Abstract
RNase L, a principal mediator of innate immunity to viral infections in higher vertebrates, is required for a complete IFN antiviral response against certain RNA stranded viruses. dsRNA produced during viral infections activates IFN-inducible synthetases that produce 5′-phosphorylated, 2′,5′-oligoadenylates (2-5A) from ATP. 2-5A activates RNase L in a wide range of different mammalian cell types, thus blocking viral replication. However, 2-5A has unfavorable pharmacologic properties; it is rapidly degraded, does not transit cell membranes, and leads to apoptosis. To obtain activators of RNase L with improved drug-like properties, high-throughput screening was performed on chemical libraries by using fluorescence resonance energy transfer. Seven compounds were obtained that activated RNase L at micromolar concentrations, and structure–activity relationship studies resulted in identification of an additional four active compounds. Two lead compounds were shown to have a similar mechanistic path toward RNase L activation as the natural activator 2-5A. The compounds bound to the 2-5A-binding domain of RNase L (as determined by surface plasmon resonance and confirmed by computational docking), and the compounds induced RNase L dimerization and activation. Interestingly, the low-molecular-weight activators of RNase L had broad-spectrum antiviral activity against diverse types of RNA viruses, including the human pathogen human parainfluenza virus type 3, yet these compounds by themselves were not cytotoxic at the effective concentrations. Therefore, these RNase L activators are prototypes for a previously uncharacterized class of broad-spectrum antiviral agents.
Keywords: high-throughput screening, interferon, 2-5A, virus
Antiviral agents that stimulate host immunity have inherent advantages over those that block viral proteins. Drugs that target specific viral proteins may be efficacious for a specific virus or type of virus, but they lack broad-spectrum antiviral activity and often lead to viral escape mutants. Antiviral strategies involving the host innate immune system are expected to provide a wider antiviral effect against unrelated types of viruses. Type I IFNs are the principal antiviral cytokines produced during the innate immune response to a wide range of different types of viral infections (1). Although IFNs are used clinically against hepatitis C virus and some other human viral pathogens, the clinical uses of IFNs as antiviral agents are limited because most, if not all, viruses have acquired or evolved mechanisms for evading IFNs. Although almost any step in the IFN antiviral response is a potential target for viral virulence genes, the early steps, in particular, induction of type I IFN genes and IFN signal transduction through the JAK-STAT pathway, are frequently blocked by viruses (2).
The 2-5A/RNase L pathway is a classical innate immunity pathway activated by IFNs and the viral pathogen-associated molecular pattern, dsRNA. When stimulated by dsRNA, IFN-inducible 2′,5′-oligoadenylate synthetases produce a series of short 5′-phosphorylated, 2′,5′-linked oligoadenylates collectively referred to as 2-5A (px5′A(2′p5′A)n; x = 1-3; n ≥ 2) from ATP (3). Because dsRNA is frequently produced during viral infections, 2-5A often accumulates in IFN-treated and virus-infected cells (4–6). The principal species of 2-5A found in such cells is the trimeric form, p3A2′p5′A2′p5′A (4). The only well established function of 2-5A is activation of RNase L (7). To do so, 2-5A must have at least one 5′-phosphoryl group, the internucleotide linkages must be 2′ to 5′, and the nucleotides must be adenylyl residues (8). RNase L is activated by subnanomolar levels of 2-5A, resulting in the cleavage of single-stranded regions of RNA, preferentially after UU and UA dinucleotides (9, 10). Because many viruses block the IFN system upstream of RNase L, for instance by sequestering dsRNA from 2′,5′-oligoadenylate synthetases (11, 12), direct activation of RNase L is expected to induce an antiviral response.
RNase L is a fascinating enzyme with an interesting arrangement of structural and functional domains. From the N to the C terminus, there are nine ankyrin repeats, several protein kinase-like motifs, and the ribonuclease domain. What distinguishes RNase L from all other ankyrin-repeat proteins is that ankyrin repeats 2 and 4 of RNase L constitute the 2-5A-binding site (13). In the absence of 2-5A, inhibitory domains in the ankyrin and protein kinase motifs suppress the ribonuclease domain while also maintaining RNase L as a monomer because of masking of the interaction sites. The binding of 2-5A to the ankyrin region induces a conformational shift that releases internal interactions, first allowing the monomers to attract and then to dimerize. In the dimer, the nuclease domains are no longer repressed by internal interactions and are thus able to cleave RNA (14). RNase L is present in a wide range of different human cell types and tissues because of the presence of several different tissue-specific promoter elements in the RNASEL gene (15). In contrast, no 2-5A-binding proteins were detected in several different organs of rnasel−/− mice (16). Therefore, the ability to bind 2-5A with high affinity (KD = 40 pM) (17) is a unique biochemical property of RNase L. As a result, 2-5A is an unambiguous signal for initiating RNA cleavage through activation of RNase L.
