carcinomas, and kidney carcinomas [18]. In keratinocytes, however, Notch1 signaling mediates terminal differentiation [16]. Neoplastic effects of Notch1 may be attributed to its role as a cell survival factor in tumor cells mainly through the inhibition of p53 through mammalian target of rapamycin (mTOR) [19]. In addition, there have been other reports of Notch-mediated inhibition of p53 functions, including apoptosis [20,21].
Although the role of Notch1 in regulating p53 function has been reported, it is not known whether p53 has any effect on Notch1 expression and/or its functions. In the present study, we demonstrate that the restoration of p53 function in human cancer cell lines deficient in p53 function upregulates the expression of Notch1. Significantly, we found that genotoxic stress, which resulted in transcriptional activation of p53, also upregulated the expression of Notch1 in human cancer cell lines. Moreover, inhibition of Notch1 activity after genotoxic stress by an inhibitor of Notch signaling increased susceptibility to apoptosis. Together, our observations provide support for the idea that induction of Notch1 expression by p53 in cells counteracts p53-mediated proapoptotic functions.
Materials and Methods
Cell Lines, Culture Conditions, and Treatments
Saos-2 cells, stably expressing the temperature-sensitive mutant (Val138 mutation) of human p53 (with Arg72) [22], were generously provided by Dr. Maureen Murphy (Fox Chase Cancer Center, Philadelphia, PA). LNCaP, DU-145, PC-3, MCF-7, and Saos-2 cell lines were purchased from the American Type Culture Collection (Manassas, VA). All cell lines (except Saos-2) used in the study were maintained in (high-glucose) Dulbecco's modified Eagle's medium (Invitrogen Life Technologies, Carlsbad, CA) supplemented with 10% (vol/vol) fetal bovine serum and antibiotics. The Saos-2 cell line was maintained in RPMI 1640 medium.
Temperature-sensitive cells were maintained at the specified temperature, and the medium was left unchanged for the duration (15–17 hours) of treatment. Etoposide (Calbiochem, San Diego, CA) was prepared as a 50-mM stock solution in dimethyl sulfoxide (DMSO) and stored at −20°C until use. Control cell plates were treated with an equal volume of DMSO.
Knockdown of p53 Expression
Subconfluent cultures of LNCaP cells were transfected with a pool of p53 siRNA (cat no. M-003557-00-05; purchased from Dharmacon, Denver, CO) or nonspecific control siRNA (cat no. D-001206-02-05) as recommended by the supplier using Lipofectamine (Invitrogen Life Technologies) transfection agent. Sixty hours after the transfection of cells, cell lysates were prepared and processed for immunoblotting.
Nucleofection
PC-3 or Saos-2 cells were nucleofected with pCMV or pCMV-p53 plasmid (2 µg) using Nucleofector-II device (Amaxa Biosystems, Nattermannallee 1, Germany). PC-3 cells were nucleofected using Nucleofection Kit VCA-1001 (Amaxa Biosystems) and the T-013 program, as suggested by the supplier. For Saos-2 cells, we used the same nucleofection kit with the program D-024. If so indicated, 24 hours after the nucleofection of cells, cells were treated with etoposide (45 µM) for 24 hours. Forty-eight hours after nucleofection, cells were processed for immunoblotting.
Reverse Transcription-Polymerase Chain Reaction
Total RNA was isolated from the indicated cells using TRIzol reagent (Invitrogen Life Technologies), as suggested by the supplier. Isolated total RNA was subjected to cDNA synthesis using SuperScript First-Stand Synthesis system (Invitrogen Life Technologies), as suggested by the supplier, followed by PCR, using a pair of primers specific to human Notch1 cDNA (forward primer: 5′-CAGGCAATCCGAGGACTATG-3′; reverse primer: 5′-CAGGCGTGTTGTTCTCACAG-3′), p21, or actin cDNA using a kit from Invitrogen Life Technologies, as suggested by the supplier. The PCR product for human Notch1 (428 bp) was analyzed by agarose gel electrophoresis after 36 PCR cycles.
Immunoblotting and Antibodies
Total cell lysates were prepared in a modified RIPA buffer (50 mM Tris-HCl pH 8.0, 250 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate plus complete mini EDTA-free protease inhibitor cocktail; Roche, Mannheim, Germany). Lysates were incubated for 30 minutes on ice and sonicated briefly. The lysates were centrifuged at 14,000 rpm in a microfuge at 4°C for 10 minutes, and equivalent protein amounts were subjected to immunoblotting as described previously [23].
