Amino acids enter into protein synthesis by virtue of their attachment to transfer RNAs. The fidelity of translation results from the summed accuracy of three distinct processes: the esterification of amino acids to their cognate tRNAs by aminoacyl-tRNA synthetases (aaRSs) (1); the binding selectivity for cognate AA-tRNA pairs by elongation factor (2); and the tRNA:mRNA decoding process mediated by the small subunit of the ribosome (3). Collectively, these processes ensure that the error frequency does not exceed, on average, ≈0.03%, and that the overall rate of protein synthesis is not unduly compromised. Although the components of the translational apparatus are highly conserved, the biosynthetic machinery responsible for producing the amino acid precursors to proteins is tremendously diverse, and the capability to produce all 20 canonical standard amino acids is not universal. This apparent contradiction presents a significant challenge to untangling the connection between amino acid and protein biosynthesis, but clarity is gradually beginning to emerge. In this issue of PNAS, Roy et al. (4) provide evidence that asparagine synthetase A (AsnA) emerged from a functional aspartyl-tRNA synthetase, and thus establish the evolutionary origin of one of the two enzymes responsible for the direct biosynthesis of the amino acid asparagine. This observation forges a new, significant link between aminoacyl-tRNA synthesis and amino acid biosynthesis. For at least some of the 20 standard amino acids, indirect pathways for amino acid biosynthesis (i.e., those occurring on the tRNA) preceded direct pathways, in which de novo biosynthesis occurs before the formation of the aminoacyl-tRNA linkage.
In his original formulation of the Adaptor hypothesis, Crick (5) proposed a separate activating enzyme (aaRS) for each amino acid. The relationship is now known to be quite a bit more complex, precisely because aminoacyl-tRNA can serve as a substrate for additional transformations of the amino acid (6). The basic aminoacylation reaction occurs in two steps: activated amino acid adenylate is created in the first step at the expense of ATP, whereas the second, transfer step, leads to formation of the aminoacyl ester at the 3′ end of the tRNA. Remarkably, there are two distinct classes of aaRSs, each with its characteristic catalytic domain fold and distinctive mechanistic features, including conformation of the ATP and regio-specificity of transfer (7). With one notable exception, the class I lysyl-tRNA synthetase, all of the orthologs across all three kingdoms that recognize a particular amino acid substrate are of the same class.
Deviations from the adaptor hypothesis arise because some organisms possess fewer than the requisite 20 aaRSs for the 20 standard amino acids, whereas others possess apparent duplications (8, 9). The ability of many taxa in the archaea and the bacteria to survive on as few as 16 of the 20 canonical enzymes is the result of their ability to synthesize amino acids by using “indirect” pathways. A de novo amino acid biosynthesis pathway can be said to be a “direct” pathway if an aaRS is available to directly attach that amino acid to its cognate tRNA. When an organism lacks asparaginylor glutaminyl-tRNA synthetase (AsnRS and GlnRS, respectively), the appropriate asparaginyl- or glutaminyl-tRNA must be generated in a two-step process featuring a mischarging reaction followed by a transamidation (10). In the case of asparagine, a “nondiscriminating” version of AspRS is responsible for aspartylating tRNAAsn, and then the resulting aspartyl-tRNAAsn is amidated by a glutamine amidotransferase (GatCAB). Notably, only cognate pairs are acceptable substrates for EF-Tu, which serves to prevent the mischarged tRNA intermediates from entering translation (2). This significant observation raises the interesting possibility that the ability of EF-Tu to discriminate against mischarged tRNAs must have either preceded or accompanied the emergence of indirect pathways of amino acid biosynthesis.
Biosynthetic machinery for producing amino acid precursors to proteins is tremendously diverse.
Indirect amino acid biosynthetic pathways, therefore, can account for the absence of GlnRS and AsnRS among the archaea. Another notable absence in the archaeal Methanococcus janaschii and Methanothermobacter thermoautotrophicus is cysteinyl-tRNA synthetase, for which a satisfactory ortholog has not yet been identified. But what of the duplicated aaRSs, which include extra versions of lysyl-, threonyl-, asparaginyl-, and histidyl-tRNA synthetases (9, 11)? In some cases, such as LysRS, fully functional homologs of the standard aaRS exist that are deployed under specialized growth conditions, such as elevated temperature or nutrient deprivation (12). Their primary role is production of aminoacyl-tRNA for protein synthesis, and they will not be considered further. Instead, I will address aaRS paralogs that contain only a portion of the tRNA synthetase polypeptide, and for which the function is not obviously aminoacylation.