2-5A itself is an intracellular mediator that does not transit the cell membrane unassisted and is rapidly degraded in serum and in cells (18). Furthermore, sustained activation of RNase L by 2-5A induces apoptosis (19). Therefore, as an alternative to 2-5A, we sought to identify small activators of RNase L by high-throughput screening (HTS) of chemical libraries with a FRET method. Here, we describe such compounds that can enter cells unassisted, are not cytotoxic, and yet suppress divergent single-stranded RNA viruses.
Results
Identification of RNase L Activators by HTS.
To identify RNase L activators with drug-like properties, we screened libraries of small molecules. Our FRET method for RNase L activation was adapted for HTS (20). The RNA sequence of the FRET probe corresponds to a segment of the intergenic region of the paramyxovirus, respiratory syncytial virus genomic RNA, chosen because it contains several cleavage sites for RNase L (UU or UA) in an optimal context for cleavage [within predicted single-stranded loop regions determined with MFOLD (21)]. We obtained an EC50 (effective concentration of activator to give 50% maximum activation of RNase L) of 0.5 nM with authentic trimer 2-5A (p3A2′p5′A2′p5′A) (Fig. 1A). The dephosphorylated trimer, A2′p5′A2′p5′A, was unable to activate RNase L, consistent with prior findings (22). There was no increase in the signal in reactions containing the FRET probe but lacking either RNase L or 2-5A. The signal-to-noise ratio was ≈10:1, and the assay was very robust. Statistical parameters [Z′-factors and coefficient of variation are provided in supporting information (SI) Table 1] (23). HTS was performed on the ChemBridge DIVERset of 30,000 small molecules and on the 1,990-compound small Diversity Set from the National Cancer Institute [National Institutes of Health (NIH)]. Compounds chosen for retesting provided signals that were at least 3-fold over background.
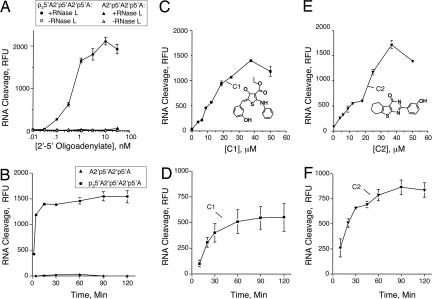
Fig. 1.
RNase L activation by pppA2′p5′A2′p5′A and A2′p5′A2′p5′A (A and B), compound 1 (C and D), and compound 2 (E and F) as determined in FRET assays. Reactions were for 30 min (A) or 90 min (C and E) at 22°C. Reactions in B contained 3 nM pppA2′p5′A2′p5′A and A2′p5′A2′p5′A; and reactions in D and F contained 25 μM compound 1 and compound 2, respectively. C1, compound 1; C2, compound 2.
Seven “hits” were obtained from the ChemBridge library (SI Fig. 6, compounds 1–7), whereas no RNase L activators were found in the NIH Diversity set library. The hits have molecular masses that range from 298 to 470 Da and were capable of activating RNase L in micromolar range (EC50 values between 22 and 99 μM). The most active compounds (1 and 2), had EC50 values of 26 and 22 μM, respectively, with peak activity at 40 μM (higher concentrations showed reduced activities because of poor solubility in aqueous solution) (Fig. 1 C and E). Therefore, although these compounds activated RNase L, they were much less active than the natural activator, 2-5A. In addition, cleavage of the FRET probe reached a maximum in 15 min with trimeric 2-5A (p3A2′p5′A2′p5′A), but not until 60–90 min with compounds 1 and 2 (Fig. 1 B, D, and F).
Compounds related in structure to compounds 1, 2, and 3 were identified in the ChemBridge repository by using a searchable database (www.hit2lead.com). Two compounds related in structure to compound 1 were either less active (compound 8) or inactive (compound 12) (SI Fig. 6). However, two compounds related to compound 2 were equally active (compounds 9 and 10), whereas two other such compounds were inactive (data not shown). One compound equally active and related in structure to compound 3 was identified (compound 11).
Validation and Characterization of the RNase L Activators.