The following antibodies used in our experiments were purchased from Santa Cruz Biotechnology (Santa Cruz, CA): p53 (horseradish peroxidase-conjugated; cat no. sc-6243), p21 (horseradish peroxidase-conjugated; no. sc-397), Notch2 (no. sc-5545), Notch4 (no. sc-5594), glyceraldehyde-3-phosphatase dehydrogenase (GAPDH) (no. sc-32233), and α-tubulin (no. sc-8035). Human total Notch1 antibody (bTan20 ascites fluid) was purchased from Developmental Studies Hybridoma Bank (Iowa City, IA). Processed/cleaved Notch1 antibody (Val1744; no. 2421) was purchased from Cell Signaling Technology (Danvers, MA). β-Actin antibody (no. A3853) was from Sigma (St. Louis, MO).
Reporter Assays
Subconfluent cultures (in 60-mm plates) of cells were transfected with FuGene6 transfection agent (Roche Applied Science, Indianapolis, IN), as suggested by the supplier, using 2 µg of total plasmid DNA per plate. Forty-two to 44 hours after transfection, cells were lysed, and the activities of firefly luciferase and Renilla luciferase were determined as described previously [24].
Flow Cytometry
Flow cytometry was performed on single-cell suspensions on adherent (after trypsin and EDTA treatment) and floating cells after pooling them. Briefly, for cell cycle analysis, cells were stained with propidium iodide (50 µg/ml; Sigma) and subjected to flow cytometry using a Coulter Epics XL-MCL flow cytometer (Coulter, Fullerton, CA), as described previously [24]. Apoptosis was measured by the accumulation of cells with sub-G0 DNA content.
Results
Recently, Notch1 gene has been identified as a potential transcriptional target of p53 in human cells [7]. Therefore, we tested whether p53 could regulate the expression of Notch1 gene in human cancer cell lines that are deficient in p53 function.
Expression of a Temperature-Sensitive Mutant of p53 in Human Saos-2 Cells Upregulates the Expression of Notch1
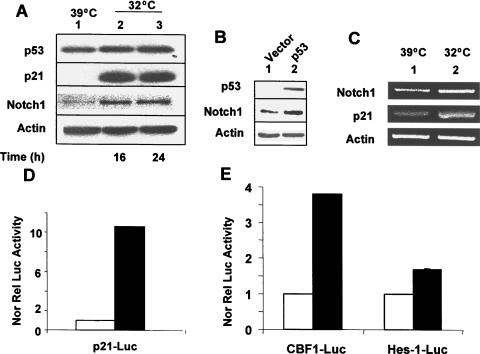
To determine whether p53 could upregulate the expression of Notch1 in human cancer cells, we chose to use the well-characterized human osteosarcoma Saos-2 cell system (Saos-2 cells are null for p53). Saos-2 cells express a temperature-sensitive mutant of p53 with the amino acid residue Arg (instead of Pro) at position 72 (cells indicated as SaosArg72; see Dumont et al. [22]). As shown in Figure 1A, incubation of SaosArg72 cells at 32°C for 16 or 24 hours resulted in upregulation of p21 protein, a transcriptional target of p53 protein. Importantly, levels of Notch1 protein also increased several fold in extracts from SaosArg72cells that were incubated at 32°C.
Figure 1.
