Abstract
Bisphosphonate drugs (e.g., Fosamax and Zometa) are thought to act primarily by inhibiting farnesyl diphosphate synthase (FPPS), resulting in decreased prenylation of small GTPases. Here, we show that some bisphosphonates can also inhibit geranylgeranyl diphosphate synthase (GGPPS), as well as undecaprenyl diphosphate synthase (UPPS), a cis-prenyltransferase of interest as a target for antibacterial therapy. Our results on GGPPS (10 structures) show that there are three bisphosphonate-binding sites, consisting of FPP or isopentenyl diphosphate substrate-binding sites together with a GGPP product- or inhibitor-binding site. In UPPS, there are a total of four binding sites (in five structures). These results are of general interest because they provide the first structures of GGPPS- and UPPS-inhibitor complexes, potentially important drug targets, in addition to revealing a remarkably broad spectrum of binding modes not seen in FPPS inhibition.
Keywords: cell wall, geranylgeranyl diphosphate synthase, undecaprenyl diphosphate synthase, x-ray structure
Isoprenoid biosynthesis involves the condensation of C5-diphosphates to form a very broad range of compounds used in cell membrane (cholesterol, ergosterol), cell wall (lipid I, II, peptidoglycan) and terpene biosynthesis, electron transfer (quinone, heme a, carotenoid, chlorophyll), and in many eukaryotes, cell signaling pathways (Ras, Rho, Rap, Rac). There has, therefore, been considerable interest in developing specific inhibitors of some of these pathways to modify cell function. For example, the bisphosphonate drugs used to treat bone resorption diseases such as osteoporosis (1) have been thought to function by targeting farnesyl diphosphate synthase (FPPS, EC 2.5.1.10) in osteoclasts, leading to dysregulation of cell-signaling pathways involving small GTPases, and in some parasitic protozoa, leading to inhibition of ergosterol biosynthesis (2). However, in recent work Goffinet et al. (3) proposed that the main biological activity of the most potent bisphosphonate zoledronate (Zometa) in humans cells is directed against protein geranylgeranylation. This opens up the intriguing possibility that it might be possible to enhance potency by developing drugs that work by inhibiting geranylgeranyl diphosphate synthase (GGPPS, EC 2.5.1.30), the enzyme that produces the geranylgeranyl diphosphate (GGPP) used to geranylgeranylate e.g., Rac, Rap, and Rho. Based on the recent observation of a previously uncharacterized (GGPP) inhibitor site in GGPPS (4), we reasoned that larger, more hydrophobic species than those in current use might bind to this site and exhibit enhanced activity, because of increased hydrophobic stabilization and, in cells, enhanced lipophilicity. Here, we thus report structures of a series of five bisphosphonates bound to GGPPS together with, for comparative purposes, the structures of five isoprenoid diphosphate–GGPPS complexes. We find three quite different binding modes, corresponding to FPP/GPP (substrate), IPP (substrate), and GGPP [product/inhibitor (4)] site occupancy.
The FPPS and GGPPS enzymes noted above belong to a class of enzymes called trans-prenyltransferases that are involved in trans double-bond addition (Fig. 1A) of isopentenyl diphosphate (IPP) to dimethylallyl diphosphate (DMAPP) to form all-trans-isoprenoid diphosphates, such as FPP and GGPP. In addition to these trans-prenyltransferases, there is also a second class of enzymes called cis-prenyltransferases. These enzymes typically use an FPP (all-trans) substrate that is then elongated via cis double-bond addition (Fig. 1A), to form mixed (E,Z) long-chain isoprenoids, such as undecaprenyl diphosphate (UPPS, EC 2.5.1.31), a C55-diphosphate of considerable interest (5, 6) as a new target for anti-microbial therapy because undecaprenyl diphosphate (UPP) is used to form the lipid-I and lipid-II species needed for peptidoglycan cell-wall biosynthesis in bacteria. We also describe herein, therefore, the x-ray structures of five UPPS inhibitors bound to UPPS, the most active having an IC50 of <600 nM. The UPPS structures obtained are unusual in that we find four distinct binding sites, one of which (seen in all structures) corresponds to the FPP-binding site seen previously (7). In addition to these crystallographic results, we also report the activities and quantitative structure-activity relationships for a larger series of bisphosphonates in UPPS inhibition, with activities being predicted within a factor of ≈2 over an ≈103× range in activity, which, combined with the crystallographic results, should facilitate the further development of these compounds.