To verify that the two most active hits (compounds 1 and 2) are, in fact, capable of activating RNase L, we performed alternative ribonuclease assays with three different RNA substrates (Fig. 2 A–C). RNase L activated by 2-5A or compounds 1 or 2 cleaved these RNA substrates on the 3′ side of the UU or UA dinucleotide sequence, consistent with the known sequence specificity of RNase L (9, 10). Cleavage of the FRET probe, analyzed by in situ fluorescence of the gel, showed multiple cleavages in the two predicted loop regions (Fig. 2A). Cleavage of two different 32P-labeled short RNA molecules produced the expected products when RNase L was activated by 2-5A, compound 1 or compound 2 (Fig. 2 B and C).
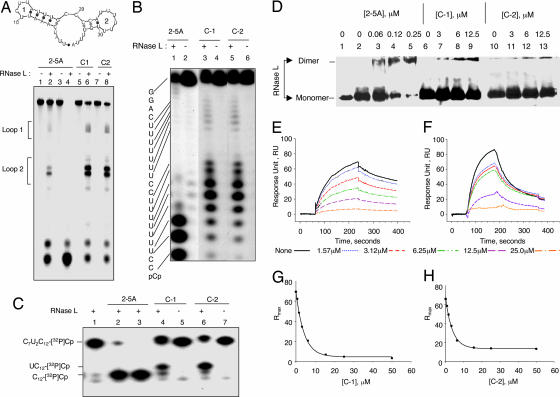
Fig. 2.
Small molecules interact with the 2-5A-binding domain inducing RNase L dimerization and activation. (A Upper) Predicted secondary structure of RNA sequence of FRET probe determined with MFOLD (21). (A Lower) Lanes 1 and 2, 0.3 nM trimeric 2-5A; lanes 3 and 4, 3 nM trimeric 2-5A; lanes 5 and 6, 25 μM compound 1; or lanes 7 and 8, 25 μM compound 2 was incubated with or without 25 nM RNase L (as indicated) with FRET RNA probe for 22°C for 30 min (with 2-5A), or for 90 min (with C1 and C2). Gel was visualized by using a fluorescence scanner (Bio-Rad, Hercules, CA) (SI Methods). (B) Lanes 1 and 2, 25 nM trimeric 2-5A; lanes 3 and 4, 25 μM compound 1; or lanes 5 and 6, 25 μM compound 2 was incubated with or without 25 nM of RNase L (as indicated) on ice for 30 min, followed by incubation with r(GGACUUUUUUUCCCUUUUUUUCC)-[32P]pCp at 22°C for 30 min. (C) Lanes 1–3, 0, 12.5, and 25 nM trimeric 2-5A, respectively; lanes 4 and 5, 25 μM compound 1; or lanes 6 and 7, 25 μM compound 2 were incubated with or without 25 nM RNase L (as indicated) on ice for 30 min, followed by incubation with C7U2C12-[32P]pCp at 22°C for 30 min. (D) RNase L in the absence (lane 1) or presence (lanes 2–13) of the cross-linking agent dimethyl suberimidate was incubated in the absence of activator (lanes 2, 6, and 10) or in the presence of trimer 2-5A (lanes 3 to 5); (lanes 7–9) compound 1; and (lanes 11–13) compound 2. (E–H) Biotinylated 2-5A (SI Methods) was immobilized on streptavidin biosensor chips. RNase L (10 nM) in the presence of varying concentration of either compound 1 (E and G) or compound 2 (F and H) was passed over the chip. Response units (RU) as a function of time (E and F) and maximum response units at equilibrium (Rmax) as a function of concentration (G and H) were calculated by using global fit and plotted. C1, compound 1; C2, compound 2; trimeric 2-5A, p3A2′p5′A2′p5′A.
RNase L Dimerizes During Activation in Response to the Small Molecules.
RNase L dimerization is a prerequisite for the nuclease activation (14). Therefore, we performed protein/protein cross-linking using dimethyl suberimidate (SI Methods) (24). The oligomeric state of RNase L was determined after SDS/PAGE in Western blots probed with monoclonal antibody against RNase L. Monomer RNase L converted to dimer in the presence of 0.06–0.25 μM 2-5A or by 3–12 μM either compound 1 or compound 2 (Fig. 2D).
The Small-Molecule Activators Interact with the 2-5A-Binding Domain of RNase L.