Restoration of p53 expression in the human Saos-2 osteosarcoma cell line upregulates the expression of Notch1. (A) Subconfluent cultures of SaosArg72 cells were either incubated at 39°C (lane 1), incubated at 32°C for 16 hours (lane 2), or incubated at 32°C for 24 hours (lane 3). After incubation, total cell lysates were analyzed by immunoblotting using antibodies specific to the indicated proteins. (B) Saos-2 cells were nucleofected with either an empty vector pCMV (lane 1) or the plasmid pCMV-p53 encoding wild-type p53 (lane 2). Sixty hours after the nucleofection of cells, cells were lysed, and cell lysates containing an equal amount of proteins were processed for immunoblotting using antibodies specific to the indicated proteins. (C) Subconfluent cultures of SaosArg72 cells were incubated either at 39°C (lane 1) or at 32°C (lane 2). Twenty-four hours after incubation, total RNA was isolated, and steady-state levels of Notch1, p21, and actin mRNA were analyzed by semiquantitative reverse transcription-polymerase chain reaction. (D and E) Two sets of SaosArg72 cell cultures (in 60-mm plates) were transfected with p21-luc, CBF1-luc, or Hes1-luc reporter plasmid (1.8 µg) along with pRL-TK plasmid (0.2 ag; plasmids in a 9:1 ratio) using FuGene6 transfection reagent. One set of plates for each reporter was incubated at 39°C, and the other sets of plates were incubated at 32°C. Forty-four hours after incubation, cells were processed for dual luciferase reporter activity assays, as described in Materials and Methods. Firefly luciferase reporter activity was normalized to Renilla luciferase activity to control for variations in nucleofection efficiencies. Luciferase activity for p21-luc (C) and for CBF1-luc and Hes1-luc (D) in control cells is shown.
To rule out the possibility that incubation of cells at a reduced (32°C) temperature could account for increases in Notch1 protein levels (independent of p53 expression), we also compared Notch1 protein levels between parental Saos-2 cells that were incubated at 39° and parental Saos-2 cells that were incubated at 32°C. We found no difference between Notch1 protein levels in extracts from cells incubated at these two temperatures (data not shown). Moreover, forced expression of wild-type p53 in Saos-2 cells (after the nucleofection of cells with pCMV-p53 plasmid encoding wild-type p53) also resulted in upregulation of Notch1 expression (Figure 1B).
We also noted that steady-state levels of Notch1 mRNA also increased about two-fold in SaosArg72 cells after their incubation at 32°C for 24 hours (Figure 1C). Although we were unable to detect the processed NIC form of Notch1 protein in extracts from SaosArg72 cells that were incubated at 32°C for 16 or 24 hours using Val1744 antibody to cleaved Notch1 protein, we found that the activity of the two Notch1- responsive reporters (CBF1-luc and Hes1-luc) was consistently stimulated in SaosArg72 cells after incubation at 32°C for 24 hours (Figure 1, D and E). Moreover, we noted that the activity of CBF1-luc reporter was stimulated robustly compared to the activity of Hes1-luc reporter. The reasons for this differential stimulation of Notch1-responsive reporters in SaosArg72 cells remain unknown. Together, these observations suggested that the restoration of p53 function in Saos-2 cells upregulated Notch1 expression and stimulation of the activity of Notch1-responsive reporters.
The Functional Status of p53 in Human Cancer Cell Lines Correlated with Expression Levels of Notch1 Protein
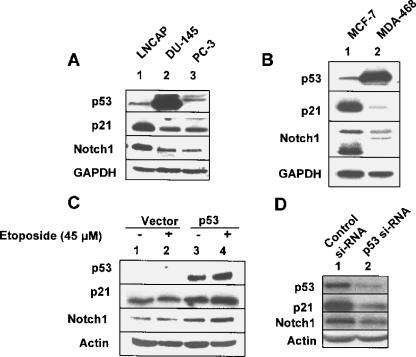
Our observations on the upregulation of Notch1 expression by functional p53 in human Saos-2 cell lines made it important to determine whether the functional status of p53 in human cancer cell lines correlates with expression levels of Notch1. Because three common human prostate cancer cell lines (LNCaP, DU-145, and PC-3) and two human breast cancer cell lines (MCF-7 and MDA-468) are known to differ with respect to the functional status of p53, we compared the expression levels of Notch1 protein in these cancer cell lines. Consistent with a previous report [25], expression of p53 was detectable in LNCaP (wild-type p53) and DU-145 (mutant p53) cells (Figure 2A). However, the expression of p53 was not detectable in PC-3 cells (null for p53). Likewise, the expression of p53 was detectable in MCF-7 (wild-type p53) and MDA-468 (mutant p53) cells (Figure 2B). Moreover, consistent with the functional status of p53 in these cell lines, expression levels of p21 were relatively high in extracts from LNCaP and MCF-7 cells. Importantly, consistent with a previous report [26], levels of the full-length Notch1 protein were also relatively high in LNCaP cells than in DU-145 and PC-3 cells (cf. lane 1 with lane 2 or lane 3). Similarly, levels of Notch1 protein were relatively higher in MCF-7 cells than in MDA-468 cells. Together, these observations suggested that the wild-type status of p53 in the above human cancer cell lines is associated with increased expression levels of Notch1 protein.