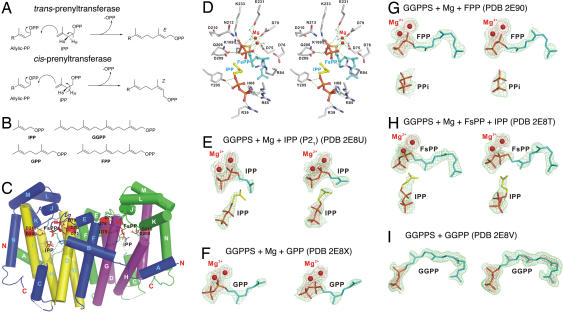
Fig. 1.
Chemistry and structures. (A) Illustration of HR,S atom abstraction in E,Z prenyltransferases. (B) Structures of isoprenoid diphosphates. (C) Structure of GGPPS-Mg–FsPP-IPP complex. (D) Substrate-binding region in C. (E–I) Electron densities (green contoured at 1σ, red at 3σ) for IPP (E), GPP (F), FPP (G), FsPP (H), and GGPP (I), bound to GGPPS.
Results and Discussion
Geranylgeranyl Diphosphate Synthase.
We first investigated the structures of four isoprenoid diphosphates: IPP, geranyl diphosphate (GPP), farnesyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP), Fig. 1B, together with thiolo-FPP (FsPP), bound to GGPPS from Saccharomyces cerevisiae, to deduce the principal binding sites for these species. Data collection and refinement statistics for five such GGPPS-diphosphate complexes are presented in supporting information (SI) Table 1, with the actual structures (Fig. 1 C–I, PDB ID codes 2E8U, 2E8X, 2E90, 2E8T, and 2E8V) providing a useful background with which to interpret the bisphosphonate-bound structures discussed below. The yeast enzyme has considerable homology to human GGPPS (43% identity, 60% similarity) and we also find that there is a good (R = 0.9, P = 0.0035) correlation between the Ki values for bisphosphonate inhibition of the yeast and human proteins (SI Table 2). So the overall results obtained here are likely to be of interest for the development of novel GGPPS inhibitors for a variety of eukaryotic species.
GGPPS crystallizes as a dimer (8) in one of two space groups (orthorhombic or monoclinic), and each monomer is composed almost entirely of α-helices. Fig. 1C shows a diagram of one representative new structure (GGPPS-Mg–FsPP-IPP) in which both IPP and FsPP bind to GGPPS and key protein–ligand interactions are illustrated in Fig. 1D. This structure closely resembles that seen with diphosphates and bisphosphonates bound to FPPS (9–13). However, when the structures of more diphosphates bound to GGPPS are investigated, we find evidence for other binding sites. For example, with IPP, Fig. 1E, we find that IPP binds to both the “normal,” homoallylic IPP site occupied by IPP in FPPS, and the allylic (GPP or FPP) binding site. The presence of two binding sites (one weak and one strong) has also been proposed for IPP binding to FPPS (9), based on isothermal titration calorimetry, and, given the results shown in Fig. 1E (PDB ID code 2E8U), it seems likely that FPPS-(IPP)2 has a similar structure, with IPP in one site chelating to Mg2+, as shown in Fig. 1E.
As the length of the isoprene side chain is increased, we see (Fig. 1 F–H) that the longer (GPP, C10; FPP, C15; FsPP, C15) side chains occupy the FPP substrate-binding site, but with GGPP (C20), this site is no longer occupied because there are three residues: Leu-67, Tyr-107, and His-139, which prevent chain elongation (i.e., there are no C25 products with GGPPS). In human GGPPS, it is known that GGPP is a GGPPS inhibitor and could be involved with negative feedback, and Kavanagh et al. (4) recently identified a so-called “inhibitor site,” occupied by GGPP. With the yeast enzyme, we find that the GGPP diphosphate group binds to the IPP site, whereas the C20 side chain binds to the FPP side chain site, as shown in Fig. 1I, similar to the orientation seen in GGPPS from Sinapis alba (14), but there is no Mg2+ present. As expected, neither IPP (Fig. 1H) nor PPi (Fig. 1G) can bind here because the IPP-diphosphate site is occupied. So, the yeast GGPP product binds with its diphosphate in the IPP diphosphate site (IPP), whereas the GGPP side chain binds to the FPP site. However, in human GGPPS, the GGPP product binds with its diphosphate in the FPP site, whereas the GGPP side chain binds to the novel (human) “inhibitor site” (GGPP). These results show a remarkably broad range of binding motifs for diphosphates in GGPPS. With the small IPP ligand, both the allylic and the homoallylic sites can be occupied, because the ligand side chains are small. This is not seen with GPP, because the presence of a C10 side chain in both sites would likely produce a steric clash, but smaller ligands (PPi or IPP) can bind in the presence of large (FPP, FsPP) side chains (Fig. 1 G and H). With the very large GGPP species, both the IPP and FPP sites are occupied, Fig. 1I, but by just one molecule, spanning both sites. Based on the similarity between the diphosphate substrates (and product) and bisphosphonate inhibitors (15), there appear therefore to be at least four likely binding modes for bisphosphonates in GGPPS: FPP-FPP, FPP-GGPP, IPP-FPP, and IPP-GGPP. That is, diphosphate moieties can bind in either the IPP or FPP sites, whereas the side chains can bind in either the FPP (substrate), GGPP (inhibitor), or, if small, the IPP site.