To determine whether the activators interact with the 2-5A-binding domain of RNase L, a 2-5A competition binding assay was performed by using surface plasmon resonance (SPR) on a model 3000 (Biacore, Uppsala, Sweden) (SI Methods). A 2-5A analog (pA2′p5′A2′p5′A2′p5′A linked through its 2′,3′-terminal ribose to biotin) was used as competitor. Mixtures of RNase L (10 nM) and varying concentrations of compounds 1 or 2 were passed over streptavidin chips precoated with 2-5A-biotin. Sensograms were recorded, and the response units (RU) and maximum resonance units (Rmax) at equilibrium were plotted as a function of time by using Bia-evaluation software (Fig. 2 E–H). A dose-dependent decrease in the resonance response was observed with both compounds 1 and 2 (Fig. 2 E and F, respectively). The data indicate that these compounds compete with 2-5A for RNase L binding. However, analysis of the data indicated that the binding constants (KD) for compounds 1 and 2 are 18 and 12 μM, respectively, as compared with 0.22 nM for 2-5A (data not shown). These results demonstrate that compounds 1 and 2 follow a similar mechanistic path as 2-5A, they interact with the ligand-binding domain, and induce catalytically active dimers of RNase L.
Computational Docking of Small Molecules with the 2-5A-Binding Domain of RNase L.
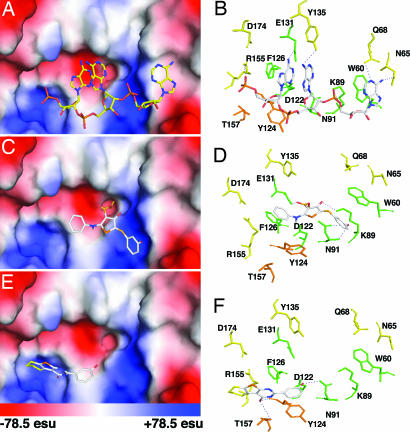
A prediction of how compounds 1 and 2 bind RNase L was obtained by using AutoDock version 3.0.5 (25) (see SI Methods). Previously reported coordinates obtained by x-ray crystallography of a portion of RNase L (amino acids 21–305, ANK) complexed with 2-5A (p5′A2′p5′A2′p5′A) were used (Fig. 3 A and B) (26). The final docked conformations for compounds 1 and 2 bound to RNase L were selected on the basis of minimum free energies of binding. The free energy ΔGbinding for compounds 1 and 2 were 9.2 kcal mol−1 and 6.8 kcal mol−1, respectively, for the best docked structure. The ΔGbinding for 2-5A is 13.2 kcal mol−1, whereas docking of the inactive compound 12 had a ΔGbinding of 3.5 kcal mol−1 (SI Fig. 7). Compounds 1 or 2 are predicted to share an overlapping binding pocket within ankyrin repeats 2 and 4 of RNase L, as does the natural activator, p5′A2′p5′A2′p5′A (13).
Fig. 3.
Compounds 1 and 2 dock with the 2-5A-binding pocket of RNase L. Surface representations and side-chain interactions of p5′A2′p5′A2′p5′A (A and B), compound 1 (C and D), and compound 2 (E and F) to the ankyrin repeat region of human RNase L (PDB ID code 1WDY). Unique interactions in RNase L with either compound 1 or 2 are depicted in orange, whereas the common interactions with RNase L between 2-5A and compounds 1 or 2 are in green. esu, electrostatic unit.
Small-Molecule Activators of RNase L Suppress Replication of Different RNA Viruses yet Are Not Cytotoxic at Effective Concentrations.