Figure 2.
The functional status of p53 and its expression levels in human cancer cell lines correlate with expression levels of Notch1 protein. (A) Total cell lysates from human prostate cancer cell lines LNCaP (lane 1), DU-145 (lane 2), or PC-3 (lane 3) were analyzed by immunoblotting using antibodies specific to the indicated proteins. (B) Total cell lysates from the human breast cancer cell line MCF-7 (lane 1) or MDA-468 (lane 2) were analyzed by immunoblotting using antibodies specific to the indicated proteins. (C) PC-3 cells were nucleofected with either pCMV vector (2 µg; lanes 1 and 2) or pCMV-p53 (wild-type) plasmid (2 µg; lanes 3 and 4), as described in Materials and Methods. Twenty-four hours after nucleofection, cells were either left untreated (lanes 1 and 3) or treated with etoposide (45 M) for 24 hours. Forty-eight hours after nucleofection, total cell extracts were analyzed by immunoblotting using antibodies specific to the indicated proteins. (D) LNCaP cells were transfected with either control siRNA (lane 1) or a pool of p53 siRNA (lane 2), as described in Materials and Methods, using Lipofectamine transfection reagent. Sixty hours after the transfection of cells, total cell lysates were analyzed by immunoblotting using antibodies specific to the indicated proteins.
Forced Expression of p53 in PC-3 Cells Upregulated Notch1 Expression
Because PC-3 cells are null for p53, we tested whether restoration of p53 expression in these cells and activation of p53 by genotoxic stress have any effect on Notch1 expression. As shown in Figure 2C, restoration of the expression of wild-type p53 in cells upregulated p21 and Notch1 protein levels (cf. lane 3 with lane 1). Interestingly, treatment of these cells expressing wild-type p53 with etoposide further upregulated the expression of both p21 and Notch1 proteins (cf. lane 4 with lane 3). These observations suggested that restoration of p53 function in PC-3 cells upregulated Notch1 expression.
Knockdown of p53 Expression in LNCaP Cells Downregulated Notch1 Expression
To further examine whether p53 regulates the expression of Notch1, we chose to knock down the expression of p53 in LNCaP cells. As shown in Figure 2D, transfection of LNCaP cells with a pool of p53 siRNA, but not control siRNA, resulted in a > 70% decrease in p53 protein levels (cf. lane 2 with lane 1). Importantly, the knockdown of p53 expression in cells resulted in decreases in p21 and Notch1 protein levels. These observations suggested that the expression of functional p53 in LNCaP cells contributes to relatively higher steady-state levels of Notch1 protein.
Activation of p53 By Genotoxic Stress Upregulates Notch1 Expression
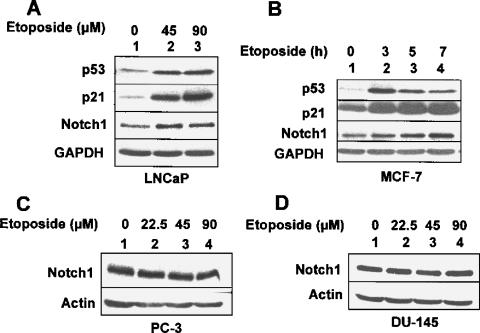
Genotoxic stress to cells due to their exposure to DNAdamaging agents, such as etoposide, is known to activate p53-mediated transcription of its target genes [27]. Therefore, we sought to determine whether exposure of LNCaP or MCF-7 cell lines, which express wild-type p53, to etoposide results in upregulation of Notch1. As shown in Figure 3A, treatment of LNCaP cells with etoposide resulted in the upregulation and activation of p53 in a dose-dependent manner. Moreover, treatment of cells resulted in upregulation of p21 and Notch1 proteins. Similarly, treatment of MCF-7 cells with etoposide resulted in upregulation of both p21 and Notch1 protein levels (Figure 3B). However, treatment of human prostate cancer cell lines PC-3 (null for p53) and DU-145 (mutant p53) with increasing concentrations (22.5, 45, or 90 µM) of etoposide for 24 hours did not result in upregulation of Notch1 protein (Figure 3,C and D, respectively). Together, these observations indicated that genotoxic stress-induced activation of p53 in human cancer cell lines also upregulates Notch1 expression.