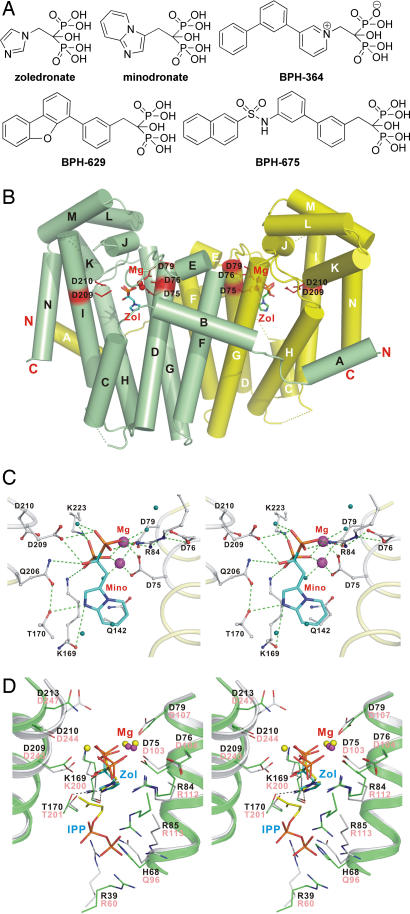
We next consider, therefore, the actual binding modes seen with bisphosphonate inhibitors and begin by investigating the small, “third generation” species zoledronate and minodronate (Fig. 2A), potent FPPS inhibitors that also inhibit GGPPS (SI Table 2). Data collection and refinement statistics for all five GGPPS–bisphosphonate complexes are given in SI Table 3. The overall structures of each of the inhibitor-bound GGPPS complexes closely resemble that seen in the apo-enzyme at 1.98-Å resolution as well as the structures of each of the diphosphate–GGPPS complexes shown in Fig. 1, as shown for example in Fig. 2B for the GGPPS–zoledronate complex. With both zoledronate (SI Fig. 7A) and minodronate, Fig. 2C, it can be seen that these bisphosphonates form GGPPS complexes containing 2 Mg2+ that are coordinated to conserved aspartate residues, with the bisphosphonate backbones also interacting with the side chains of Arg-84, Lys-169, Asp-209, and Lys-223 via hydrogen bonds. The distal N-atoms in the rings are also hydrogen bonded to the side-chain oxygen of Thr-170 and the main-chain carbonyl oxygen of Lys-169 (Fig. 2C and SI Fig. 7A). This binding pattern is remarkably similar to that seen with zoledronate and minodronate (and, indeed, many other bisphosphonates) binding to a variety of FPPS enzymes (9–13) and strongly suggests, at least for these compounds, that they could act as carbocation transition state/reactive intermediate analogs (15). This similarity is seen more graphically in Fig. 2D in which we superimpose the structure of zoledronate bound to human FPPS onto that of GGPPS–zoledronate. For example, N3 in the imidazole ring of zoledronate is within hydrogen-bond distance of Thr-170 in GGPPS as well as the corresponding Thr-201 in human FPPS. One difference, however, is that in GGPPS we (again) find evidence for only two and not three Mg2+, the same situation as found with GGPP binding to human GGPPS (4) as well as in most of the diphosphate complexes shown in Fig. 1. This may be related to buffer pH effects, resulting in a protonated Asp-209. The third-generation bisphosphonates zoledronate and minodronate thus bind to GGPPS in a basically similar way as they do to human, Escherichia coli, Trypanosoma brucei, and Trypanosoma cruzi FPPS. However, as can be seen in SI Table 2, these compounds are not the most active ones in GGPPS inhibition, although they are the most potent FPPS inhibitors.
Fig. 2.
Bisphosphonates and GGPPS structures. (A) Structures of bisphosphonates investigated as GGPPS inhibitors. (B) GGPPS structure containing zoledronate (PDB ID code 2E91) showing dimer structure. (C) Stereoview of minodronate bound to GGPPS (PDB ID code 2E92). (D) Stereoview of zoledronate/GGPPS (PDB ID code 2E91) superimposed on zoledronate/IPP/FPPS structure (PDB ID code 2F8C).