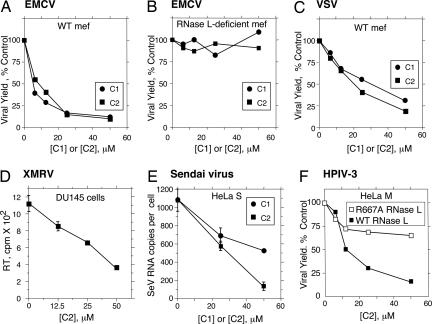
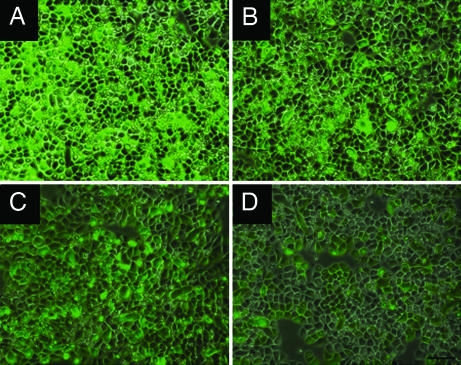
To determine whether compounds 1 and 2 possessed antiviral activity, virus growth assays were performed with a range of different types of RNA viruses. Fibroblasts (mef) from WT and rnasel−/− (RNase L-deficient) mouse embryos were infected with the picornavirus, encephalomyocarditis virus (EMCV) (a positive RNA strand virus). Either compound 1 or compound 2 caused a 7- to 8-fold reduction in viral growth in WT mef; however, there was no effect in RNase L-deficient mef (Fig. 4 A and B, respectively). Similar findings were observed when using the negative RNA strand rhabdovirus, vesicular stomatitis virus (Fig. 4C and data not shown). Compound 2 was effective in suppressing an acute infection of human DU145 prostate cancer cells with the human retrovirus, XMRV (Fig. 4D) (27, 28). Compound 2 was more effective than compound 1 in suppressing Sendai virus, a negative RNA strand paramyxovirus that evades IFN action (29) (Fig. 4E). We further demonstrate antiviral activity of compound 2 against a human pathogenic virus, human parainfluenza virus 3 (HPIV3) in which a GFP reporter gene was inserted between the P and M genes (H.M. and A.K.B., unpublished work). Compound 2 suppressed HPIV3 replication ≈7-fold in HeLa M cells expressing WT RNase L, whereas virus growth was reduced <2-fold in the same cells expressing a nuclease-dead RNase L mutant (R667A) cDNA (30). A dose-dependent 5- to 6-fold decrease in fluorescence generated by HPIV3/GFP was visible in infected HeLa S cells (expressing endogenous WT RNase L) at 24 h after infection with increasing concentrations of compound 2 (Fig. 5, panels B to D). Neither compound 1 nor compound 2 reduced cell viability of mef or HeLa S cells at concentrations up to 50 μM (SI Fig. 8). These results show that activators of RNase L produce relatively broad antiviral effects, effectively by-passing viral evasion of innate immunity, without causing significant cytotoxicity.
Fig. 4.
Small-molecule activators of RNase L suppress diverse RNA viruses. Effects of compounds 1 or 2 on EMCV replication in rnasel+/+ (WT) (A) and rnasel−/− (RNase L-deficient) mef infected with EMCV (B); compounds 1 or 2 on VSV replication in WT mef (C); compound 2 on acute XMRV infection of DU145 cells as determined by reverse transcriptase (RT) released in the conditioned media (D); compounds 1 or 2 on Sendai virus replication in HeLa S cells as determined by real-time RT-PCR of the viral genomic RNA (SI Methods) (E); and compound 2 on replication of HPIV3 in HeLa M cells expressing either a nuclease-dead mutant of RNase L (R667A) or WT RNase L (F). All assays were performed in triplicate and represent results of plaque assays on indicator cells except for D and E.
Fig. 5.
RNase L activators suppress HPIV3 replication. HeLa S cells were infected with HPIV3/GFP at an MOI of 1.0. At 90 min after infection, media were removed and cells were washed twice with PBS. Cells were incubated with complete media in the absence (A) or containing 10, 50, and 100 μM of compound 2 (B–D), respectively. At 24 h after infection, fluorescence images were captured by using a Leica DM IRB fluorescence microscope.
Discussion
These HTS efforts have identified several novel small molecules that activate RNase L, the first compounds other than the natural activators, 2-5A, known to do so. Seven activators were identified from screening ≈32,000 compounds, a hit rate of 1 in 4,571. A previous HTS effort that identified inhibitors of the ribonucleolytic function of angiogenin resulted in a “hit rate” ≈4-fold higher (31). Comparison of these reports suggests that the occurrence of ribonuclease activators in small molecule libraries may be much less frequent than inhibitors. In both of these studies, however, minor chemical modifications resulted in loss of activity, demonstrating specificity in the mode-of-action of the compounds (e.g., compare compounds 1 and 12 in (SI Fig. 6).