Figure 3.
Etoposide treatment-induced genotoxic stress in p53-positive human cancer cell lines upregulates Notch1 expression. (A) Total cell lysates from the human prostate cancer cell line LNCaP, treated with either DMSO alone (lane 1), 45 µM etoposide (final concentration; lane 2), or 90 µM etoposide (final concentration; lane 3) for 15 hours, were analyzed by immunoblotting using antibodies specific to the indicated proteins. (B) Total cell lysates from the human breast cancer cell line MCF-7, either treated with DMSO alone (lane 1) or treated with 45 µM etoposide (final concentration) for the indicated time (hours), were analyzed by immunoblotting using antibodies specific to the indicated proteins. (C and D) Total cell lysates from the human prostate cancer cell line PC-3 (C) or DU-145 (D), either treated with DMSO alone (lane 1) or treated with indicated concentrations of etoposide for 24 hours, were analyzed by immunoblotting using antibodies specific to the indicated proteins.
p53 Differentially Regulates the Expression of Notch Family Members
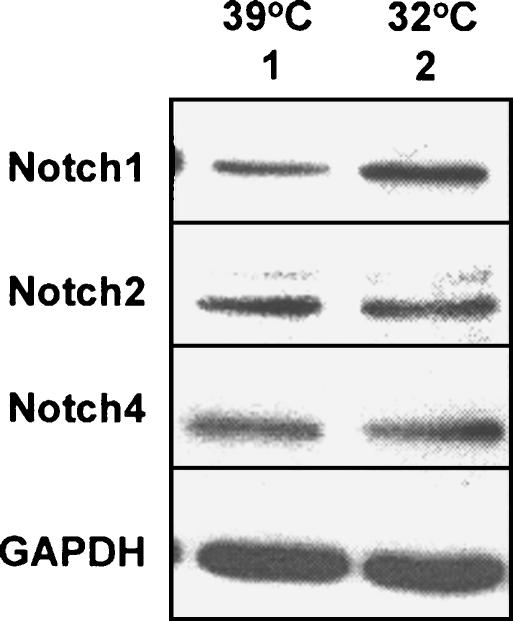
p53-mediated upregulation of Notch1 mRNA and protein in Saos-2 cells (Figure 1) made it interesting to determine whether p53 also regulates the expression of other members of the Notch receptor family. For this purpose, we compared the expression levels of Notch1, Notch2, and Notch4 proteins in extracts from SaosArg72 cells that were incubated at 39° or 32°. Consistent with our previous observations (Figure 1A), we found that incubation of cells at 32°C resulted in upregulation of Notch1 proteins (Figure 4; cf. lane 2 with lane 1). Interestingly, incubation of cells at 32°C did not result in upregulation of Notch2 or Notch4 protein levels (cf. lane 2 with lane 1). These observations suggested that functional p53 differentially regulates the expression of Notch receptor family members in human SaosArg72 cells.
Figure 4.
p53 differentially regulates the expression of Notch receptor family members. Subconfluent cultures of SaosArg72 cells were incubated at either 39°C (lane 1) or 32°C (lane 2) for 24 hours. After incubation, total cell lysates were analyzed by immunoblotting using antibodies specific to the indicated proteins.
Inhibition of Notch1 Activity and Genotoxic Stress Cooperate in MCF7 Cells to Increase Susceptibility to Apoptosis
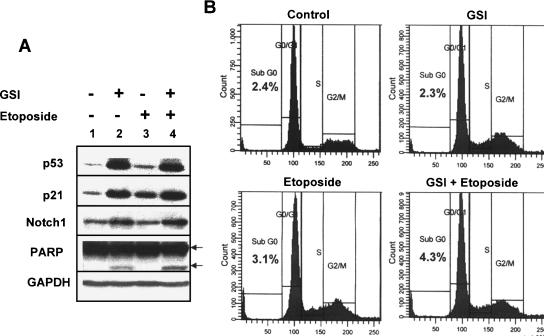
Notch signaling is known to play an important role in mammary gland tumorigenesis [28]. Moreover, increased expression of Notch1 protein in human breast cancer cell lines is associated with their resistance to apoptosis [29]. Therefore, we tested whether genotoxic stress-induced upregulation of Notch1 expression in the MCF-7 breast cancer cell line contributes to increases in resistance to apoptosis. As shown in Figure 5, treatment of MCF-7 cells with GSI (an inhibitor of Notch1 activity) [13,15] or etoposide resulted in increases in p53, p21, and Notch1 protein levels. Consistent with increases in p53 protein levels after GSI treatment of cells, we noted some cleavage of PARP protein, an indication of apoptosis. However, flow cytometry analysis of GSI-treated cells (after propidium iodide staining of DNA) for the accumulation of cells in sub-G0 phase indicated no increase in sub-G0 cells (compared to control cells; Figure 5B). Moreover, treatment of cells with etoposide alone did not result in detectable cleavage of PARP protein. Interestingly, treatment of cells with both GSI and etoposide resulted in measurable increases in the cleavage of PARP protein (cf. lane 4 with lane 2) and measurably increased accumulation of cells in the sub-G0 phase, as determined by flow cytometry (Figure 5B). These observations indicated that inhibition of Notch1 activation in MCF-7 cells and etoposide treatment of cells together increase susceptibility to apoptosis.