Roy et al. (4) set out to investigate the apparent duplication of asparaginyl-tRNA synthetase in Pyrococcus abyssi, an organism that inhabits deep ocean thermal vents. The first asparaginyl-tRNA synthetase gene encodes a 434-residue polypeptide (herein AsnRS) that is closely related (47% identical) to AsnS from Bacillus subtilis and is the appropriate candidate for the standard AsnRS. The second ORF (AsnRS2) encodes a 294-residue polypeptide that is 39% identical to the first gene, yet lacks the first ≈100 N-terminal residues. In the full-length protein, this domain is responsible for binding the tRNA anticodon (13). A priori, AsnRS2 might serve as a bona fide AsnRS, a nondiscriminating AspRS, or provide a function unrelated to aminoacylation. The identification of AsnRS2 as a functional aaRS would have provided support to models invoking a primordial single chain, single domain class II tRNA synthetase (14). This was not the case.
Roy et al. (4) found that AsnRS2 is the P. abyssi homolog of asparagine synthetase A (AsnA), an enzyme previously described in bacteria that carries out ammonia-dependent synthesis of asparagine (15). Several key observations support this conclusion. First, AsnRS2 carries out pyrophosphate exchange in the presence of aspartate and ATP, but not with asparagine, which acts as a competitive inhibitor. When incubated with ammonium chloride, [3H]aspartate, and ATP, radioactive asparagine is formed and could be transferred to tRNAAsn by an authentic AsnRS from Thermus thermophilus. Moreover, the gene for AsnRS2 conferred asparagine prototrophy on an Escherichia coli strain in which both asparagine synthetase genes (AsnA and AsnB) were disrupted. These studies establish unambiguously that P. abyssi possesses an AsnA homolog that is based on a class II catalytic fold related to asparaginyl-tRNA synthetase. Accordingly, the combination of AsnRS and AsnRS2 constitute a direct pathway for asparagine synthesis and aminoacyl-tRNA synthesis, rationalizing the absence of a nondiscriminating AspRS and GatCAB transamidase in the P. abyssi genome (16). This direct pathway for asparagine synthesis/incorporation is limited to the Pyrococcus, Thermoplasma, and Pyrobaculum genera; other archaea employ indirect pathways for asparaginyl-tRNA synthesis (16).
The relationship between AsnA and AsnRS2 is not entirely unexpected, as previous work on the E. coli version of AsnA had already confirmed that it is related to aspartyl-tRNA synthetase. Although the overall sequence identity between AsnA and aspartyl-tRNA synthetase is <25%, several groups detected sequences in AsnA that were a match to Motif3, one of the class II aaRS signature sequences (15, 17). Subsequently, Nakatsu et al. (18) determined the structure of E. coli AsnA and showed that it is remarkably similar to the catalytic domain of AspRS. Like other functional class II aaRSs, AsnA is a dimer of identical subunits, with a catalytic domain consisting of an eight-stranded antiparallel β-sheet flanked by two long and eight short α helices. The structure determined was of a complex containing asparagine and AMP, allowing important specificity-determining interactions to be identified. Along with information derived from the Pyrococcus furiosis AspRS aspartyl-adenylate complex, Roy et al. (4) were able to identify with precision those interactions with the aspartyl and AMP moieties that are conserved in all three complexes, as well as those evolutionary changes in the active site of AsnA that permit asparagine synthesis and block aminoacylation. These changes lead to opposite orientations of aspartate in the active sites of AsnA and AspRS, because amidation (as well as the adenylation that proceeds it) must occur on the β-carboxyl of aspartate, whereas aminoacylation demands formation of the adenylate on the α-carboxylate. Accordingly, loss of the anticodon binding domain likely occurred to eliminate tRNA binding, and thus the formation of the potentially toxic β-activated aspartate.