The ability of compounds 1 and 2 to specifically bind with and activate RNase L was validated in several different assays systems. Furthermore, the compounds produced a similar mechanistic pathway to RNase L activation as 2-5A, binding to the ligand interaction domain inducing enzyme dimerization and ribonucleolytic activity. The docking models show both similarities and differences between how compounds 1 and 2 interact with RNase L as compared with 2-5A (Fig. 3). For example, the aromatic rings of Trp-60 and Phe-126 that stack with adenine ring 1 and 3 of 2-5A also show similar interactions with phenyl ring 1 and 3 of compound 1; with comparable average stacking distances of ∼4.0 Å (Fig. 3 B and D). In addition, the hydroxyl group on phenyl ring 1 is in a two-way H-bond formation with Asn-91 and the carbonyl group of ring 2 is in close contact with Lys-89. A related structure, compounds 12, which lacks activity, docks in the same binding pocket, but with low binding energy (ΔGbinding = 3.5 kcal mol−1) (SI Fig. 7). Because of the bulky group substitution on phenyl ring 1, compound 12 fails to stack with Phe-126. Compound 2 has unique interactions within the 2-5A binding domain (Fig. 3 E and F). The hydroxyl group on ring 1 of compound 2 can potentially form a two-way H-bond with Asn-91 and Asp-122 (latter not shown) whereas the carbonyl group at ring 2 is in close proximity with Thr-157 and the main chain oxygen of Tyr-124. In addition, the aromatic ring 1 shows stacking interaction with phenolyl ring of Tyr-124.
Our current lead compounds require ≈105 fold higher concentrations to activate RNase L compared with the natural ligand, 2-5A. Clearly, therefore, there is a large scope for improvement. However, the cell culture data are remarkable in that five divergent RNA viruses (a picornavirus, a rhabdovirus, two paramyxoviruses, and a retrovirus) were similarly inhibited. It is necessary only for RNase L to cleave a viral RNA genome once to inactivate it. In contrast, the host cell is resilient to a low level of RNA decay. Therefore, it should be possible to increase the potency of RNase L activation although retaining relatively low toxicity. Future HTS efforts, coupled with structure-based drug design by docking to the ligand-binding domain of RNase L, are expected to lead to compounds with improved antiviral activities. A significant advantage of targeting RNase L with small molecule activators, is that most viruses interfere with the IFN system at steps in innate immunity that precede RNase L. Therefore, activators of RNase L will circumvent many mechanisms viruses have for evading host defense pathways. Our results demonstrate that RNase L is a bona fide target for development of small molecule activators with the potential to provide a wide range of activities against different human viral pathogens.
Materials and Methods
Chemicals and 2-5A.
Compounds (≈30,000) were from the Diverset collection (Chembridge Corp., San Diego, CA) and 1,990 compounds were from the NCI Diversity Set. 2-5A was prepared enzymatically from ATP by using hexahistidine tagged and purified, recombinant porcine 42 kDa 2-5A Synthetase (OAS) (32). The 2-5A synthetase was activated by using immobilized poly(I):poly(C)-agarose (33). Separation of 2-5A oligomers was by HPLC (Beckman Systems Gold, Fullerton, CA) on a Dionex semipreparative column (DNA PacPA-100, 4 × 250, Dionex, Sunnyvale, CA) or by FPLC (Amersham Pharmacia, Piscataway, NJ) on a mono-Q HR10/10 column (Amersham Bioscience).
HTS.
HTS was performed by a modification of our previously described FRET assay for RNase L activity (20). The RNA substrate for the FRET assays was a 36 nucleotide synthetic oligoribonucleotide, 6-FAM-UUA UCA AAU UCU UAU UUG CCC CAU UUU UUU GGU UUA-BHQ-1 (Integrated DNA Technologies, Inc., Coralville, IA). Recombinant human RNase L expressed from a baculovirus vector in insect cells was purified by FPLC as described previously (20, 22). HTS was performed in black polypropylene 96-well round bottom Costar plates (Fisher Scientific, Newark, DE). Reaction mixtures contained 25 nM RNase L and 100 nM FRET probe (Km = 75 nM) in a final volume of 50 μl cleavage buffer [25 mM Tris·HCl (pH 7.4), 100 mM KCl, 10 mM MgCl2, 50 μM ATP, 7 mM 2-mercaptoethanol]. Test compounds or 2-5A were added with a 96-well standard replicator (Incyte, Wilmington, DE). Stock concentrations of compounds were 10 mM (NCI library) or 5 mg/ml (ChemBridge compounds) in 100% DMSO and the final concentrations of compounds during HTS were 50 to 65 μM. Trimeric 2-5A (10 nM), Z'-factor >0.8, was used as a positive control in every screening plate. The assay plates were gently agitated, centrifuged briefly at 150 × g and incubated at 22°C for up to 2 h. Fluorescence was measured with a Wallac 1420 Victor2 multilabel counter (PerkinElmer Life Sciences, Shelton, CT) (excitation 485 nm; emission 535 nm). False positives were eliminated by rescreening compounds in the absence of RNase L.