Figure 5.
Inhibition of Notch1 activity and genotoxic stress increase susceptibility to apoptosis. (A) Subconfluent cultures of MCF-7 cell line were either left untreated (lane 1) or treated with GSI (lane 2; 25 µM), etoposide (lane 3; 45 µM), or both GSI and etoposide (lane 4). Cells were incubated for 20 hours, and total cell lysates were analyzed by immunoblotting using antibodies specific to the indicated proteins. Two arrows indicate two forms of PARP protein: the precursor form (113 kDa) and the cleaved form (85 kDa). (B) Subconfluent cultures of the MCF-7 cell line were either left untreated (control; top left panel) or treated with GSI (25 µM; top right panel), etoposide (45 µM; bottom left panel), or both GSI and etoposide (bottom right panel). After incubation of cells for 20 hours, floating and attached cells were collected and processed for propidium iodide staining followed by flow cytometry. The percentage of cells in sub-G0 phase is indicated in each panel.
Discussion
Inhibition of cell proliferation by p53 is largely attributable to its ability to transcriptionally activate the expression of genes that encode proteins, which determine cell fate [4,30]. Depending on cellular context, wild-type p53 limits cell proliferation in response to DNA damage and other cellular stresses by inducing cell cycle arrest, apoptosis, or senescence [2–5].
The human Notch1 gene has been identified as a direct transcriptional target of p53 [7]. Based on this observation and accumulating evidence for crosstalks between Notch1 and p53 signaling pathways to regulate cell fate [20,21], we investigated whether p53 could regulate Notch1 gene expression in human cancer cell lines that lack p53 function. Our experiments revealed that: 1) expression of functional p53 in human cancer cell lines upregulated the expression of Notch1 mRNA and protein (Figure 1); 2) p53-mediated increased levels of Notch1 protein in cell lines stimulated the activity of Notch-responsive reporters (Figure 1); 3) knockdown of p53 expression in LNCaP cells reduced the expression levels of Notch1 protein (Figure 2); 4) in response to genotoxic stress by etoposide treatment of human prostate (LNCaP) and breast (MCF-7) cancer cell lines, transcriptional activation of p53 also upregulated Notch1 expression (Figure 3); 5) p53 did not upregulate the expression of Notch2 and Notch4 in Saos-2 cells (Figure 4); and 6) treatment of MCF-7 cells with GSI, an inhibitor of Notch signaling, increased susceptibility to apoptosis induced by etoposide treatment of cells (Figure 5). Together, these observations provide support for the idea that p53-mediated upregulation of Notch1 expression in human cancer cell lines counteracts p53-mediated proapoptotic functions.
Consistent with our observations, activated Notch1 is shown to exert antiapoptotic effects on cells by inhibiting p53-mediated transactivation [20]. Moreover, survival signaling by Notch1 is mediated, in part, through the PI3 kinase/AKT/mTOR axis [19]. In contrast to the antiapoptotic role of activated Notch1 in the above cell systems, conditional expression of a constitutively active form of Notch1 in early progenitor cells, but not in postmitotic neurons, selectively induced apoptosis [31]. Together, these observations provide support for the idea that Notch1-mediated determination of cell fate depends on cellular context.
p53 has been reported to negatively regulate Notch1 expression and activation in several mouse thymoma cell lines [21]. Moreover, the study also showed that expression of activated Notch1 (NIC) was elevated in Trp53-/- thymocytes compared to that in Trp53+/+ thymocytes. Because activated p53 is shown to differently activate the transcription of genes in different tissues in vivo [32], further work will be needed to determine whether p53-mediated regulation of Notch1 expression varies among different mouse and human tissues and cell lines derived from these tissues.