Contemporary organisms therefore use three distinct paths for asparagine synthesis: an indirect pathway, employing mischarged Asp-tRNAAsn and the GatCAB transamidase; a direct pathway employing the glutamine-dependent AsnB and canonical AsnRS; and a second direct pathway that utilizes AsnA and AsnRS. The three pathways appear to be more or less functionally equivalent, as the E. coli AsnA, AsnB mutant can be rescued either by P. abyssi AsnRS2/AsnA (the direct pathway) or by a nondiscriminating AspRS from Deinnococcus radiodurans and GatCAB amidotransferase (19), representing an indirect pathway. The selection of pathway likely represents an adaptive response to the availability of different nitrogen sources (e.g., glutamine versus ammonium salts). Moreover, pathway choice may also reflect dissimilar rates of evolution among different species in different environments. In general, archaeal genes may be evolving more slowly than those of bacteria and eukaryotes (20). A slower evolutionary rate could account for the unique position of P. abyssi AsnRS2/AsnA as an evolutionary intermediate between the archaeal AspRS clade and the bacterial AsnRS clade. The phylogenetic analysis presented by Roy et al. (4) indicates that the AsnA family arose as a consequence of two separate duplications of the archaeal/eukaryotic AspRS, and that AsnA split off from the second duplication before the differentiation of the canonical AsnRS family. If this argument concerning the rates of evolution is true, one may infer from the abundance of indirect pathways for amino acid biosynthesis in archaea and their relative absence in γ-proteobacteria and eukaryotes that they preceded direct pathways, a possibility originally suggested by Söll and colleagues (19).
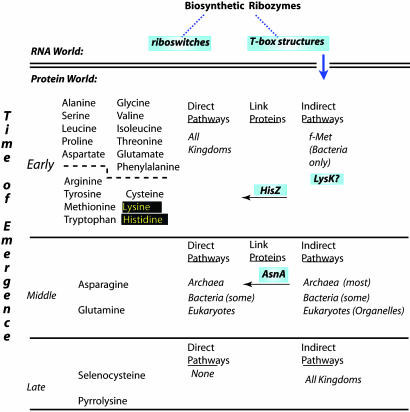
The conclusion that indirect pathways preceded direct pathways is a key feature of the model that suggests the organization of the genetic code reflects biosynthetic relationships among the amino acids (21, 22). In this model (Fig. 1), the 22 amino acids that are inserted during translation can be divided into three groups: the “early” amino acids (Leu, Ala, Ile, Phe, Val, Gly, Arg, Ser, Glu, Pro, Tyr, Thr, Trp, Asp Cys, Lys, Met, His), which are synthesized in extant organisms by using only direct pathways; “middle” amino acids (glutamine and asparagine), which are synthesized by using a combination of indirect and direct pathways (23); and “late” amino acids (selenocysteine and pyrrolysine), which are synthesized by using only indirect pathways. Thus, contemporary organisms provide a picture of evolution-in-progress: the most recently emerging amino acids, selenocysteine (24) and pyrrolysine (25), are the only ones that require biosynthesis on a tRNA scaffold, whereas the middle amino acids exhibit both routes. Early amino acids are likely to be a mixture of those that have graduated from indirect to tRNA-independent synthesis and those that were always synthesized by direct routes.
Fig. 1.
A model of the temporal order in which amino acids emerged, emphasizing the relationship between direct and indirect pathways. The amino acids are arranged in three groups, which reflect their order of emergence (21). The dotted line among the early amino acids highlights the possibility that some of the smaller, simpler amino acids might have always been synthesized by using direct routes. The double line demarcates the transition from the RNA world to protein world, and the downward arrow signifies emergence of indirect pathways. HisZ and AsnA, the putative evolutionary links between direct and indirect pathways, are highlighted above arrows showing the direction of evolution from indirect to direct pathways.