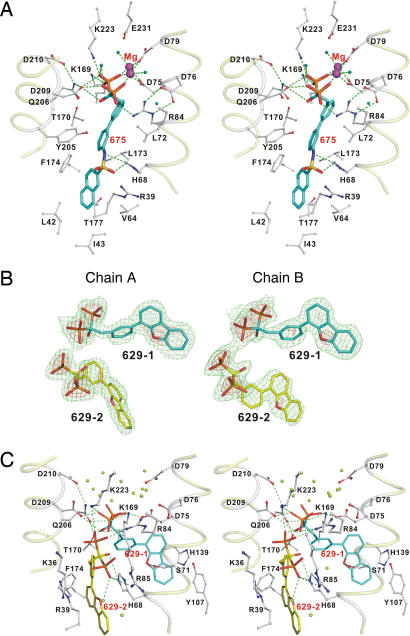
We next consider the binding of three more potent GGPPS inhibitors: BPH-364, BPH-629, and BPH-675 (Fig. 2A, BPH, bisphosphonate), compounds that appear to be more potent GGPPS inhibitors because of the addition of a large, hydrophobic moiety. We show in Fig. 3 and SI Fig. 7B the structures of each of these inhibitors bound to GGPPS. With BPH-364 and BPH-675, the bisphosphonate backbones bind via 1 or 2 Mg2+ to the protein into the same sites as seen with GGPP bound to human GGPPS: FPP-GGPP. The ligand interactions seen with BPH-675 are shown in Fig. 3A and SI Fig. 8 and clearly place it in the FPP-GGPP site, shown more graphically in Fig. 4A superimposed on BPH-364 and GGPP (human structure, blue shading). These results are of interest because they clearly show that large, hydrophobic inhibitors bind to the GGPP site seen in the human enzyme, where they can chelate to Mg2+, absent in the IPP site. In the case of BPH-629 (Figs. 2A and 3 B and C), a neutral side chain (dibenzofuran) containing bisphosphonate, the results are surprising in that, as can be seen from the electron density shown in Fig. 3B, there are two (as opposed to one) bisphosphonate binding sites, and there is no Mg2+ observable. In the first structure (BPH-629-1, Fig. 3C), the bisphosphonate backbone forms H bonds with Arg-84, Lys-169, Gln-206, Asp-209, and Lys-223, with the O atom on the dibenzofuran ring forming an H bond with the side chain of Ser-71. This represents the FPP-FPP-binding site location seen with zoledronate, minodronate, and FsPP and is shown in orange in Fig. 4B. The other conformer (BPH-629-2) binds with its bisphosphonate in the IPP site, with one phosphonate group binding to Arg-85 and His-68, the same residues that are responsible for binding to the diphosphate of IPP. As expected, there is no Mg2+ present in this IPP site. However, in this instance, the bulky aromatic moiety extends into the (human) GGPP site, so binding is IPP-GGPP, using the nomenclature suggested above, and this site is shown in pink in Fig. 4C.
Fig. 3.
Structures of hydrophobic bisphosphonates bound to GGPPS. (A) Stereoview of BPH-675 (PDB ID code 2E95). (B) Electron density (green contoured at 1σ, red at 3σ) for the two BPH-629 conformers (PDB ID code 2E93). (C) Structure of BPH-629 bound to GGPPS (monomer A model is shown).
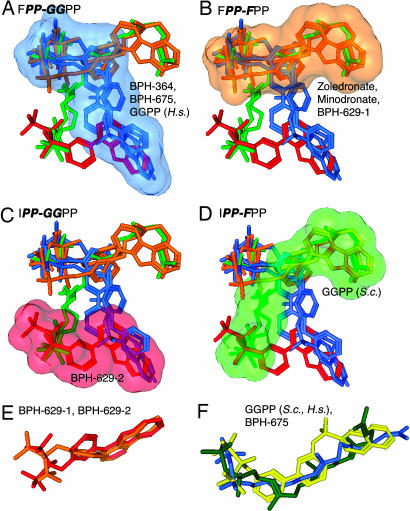
Fig. 4.
Binding site motifs for GGPPS inhibitors. (A) FPP-GGPP site (in blue) occupied by BPH-364, BPH-675, and GGPP (human protein, PDB ID code 2FVI). (B) FPP-FPP site (in orange) occupied by zoledronate, minodronate, and BPH-629-1. (C) IPP-GGPP site (in pink) occupied by BPH-629-2. (D) IPP-FPP site (in green) occupied by GGPP in the yeast enzyme. (E) Superimposition of both BPH-629 conformers from chain A model. (F) Superimposition of yeast, human GGPP structures with BPH-675.