It has been reported that SV40 virus-encoded proteins (large T antigen and small t antigen) that inactivate p53 function can upregulate the expression of Notch1 mRNA and protein in human mesothelial cells [33]. Therefore, it is likely that signaling pathways independent of p53 also activate the transcription of Notch1 gene in SV40 virus-infected mesothelial cells. Furthermore, it is important to note that p53 knockout mice are viable with no obvious developmental abnormalities [34]. However, these mice are prone to developing certain types of cancers later in life [34]. These observations are consistent with the idea that, in addition to p53, other factors also contribute to the regulation of Notch1 expression in certain mouse and human cells.
While this manuscript was under review, two reports [35,36] provided evidence that the human Notch1 gene is a direct transcriptional target of p53 tumor suppressor. In one of these studies [35], the authors identified two potential p53 DNA-binding sites (p53-A and p53-B) in the promoter region (about 3.5 and 0.8 kb, respectively) of Notch1 gene. Moreover, they reported that p53 associated with these p53 DNAbinding sites in chromatin immunoprecipitation assays and upregulated the expression of Notch1 in normal primary human keratinocytes. Similarly, Yugawa et al. [36] reported the presence of at least two (proximal and distal) putative p53 DNA-binding sites in the promoter region of the human Notch1 gene. Importantly, mutations in the distal p53 DNAbinding site resulted in reduction in the basal activity of the promoter and lack of response to ionizing radiation. Moreover, this study also indicated that inactivation of p53 by human papillomavirus-encoded E6 protein in human keratinocytes and epithelial cells resulted in downregulation of Notch1 expression. Therefore, taken together, the observations described here and in the two studies [35,36] are consistent with the idea that the human Notch1 gene is a transcriptional target of p53 in a variety of human cells. More importantly, these studies provide support for the idea that, depending on cellular context, Notch1 expression contributes to a different cell fate.
Aberrant Notch1 signaling has been cited as causative in T-cell lymphoblastic leukemia, certain lymphomas, breast carcinomas, and kidney carcinomas [18]. Significantly, our observations raise the possibility that Notch1 protein can function as a survival effector in wild-type p53-containing human cancer cells exposed to genotoxic stress. Because many human tumors maintain wild-type p53 status, inhibition of Notch1 function provides a logical approach to selectively targeting such tumors for effective cancer therapy.
Acknowledgements
We thank A. Levine and M. Murphy for generously providing the cell lines.
Abbreviations
- GSI
gamma secretase inhibitor
- NIC
notch intracellular domain
Footnotes
This research was supported by an award from the Department of Veteran Affairs, Medical Research, and Development Service to D.C.
References
- 1.Ko J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 5.Ljungman M. Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress. Neoplasia. 2000;2:208–225. doi: 10.1038/sj.neo.7900073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pucci B, Kasten M, Giordano A. Cell cycle and apoptosis. Neoplasia. 2000;2:291–299. doi: 10.1038/sj.neo.7900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei C-L, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 8.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 9.Wilson A, Radtke F. Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett. 2006;580:2860–2868. doi: 10.1016/j.febslet.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 10.Katsube K, Sakamoto K. Notch in vertebrates—molecular aspects of the signal. Int J Dev Biol. 2005;49:369–374. doi: 10.1387/ijdb.041950kk. [DOI] [PubMed] [Google Scholar]
- 11.Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- 12.Koo EH, Kopan R. Potential role of presenilin-regulated signaling pathways in sporadic neurodegeneration. Nat Med. 2004;10:S26–S33. doi: 10.1038/nm1065. [DOI] [PubMed] [Google Scholar]