If the model proposed by Wong (21) and others (22) is true, extant genomes are likely to contain ORFs that serve as evolutionary links to mark the conversion of indirect pathways to direct pathways. The study by Roy et al. (4) provides a confirmation of the prediction, because AsnRS2/AsnA is directly related to the archaeal nondiscriminating AspRS and can clearly perform de novo asparagine synthesis. Are there other such examples? HisZ, the histidyl-tRNA synthetase paralog that collaborates with the HisG ATP-phosphoribosyl transferase to catalyze the first step in histidine biosynthesis, represents such a candidate (26). It possesses neither adenylation nor aminoacylation function but is required for proper regulation and assembly of a functional HisZ-HisG complex. Like AsnA, HisZ also lacks an anticodon binding domain, and phylogenetic analysis of HisZ suggests that it diverged from a functional class II aaRS early on in the separation of the three principal kingdoms (27). It is therefore tempting to speculate that HisZ represents the evolutionary link between an early, tRNA-dependent version of histidine biosynthesis and a later, tRNA-independent de novo histidine biosynthesis pathway.
Another aaRS paralog that could serve as a link between direct and indirect amino acids biosynthetic pathways is the class I lysyl-tRNA synthetase LysK (28). Unlike class II lysyl-tRNA synthetases, LysK is restricted to a limited range in the archaea and a few selected bacteria and requires tRNA for adenylate synthesis. The fact that lysine is apparently the precursor of pyrrolysine is intriguing (25). A further echo of the early importance of indirect pathways is the role of aminoacyl-tRNA as an intermediate in reactions that are distinct from protein synthesis, including the synthesis of 5-aminolevulinc acid, a key precursor of tetrapyrroles, from glutamyl-tRNA (29), and the transfer of arginine to N termini of proteins as part of the N-end rule-mediated degradation (30).
Ultimately, any model that seeks to address the origin of the aminoacylation system must confront the inherent paradox that protein-based machinery is required to synthesize proteins. An RNA world (31) has been invoked to resolve this paradox, populated with RNA catalysts capable of performing a variety of key metabolic reactions, including the synthesis of at least some of 20 different groups of aminoacyl-tRNA. As an elegant proof of principle that this scenario is not far fetched, RNAs capable of aminoacylating themselves and tRNAs (in trans) have been selected in the laboratory (32, 33). In the biological realm, it is highly significant that a number of bacterial (and potentially eukaryotic) genera employ RNA-based molecular switches as control devices. These include T-box-mediated control, in which a complex fold in the mRNA has been shown to sense the aminoacylation status of a given tRNA (34), and “riboswitches,” a recent class of mRNA structures that regulate termination/antitermination through the binding of small molecule cofactors that require their own biosynthetic pathway (35). Arguably, such molecules could be the contemporary descendents of early RNAs that transformed amino acids on their tRNAs scaffolds, in a process that anticipated the rich diversity of extant metabolism.
See companion article on page 9837.
References
- 1.Ibba, M. & Soll, D. (2000) Annu. Rev. Biochem. 69, 617–650. [DOI] [PubMed] [Google Scholar]
- 2.LaRiviere, F. J., Wolfson, A. D. & Uhlenbeck, O. C. (2001) Science 294, 165–168. [DOI] [PubMed] [Google Scholar]
- 3.Ogle, J. M., Murphy, F. V., Tarry, M. J. & Ramakrishnan, V. (2002) Cell 111, 721–732. [DOI] [PubMed] [Google Scholar]
- 4.Roy, H., Becker, H. D., Reinbolt, J. & Kern, D. (2003) Proc. Natl. Acad. Sci. USA 100, 9837–9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crick, F. H. C. (1958) Symp. Soc. Exp. Biol. 12, 138–163. [PubMed] [Google Scholar]