The first binding site (Fig. 4A), occupied by BPH-364 and BPH-675, thus corresponds to the FPP-GGPP site identified as the GGPP “inhibitor” site in the human GGPPS-GGPP structure by Kavanagh et al. (4). The pocket is 19.3 Å long and can readily accommodate long bisphosphonates, such as BPH-675, that have about the same length as GGPP itself (16.2 Å, BPH-675; 17.1 Å, GGPP). Clearly, longer bisphosphonates will experience enhanced van der Waals dispersion interactions in this site and an enhanced hydrophobic effect, resulting in enhanced potency as GGPPS inhibitors, plus, in some cases, we find evidence for Mg2+ binding as well, which can also enhance potency, because of bidentate chelation. However, much longer analogs may have diminished activity, because they are likely to clash sterically with Leu-42 and Thr-177 at the distal end of the GGPP-binding pocket. In this binding mode, the presence of a carbocation positive-charge feature would be unlikely to be a factor in potency, because the GGPP inhibitor site has no obvious requirement for carbocation character, and, indeed, earlier quantitative structure activity relationship (QSAR) investigations revealed no requirement for a positive charge feature in GGPPS inhibition (16), unlike the situation found with FPPS inhibition (17).
We show in Fig. 4B (in orange) the FsPP, zoledronate, minodronate, and BPH-629-1 conformers located in the FPP-FPP-binding site, that is, the one analogous to that seen in FPPS. This site is ≈13 Å long (about the same length as the substrate, FPP, 12 Å) and the bisphosphonates zoledronate and minodronate bind here to the protein via two Mg2+. However, the smaller bisphosphonates are less potent inhibitors of both the Saccharomyces cerevisiae and human enzymes (SI Table 2). The bisphosphonate BPH-629 also binds to this site in one conformation, however, no Mg2+ is found. As noted above, the second conformer of BPH-629 binds IPP-GGPP (pink in Fig. 4C), and again reflects the smaller size (≈13 Å) of the pocket and the inhibitor. In the yeast enzyme, GGPP binds IPP-FPP (shown in green, Fig. 4D), but no bisphosphonates examined so far bind in this manner.
We also find, as shown in Fig. 4E, that the backbone and side-chain conformations of BPH-629-1 and -2 are very similar, with only a 1.47-Å rms deviation for the heavy atoms, and BPH-364 adopts a similar conformation (SI Fig. 7B). Also of interest is the observation that the GGPP structures seen in the human and S. cerevisiae enzymes are very similar (a 2.3-Å rmsd), plus, the “curved” conformation of the longest inhibitor (BPH-675) is clearly quite similar to that seen with the GGPP inhibitors, as illustrated in Fig. 4F. It is, of course, intriguing to speculate that even more potent compounds can be designed that occupy both hydrophobic sites [perhaps, for example, species such as the recently reported bis-prenyl bisphosphonates (18)], enhancing potency and specificity, although it is also possible that optimum cellular activity may be obtained with compounds that can inhibit both FPPS and GGPPS.
Crystal Structures of UPPS–Bisphosphonate Complexes.
We next consider the x-ray crystallographic structures of five UPPS–bisphosphonate complexes. We first screened a library of 29 bisphosphonates for activity against E. coli UPPS. The structures of the compounds investigated are shown in SI Fig. 9, and their IC50 and pIC50 values are shown in SI Table 4. Based on these results, we selected the three compounds shown in Fig. 5A for crystallographic investigation, in addition to BPH-629 and BPH-675 (Fig. 2A). Data collection and refinement statistics for all UPPS complexes are given in SI Table 5 (PDB ID codes 2E98, 2E99, 2E9A, 2E9C, and 2E9D).
Fig. 5.
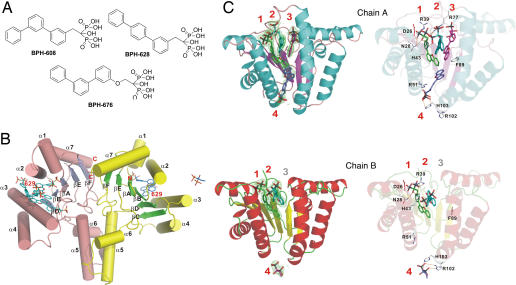
Bisphosphonate and UPPS structures. (A) Structures of additional UPPS inhibitors. (B) Structure of UPPS-BPH-629 complex (PDB ID code 2E98). (C) Structures of BPH-629 bound to four different sites (A and B molecules in the dimer are both shown).