- 13.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Rastransformed cells. Nat Med. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 14.Curry CL, Reed LL, Golde TE, Miele L, Nickoloff BJ, Foreman KE. Gamma secretase inhibitor blocks Notch activation and induces apoptosis in Kaposi's sarcoma tumor cells. Oncogene. 2005;24:6333–6344. doi: 10.1038/sj.onc.1208783. [DOI] [PubMed] [Google Scholar]
- 15.Qin JZ, Stennett L, Bacon P, Bonder B, Hendrix MJ, Seftor RE, Seftor EA, Margaryan NV, Pollock PM, Curtis A, et al. p53-independent NOXA induction overcomes apoptotic resistance of malignant melanomas. Mol Cancer Ther. 2004;3:895–902. [PubMed] [Google Scholar]
- 16.Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iso T, Hamamori Y, Kedes L. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Arterioscler Thromb Vasc Biol. 2003;23:543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- 18.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 19.Mungamuri SK, Yang X, Thor AD, Somasunderam K. Survival signaling by Notch1: mammalian target of rapamycin (mTOR)-dependent inhibition of p53. Cancer Res. 2006;66:4715–4724. doi: 10.1158/0008-5472.CAN-05-3830. [DOI] [PubMed] [Google Scholar]
- 20.Nair P, Somasundaram K, Krishna S. Activated Notch1 inhibits p53-induced apoptosis and sustains transformation by human papillomavirus type 16 E6 and E7 oncogenes through a PI3K-PKB/Akt-dependent pathway. J Virol. 2003;77:7106–7112. doi: 10.1128/JVI.77.12.7106-7112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laws AM, Osborne BA. p53 regulates thymic Notch1 activation. Eur J Immunol. 2004;34:726–734. doi: 10.1002/eji.200324772. [DOI] [PubMed] [Google Scholar]
- 22.Dumont P, Leu JI, Pietra AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 23.Alimirah F, Chen J, Basrawala Z, Xin H, Choubey D. DU-145 and PC-3 human prostate cancer cell lines express androgen receptor: implications for the androgen receptor functions and regulation. FEBS Lett. 2006;580:2294–2300. doi: 10.1016/j.febslet.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 24.D'souza S, Xin H, Walter S, Choubey D. The gene encoding p202, an interferon-inducible negative regulator of the p53 tumor suppressor, is a target of p53-mediated transcriptional repression. J Biol Chem. 2001;276:298–305. doi: 10.1074/jbc.M007155200. [DOI] [PubMed] [Google Scholar]
- 25.Van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, Nordeen SK, Miller GJ, Lucia MS. Molecular characterization of human prostate carcinoma cell lines. Prostate. 2003;57:205–225. doi: 10.1002/pros.10290. [DOI] [PubMed] [Google Scholar]
- 26.Shou J, Ross S, Koeppen H, de Sauvage FJ, Gao WQ. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 2001;61:7291, 7297. [PubMed] [Google Scholar]
- 27.Meek DW. The p53 response to DNA damage. DNA Repair. 2004;3:1049–1056. doi: 10.1016/j.dnarep.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 28.Callahan R, Raafat A. Notch signaling in mammary gland tunorigenesis. Neoplasia. 2001;6:23–36. doi: 10.1023/a:1009512414430. [DOI] [PubMed] [Google Scholar]
- 29.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 30.McKay BC, Ljungman M. Role for p53 in the recovery of transcription and protection against apoptosis induced by ultraviolet light. Neoplasia. 1999;1:276–284. doi: 10.1038/sj.neo.7900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, Klein R, Tian X, Cheng HT, Kopan R, Shen J. Notch activation induces apoptosis in neural progenitor cells through a p53- dependent pathway. Dev Biol. 2004;269:81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Fei P, Bernhard EJ, El-Deiry WS. Tissue-specific induction of p53 targets in vivo. Cancer Res. 2002;62:7316–7327. [PubMed] [Google Scholar]
- 33.Bocchetta M, Miele L, Pass HI, Carbone M. Notch-1 induction, a novel activity of SV40 required for growth of SV40-transformed human mesothelial cells. Oncogene. 2003;22:81–89. doi: 10.1038/sj.onc.1206097. [DOI] [PubMed] [Google Scholar]
- 34.Kemp CJ, Donehower LA, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74:813–822. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
- 35.Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, Devgan V, Lieb J, Raffoul W, Hohl D, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCK-alpha kinases. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yugawa T, Handa K, Narisawa-Saito M, Ohno SI, Fujita M, Kiyono T. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol. 2007 doi: 10.1128/MCB.02119-06. [Epub March 12]. [DOI] [PMC free article] [PubMed] [Google Scholar]