- 6.Ibba, M., Becker, H. D., Stathopoulos, C., Tumbula, D. L. & Soll, D. (2000) Trends Biochem. Sci. 25, 311–316. [DOI] [PubMed] [Google Scholar]
- 7.Martinis, S. A., Plateau, P., Cavarelli, J. & Florentz, C. (1999) EMBO J. 18, 4591–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stathopoulos, C., Ahel, I., Ali, K., Ambrogelly, A., Becker, H., Bunjun, S., Feng, L., Herring, S., Jacquin-Becker, C., Kobayashi, H., et al. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 175–183. [DOI] [PubMed] [Google Scholar]
- 9.Schimmel, P. & Ribas De Pouplana, L. (2000) Trends Biochem. Sci. 25, 207–209. [DOI] [PubMed] [Google Scholar]
- 10.Ibba, M. & Soll, D. (2001) EMBO Rep. 2, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner, A. M. (1999) Curr. Biol. 9, R842–R844. [DOI] [PubMed] [Google Scholar]
- 12.Brevet, A., Chen, J., Leveque, F., Blanquet, S. & Plateau, P. (1995) J. Biol. Chem. 270, 14439–14444. [DOI] [PubMed] [Google Scholar]
- 13.Berthet-Colominas, C., Seignovert, L., Hartlein, M., Grotli, M., Cusack, S. & Leberman, R. (1998) EMBO J. 17, 2947–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schimmel, P. & Ribas de Pouplana, L. (1995) Cell 81, 983–986. [DOI] [PubMed] [Google Scholar]
- 15.Hinchman, S. K., Henikoff, S. & Schuster, S. M. (1992) J. Biol. Chem. 267, 144–149. [PubMed] [Google Scholar]
- 16.Cohen, G. N., Barbe, V., Flament, D., Galperin, M., Heilig, R., Lecompte, O., Poch, O., Prieur, D., Querellou, J., Ripp, R., et al. (2003) Mol. Microbiol. 47, 1495–1512. [DOI] [PubMed] [Google Scholar]
- 17.Gatti, D. L. & Tzagloff, A. (1991) J. Mol. Biol. 218, 557–568. [DOI] [PubMed] [Google Scholar]
- 18.Nakatsu, T., Kato, H. & Oda, J. (1998) Nat. Struct. Biol 5, 15–19. [DOI] [PubMed] [Google Scholar]
- 19.Min, B., Pelaschier, J. T., Graham, D. E., Tumbula-Hansen, D. & Soll, D. (2002) Proc. Natl. Acad. Sci. USA 99, 2678–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kollman, J. M. & Doolittle, R. F. (2000) J. Mol. Evol. 51, 173–181. [DOI] [PubMed] [Google Scholar]
- 21.Wong, J. T. (1975) Proc. Natl. Acad. Sci. USA 72, 1909–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Giulio, M. (1997) J. Mol. Evol. 45, 571–578. [DOI] [PubMed] [Google Scholar]
- 23.Woese, C. R., Olsen, G. J., Ibba, M. & Soll, D. (2000) Microbiol. Mol. Biol. Rev. 64, 202–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bock, A. (2000) Biofactors 11, 77–78. [DOI] [PubMed] [Google Scholar]
- 25.Srinivasan, G., James, C. M. & Krzycki, J. A. (2002) Science 296, 1459–1462. [DOI] [PubMed] [Google Scholar]
- 26.Sissler, M., Delorme, C., Bond, J., Ehrlich, S. D., Renault, P. & Francklyn, C. (1999) Proc. Natl. Acad. Sci. USA 96, 8985–8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bond, J. P. & Francklyn, C. (2000) J. Mol. Evol. 50, 339–347. [DOI] [PubMed] [Google Scholar]
- 28.Ibba, M., Morgan, S., Curnow, A. W., Pridmore, D. R., Vothknecht, U. C., Gardner, W., Lin, W., Woese, C. R. & Soll, D. (1997) Science 278, 1119–1122. [DOI] [PubMed] [Google Scholar]
- 29.Moser, J., Schubert, W. D., Beier, V., Bringemeier, I., Jahn, D. & Heinz, D. W. (2001) EMBO J. 20, 6583–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon, Y. T., Kashina, A. S. & Varshavsky, A. (1999) Mol. Cell. Biol. 19, 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert, W. (1986) Nature 320, 485–486. [DOI] [PubMed] [Google Scholar]
- 32.Illangasekare, M., Sanchez, G., Nickles, T. & Yarus, M. (1995) Science 267, 643–647. [DOI] [PubMed] [Google Scholar]
- 33.Lee, N., Bessho, Y., Wei, K., Szostak, J. W. & Suga, H. (2000) Nat. Struct. Biol. 7, 28–33. [DOI] [PubMed] [Google Scholar]
- 34.Grundy, F. J. & Henkin, T. M. (1993) Cell 74, 475–482. [DOI] [PubMed] [Google Scholar]
- 35.Winkler, W., Nahvi, A. & Breaker, R. R. (2002) Nature 419, 952–956. [DOI] [PubMed] [Google Scholar]