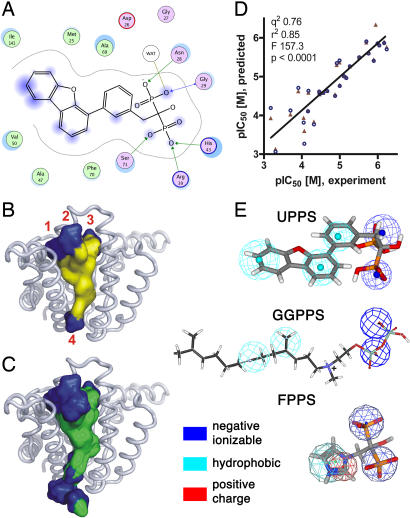
The UPPS–bisphosphonate complex structures each contain a central β-sheet with six parallel strands and seven surrounding α-helices in each monomer of the dimeric protein, and two views of the structure of one dimeric complex (BPH-629) are shown in Fig. 5 B and C (the four other structures are shown in SI Figs. 10–13). In each of the five structures, we found that there were up to four binding sites per monomer, labeled 1–4 in Fig. 5C. Three of the binding sites occupy the top of a “funnel” region, whereas the fourth site is situated at the bottom of the funnel, as shown in Fig. 5C. When all bisphosphonate complex structures are superimposed on the UPPS-Mg–FsPP-IPP structure (SI Fig. 14), the width (10.3–12.9 Å) and length (21–25.2 Å) of the funnel-shaped hydrophobic tunnel are well defined, and it is clear that site 1 corresponds to the FPP (FsPP) substrate-binding site (shown in yellow in SI Fig. 14). Ligand interactions with Asp-26, Asn-28, Arg-39, His-43, Arg-51, Arg-77, Phe-89, Arg-102, and His-103 are common to all five structures (all four sites), and detailed interactions for site 1 are shown in terms of a ligand-interaction map for the most potent inhibitor, BPH-629, in Fig. 6A. The interactions of other ligands are shown in SI Figs. 15–18. The presence of a large hydrophobic cavity is not unexpected in a C55 prenyl synthase, and with the bisphosphonates investigated here, enables up to four inhibitors to bind to a central cavity, illustrated by the surface representation for BPH-629 (all four sites) in Fig. 6B and for all bisphosphonates in Fig. 6C; stereo views are shown in SI Fig. 19.
Fig. 6.
Binding-site interactions and computational modeling. (A) BPH-629 protein–ligand interactions. (B) Diagram showing surface structure of all four BPH-629 molecules (from Fig. 5C Upper). (C) Diagram showing surface structure of all five inhibitors bound to UPPS (all crystal structures). (D) Graph showing correlation between experimental and 2D-QSAR predicted pIC50 values for UPPS inhibition, (see SI Table 7). (E) Pharmacophores for UPPS, GGPPS (adapted from ref. 16), and FPPS (data from ref. 17).
So, in FPPS, there is just a single inhibitor site, in GGPPS there are at least three, and in UPPS, there are four. This raises the question: is it possible to predict the activity of a given bisphosphonate by using QSAR methods? This might be expected to be rather difficult, given the multiple site binding seen crystallographically in GGPPS and UPPS. However, in previous work, we found that GGPPS inhibition by diphosphates and bisphosphonates could, in fact, be quite well predicted, perhaps because most of the potent compounds that dominated the QSAR were GGPP-like (16). This ability to predict GGPPS activity suggests that it might also be possible to predict UPPS inhibition, especially if the inhibitors bind more tightly to one particular site, and, upon detailed inspection of the UPPS structures, we find that, on average, there are only ≈9 protein contacts for sites 2–4, to be compared with ≈14 for site 1, and 15 for FsPP, strongly suggesting that inhibition at the FPP site, site 1, might well dominate any SAR. The results of 2D and hologram QSAR (HQSAR) in addition to a pharmacophore model (see SI Methods for details) are shown in Fig. 6 D and E, SI Tables 6–10, and SI Fig. 20 and clearly show that excellent predictions of the experimental pIC50 values can be made. For example, R2 = 0.85, q2 = 0.76, F test: 157.3, P < 0.0001, n = 29 for the 2D QSAR method, Fig. 6D, with a prediction error of ≈2 over a ≈103× range in activity. The HQSAR and pharmacophore modeling results gave similarly good accord, and a representative top-scoring UPPS pharmacophore is shown in Fig. 6E compared with previous results for FPPS and GGPPS inhibition (16). In the UPPS pharmacophore, there are two negative ionizable groups (blue) and three hydrophobic features (cyan) shown superimposed on the structure of BPH-629, the most potent UPPS inhibitor, Fig. 6E. The pharmacophore for GGPPS inhibition is similar, but neither pharmacophore gives evidence of the importance of the cationic feature present in the FPPS result, Fig. 6E. In fact, the UPPS 2D-QSAR descriptors involving positive charge actually indicate a slight reduction in activity with positive charge (SI Table 6). Although perhaps at first surprising, it now seems clear that FPPS inhibition requires a positive-charge feature, as deduced by QSAR (17) and observed by NMR (13), but GGPPS inhibition with the compounds investigated so far does not because, especially for the larger inhibitors, the combination of a large hydrophobic feature together with, in some cases, Mg2+ binding, provides good potency. That is, because the more potent GGPPS inhibitors are not transition state/reactive intermediate analogs, there is no charge required.