RNA Cleavage Assays.
RNA cleavage assays were performed by using r(C7U2C12) (34) and r(GGACUUUUUUUCCCUUUUUUUCC) (ITD) 3′end [32P]-pCp labeled RNA with T4 RNA ligase (Cat. M1051; Promega, Madison, WI). Purified RNase L (25 nM per reaction) was incubated with 25 nM trimeric 2-5A, or lead compounds in cleavage buffer for 30 min on ice, followed by incubation with labeled RNA at 22°C. Gel loading buffer II (Ambion; Cat. 8546G) was added, samples were heated before load onto 20% acrylamide/7 M urea/TBE sequencing gels. Results were visualized by exposing x-ray film.
Cell Culture and Viral Growth.
WT and rnasel−/− mef cell lines (16) transformed with SV40 T antigen were grown in RPMI medium 1640 supplemented with 10% FBS (Invitrogen, Carlsbad, CA) and penicillin/streptomycin. HeLa S, HeLa M, and CV1 cells were grown in DMEM supplemented with 10% FBS and penicillin/streptomycin. Mef were seeded in 12-well plates at 2.5 × 105 cells per well, and, after 1 h, were infected with EMCV (strain K) (MOI = 0.5) or VSV (Indiana) (MOI = 0.1) in media without serum. After 1 h, media were removed, cells were washed with PBS, and complete media were added containing different amounts of compound 1 or compound 2. At 12 h after infection, media containing virus was collected from each well, and virus titer were determined by plaque assays on indicator cells of mouse L929 fibroblast cell line for EMCV or CV1 for VSV. HeLa M cells expressing WT or R667A mutant RNase L (30) were seeded in six-well plates and infected with HPIV3/GFP at an MOI of 1.0 in media without serum. After 90 min, media were removed, cells were washed with PBS, and complete media were added with different amounts of compound 1 or compound 2. At 24 h after infection, virus was collected by freeze–thawing twice, and plaque was assayed on CV1 cells. Alternatively, HeLa S cells were infected with HPIV3/GFP at an MOI of 1.0, and, subsequently, fluorescence images were captured by using a fluorescence microscope (DM IRB; Leica, Wetzlar, Germany). DU145 cells were infected with XMRV in FBS-free RPMI medium 1640 with 8 μg/ml polybrene for 3 h, followed by washing with PBS. Cells were treated in triplicate with compound 2 with fresh complete RPMI medium 1640 for 2 d. Reverse transcriptase activities in cell supernatants were measured as described (28). Sendai virus (Cantell strain) at 20 HA units/ml was used to infect HeLa S cells, seeded in six-well plates at 50 × 104 cells per well, in medium without serum. After 90 min, media were removed, cells were washed with PBS, and complete media were added with different doses of compounds 1 and 2. At 24 h after infection, virus in supernatant was collected from each well, and viral genomic RNA strands were determined by real-time RT-PCR (SI Methods). Viability of cells was determined by using the colorimetric CellTiter 96 Aqueous Cell Proliferation Assay (MTS assay) (Promega). Cells (104 per well) in 96-well culture plates were treated with compound for 24 h. MTS reagent was added to each well, and incubation was continued at 37°C for 3 h. Absorbance was measured at 490 nm with a 96-well plate reader (Wallac 1420 Victor2 multilabel counter; PerkinElmer Life Sciences, Boston, MA). Experiments were performed in triplicate, and SDs were calculated.
Supplementary Material
Acknowledgments
We thank Jayashree M. Paranjape, Anatoliy Prokvolit, and Greggory Wroblewski for expert technical assistance and Jonathan Karn, Zan Xu, and Malathi Krishnamurthy (Cleveland) for valuable discussions, and to Rune Hartmann (Aarhus, Denmark) for the gift of OAS cDNA. Biacore 3000 use was in the Molecular Biotechnology Core facility, Cleveland Clinic (Satya Yadav, Director). These studies were supported by National Institutes of Health, National Cancer Institute Grant CA044059 and National Institute of Allergy and Infectious Disease Grant U54-A1057160.
Abbreviations
- 2-5A
5′-phosphorylated 2′,5′-linked oligoadenylates
- HTS
high throughput screening
- RU
response units
- Rmax
maximum resonance units
- SPR
surface plasmon resonance.
Footnotes
Conflict of interest statement: R.H.S. is a member of the Scientific Advisory Board of Alios BioPharma, San Francisco, CA.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700590104/DC1.