Finally, we should note that the observation of the lack of importance of a positive-charge feature in GGPPS inhibitors might be the exception and not the rule for trans-prenyltransferase inhibitors. For example, geranyl (C10) diphosphate synthase and FPPS are potently inhibited by bisphosphonates (9–13, 19). Moreover, other long-chain prenyltransferases are also potently inhibited by bisphosphonates, such as zoledronate, and we show in SI Table 11 a compilation of IC50 values for GGPPS, hexaprenyl diphosphate synthase (HPPS, from Sulfolobus solfataricus) and octaprenyl diphosphate synthase (OPPS, from E. coli) for each of the inhibitors investigated here. The results obtained clearly show that some bisphosphonates can have potent activity against other, long-chain trans-prenyltransferases, in particular those used in quinone biosynthesis, opening up further possibilities for inhibitor or drug design.
Overall then, the results we have obtained above are of general interest because they show that bisphosphonates can be potent inhibitors of both cis- and trans-prenyltransferases. In GGPPS, there are potentially four binding modes based on two polar (the FPP and IPP diphosphate) and two hydrophobic (farnesyl, geranylgeranyl side-chain) binding sites, with bisphosphonates binding to three of these sites. In UPPS, four binding sites are seen crystallographically, but only the FPP-site (site 1) is occupied in all structures, and, because it has the largest number of protein-ligand contacts, we propose that occupancy of this site dominates enzyme inhibition. Indeed, the results of a variety of QSAR methods give excellent predictions of UPPS inhibition with a factor of ≈2 error in activity prediction over a ≈103× range in activity. The observation of multiple binding sites in GGPPS and UPPS is in sharp contrast to the observation of only one bisphosphonate-binding site in FPPS, and the availability of these structures opens up new avenues for the design of novel inhibitors (drugs) that target these important enzymes, including the possibility of using them in a “fragment-based” approach to the development of more potent and specific species.
Methods
GGPPS and UPPS crystals were obtained as described previously with some modifications and were soaked with various diphosphates and bisphosphonates to obtain the complex structures. Full details are given in SI Methods. Structure determination and refinement were carried out as described previously for ligand-free GGPPS and ligated UPPS, with full details given in SI Methods.
Supplementary Material
Acknowledgments
We thank M.-F. Hsu for technical assistance; H. Sagami (Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Sendai, Japan) for providing the human GGPPS expression system; and K. L. Kavanagh, D. Hosfield, and R. Docampo for valuable suggestions. Portions of this research were carried out at the National Synchrotron Radiation Research Center, a national user facility supported by the National Science Council of Taiwan, Republic of China. This work was supported by Academia Sinica and the National Core Facility of High-Throughput Protein Crystallography Grant NSC95-3112-B-001-015-Y (to A.H.-J.W.) and in part by the U.S. Public Health Service [National Institutes of Health Grants GM-50694, GM-65307, and GM-073216 (to E.O.)]. Y.Z. was supported by an American Heart Association, Midwest Affiliate, Postdoctoral Fellowship, and Y.S. was supported by a Leukemia and Lymphoma Society Special Fellowship.
Abbreviations
- FPP
farnesyl diphosphate
- FPPS
FPP synthase
- GGPP
geranylgeranyl diphosphate
- GGPPS
geranylgeranyl diphosphate synthase
- IPP
isopentenyl diphosphate
- QSAR
quantitative structure activity relationship
- UPP
undecaprenyl diphosphate
- UPP
undecaprenyl diphosphate
- UPPS
undecaprenyl diphosphate synthase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank, www.pdb.org [PDB ID codes for GGPPS complexed with: IPP (2E8U), GPP (2E8X), FPP (2E90), FsPP/IPP (2E8T), GGPP (2E8V), zoledronate (2E91), minodronate (2E92), BPH-629 (2E93), BPH-364 (2E94), and BPH-675 (2E95) and for UPPS complexed with BPH-629 (2E98), BPH-608 (2E99), BPH-628 (2E9A), BPH-675 (2E9C), and BPH-676 (2E9D)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0702254104/DC1.