References
- 1.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Horvath CM. Eur J Biochem. 2004;271:4621–4628. doi: 10.1111/j.1432-1033.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- 3.Kerr IM, Brown RE. Proc Natl Acad Sci USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knight M, Cayley PJ, Silverman RH, Wreschner DH, Gilbert CS, Brown RE, Kerr IM. Nature. 1980;288:189–192. doi: 10.1038/288189a0. [DOI] [PubMed] [Google Scholar]
- 5.Rice AP, Kerr SM, Roberts WK, Brown RE, Kerr IM. J Virol. 1985;56:1041–1044. doi: 10.1128/jvi.56.3.1041-1044.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cayley PJ, Davies JA, McCullagh KG, Kerr IM. Eur J Biochem. 1984;143:165–174. doi: 10.1111/j.1432-1033.1984.tb08355.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhou A, Hassel BA, Silverman RH. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 8.Player MR, Torrence PF. Pharmacol Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 10.Floyd-Smith G, Slattery E, Lengyel P. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 11.Beattie E, Denzler KL, Tartaglia J, Perkus ME, Paoletti E, Jacobs BL. J Virol. 1995;69:499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Min JY, Krug RM. Proc Natl Acad Sci USA. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka N, Nakanishi M, Kusakabe Y, Goto Y, Kitade Y, Nakamura KT. EMBO J. 2004;23:3929–3938. doi: 10.1038/sj.emboj.7600420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong B, Silverman RH. J Biol Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 15.Zhou A, Molinaro RJ, Malathi K, Silverman RH. J Interferon Cytokine Res. 2005;25:595–603. doi: 10.1089/jir.2005.25.595. [DOI] [PubMed] [Google Scholar]
- 16.Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman RH. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverman RH, Jung DD, Nolan-Sorden NL, Dieffenbach CW, Kedar VP, SenGupta DN. J Biol Chem. 1988;263:7336–7341. [PubMed] [Google Scholar]
- 18.Kubota K, Nakahara K, Ohtsuka T, Yoshida S, Kawaguchi J, Fujita Y, Ozeki Y, Hara A, Yoshimura C, Furukawa H, et al. J Biol Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- 19.Malathi K, Paranjape JM, Ganapathi R, Silverman RH. Cancer Res. 2004;64:9144–9151. doi: 10.1158/0008-5472.CAN-04-2226. [DOI] [PubMed] [Google Scholar]
- 20.Thakur CS, Xu Z, Wang Z, Novince Z, Silverman RH. Methods Mol Med. 2005;116:103–113. doi: 10.1385/1-59259-939-7:103. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong B, Xu L, Zhou A, Hassel BA, Lee X, Torrence PF, Silverman RH. J Biol Chem. 1994;269:14153–14158. [PubMed] [Google Scholar]
- 23.Zhang JH, Chung TD, Oldenburg KR. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 24.Davies GE, Stark GR. Proc Natl Acad Sci USA. 1970;66:651–656. doi: 10.1073/pnas.66.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 26.Tanaka N, Nakanishi M, Kusakabe Y, Goto Y, Kitade Y, Nakamura KT. Protein Pept Lett. 2005;12:387–389. doi: 10.2174/0929866053765734. [DOI] [PubMed] [Google Scholar]
- 27.Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, et al. PLoS Pathol. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Dong B, Kim S, Hong S, Das Gupta J, Malathi K, Klein EA, Ganem D, Derisi JL, Chow SA, Silverman RH. Proc Natl Acad Sci USA. 2007;104:1655–1660. doi: 10.1073/pnas.0610291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato A, Kiyotani K, Kubota T, Yoshida T, Tashiro M, Nagai Y. J Virol. 2007;81:3264–3271. doi: 10.1128/JVI.02590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han JQ, Townsend HL, Jha BK, Paranjape JM, Silverman RH, Barton DJ. J Virol. 2007 doi: 10.1128/JVI.01857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kao RY, Jenkins JL, Olson KA, Key ME, Fett JW, Shapiro R. Proc Natl Acad Sci USA. 2002;99:10066–10071. doi: 10.1073/pnas.152342999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Mol Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 33.Rusch L, Dong B, Silverman RH. Methods Enzymol. 2001;342:10–20. doi: 10.1016/s0076-6879(01)42531-6. [DOI] [PubMed] [Google Scholar]
- 34.Carroll SS, Chen E, Viscount T, Geib J, Sardana MK, Gehman J, Kuo LC. J Biol Chem. 1996;271:4988–4992. doi: 10.1074/jbc.271.9.4988. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.