References
- 1.Russell RG. Ann NY Acad Sci. 2006;1068:367–401. doi: 10.1196/annals.1346.041. [DOI] [PubMed] [Google Scholar]
- 2.Martin MB, Grimley JS, Lewis JC, Heath HT, III, Bailey BN, Kendrick H, Yardley V, Caldera A, Lira R, Urbina JA, et al. J Med Chem. 2001;44:909–916. doi: 10.1021/jm0002578. [DOI] [PubMed] [Google Scholar]
- 3.Goffinet M, Thoulouzan M, Pradines A, Lajoie-Mazenc I, Weinbaum C, Faye JC, Seronie-Vivien S. BMC Cancer. 2006;6:60. doi: 10.1186/1471-2407-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kavanagh KL, Dunford JE, Bunkoczi G, Russell RG, Oppermann U. J Biol Chem. 2006;281:22004–22012. doi: 10.1074/jbc.M602603200. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Huang J, Jiang X, Seefeld M, McQueney M, Macarron R. J Biomol Screen. 2003;8:712–715. doi: 10.1177/1087057103258185. [DOI] [PubMed] [Google Scholar]
- 6.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Nat Rev Drug Dis. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 7.Guo RT, Ko TP, Chen AP, Kuo CJ, Wang AH-J, Liang PH. J Biol Chem. 2005;280:20762–20774. doi: 10.1074/jbc.M502121200. [DOI] [PubMed] [Google Scholar]
- 8.Chang TH, Guo RT, Ko TP, Wang AH-J, Liang PH. J Biol Chem. 2006;281:14991–15000. doi: 10.1074/jbc.M512886200. [DOI] [PubMed] [Google Scholar]
- 9.Kavanagh KL, Guo K, Dunford JE, Wu X, Knapp S, Ebetino FH, Rogers MJ, Russell RG, Oppermann U. Proc Natl Acad Sci USA. 2006;103:7829–7834. doi: 10.1073/pnas.0601643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosfield DJ, Zhang Y, Dougan DR, Broun A, Tari LW, Swanson RV, Finn J. J Biol Chem. 2004;279:8526–8529. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 11.Gabelli SB, McLellan JS, Montalvetti A, Oldfield E, Docampo R, Amzel LM. Proteins. 2006;62:80–88. doi: 10.1002/prot.20754. [DOI] [PubMed] [Google Scholar]
- 12.Rondeau JM, Bitsch F, Bourgier E, Geiser M, Hemmig R, Kroemer M, Lehmann S, Ramage P, Rieffel S, Strauss A, et al. Chem Med Chem. 2006;1:267–273. doi: 10.1002/cmdc.200500059. [DOI] [PubMed] [Google Scholar]
- 13.Mao J, Mukherjee S, Zhang Y, Cao R, Sanders JM, Song Y, Zhang Y, Meints GA, Gao YG, Mukkamala D, et al. J Am Chem Soc. 2006;128:14485–14497. doi: 10.1021/ja061737c. [DOI] [PubMed] [Google Scholar]
- 14.Kloer DP, Welsch R, Beyer P, Schulz GE. Biochemistry. 2006;45:15197–15204. doi: 10.1021/bi061572k. [DOI] [PubMed] [Google Scholar]
- 15.Martin MB, Arnold W, Heath HT, III, Urbina JA, Oldfield E. Biochem Biophys Res Commun. 1999;263:754–758. doi: 10.1006/bbrc.1999.1404. [DOI] [PubMed] [Google Scholar]
- 16.Szabo CM, Matsumura Y, Fukura S, Martin MB, Sanders JM, Sengupta S, Cieslak JA, Loftus TC, Lea CR, Lee HJ, et al. J Med Chem. 2002;45:2185–2196. doi: 10.1021/jm010412y. [DOI] [PubMed] [Google Scholar]
- 17.Sanders JM, Gomez AO, Mao J, Meints GA, Van Brussel EM, Burzynska A, Kafarski P, Gonzalez-Pacanowska D, Oldfield E. J Med Chem. 2003;46:5171–5183. doi: 10.1021/jm0302344. [DOI] [PubMed] [Google Scholar]
- 18.Wiemer AJ, Tong H, Swanson KM, Hohl RJ. Biochem Biophys Res Commun. 2007;353:921–925. doi: 10.1016/j.bbrc.2006.12.094. [DOI] [PubMed] [Google Scholar]
- 19.Burke C, Klettke K, Croteau R. Arch Biochem Biophys. 2004;422:52–60. doi: 10.1016/j.abb.2003.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.