The most common neurodegenerative diseases are characterized by the presence of abnormal filamentous protein inclusions in nerve cells of the brain. In Alzheimer's disease, these inclusions are made of hyperphosphorylated tau protein (1). Together with the extracellular β-amyloid deposits, they constitute the defining neuropathological characteristics of Alzheimer's disease. Tau inclusions, in the absence of extracellular deposits, are characteristic of progressive supranuclear palsy, corticobasal degeneration, Pick's disease, argyrophilic grain disease, and frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) (1). The identification of mutations in Tau in FTDP-17 (2–4) has established that dysfunction of tau protein is central to the neurodegenerative process. At an experimental level, the expression of mutant human tau in nerve cells is leading to improved models of neurodegeneration. In this issue of PNAS, Kraemer et al. (5) describe lines of Caenorhabditis elegans expressing transgenic wild-type and mutant human tau protein. They represent an important addition to existing transgenic models for the human tauopathies.
Tau protein is widely expressed in the mammalian nervous system, where it plays a role in the assembly and stabilization of microtubules (1). In adult human brain, there are six isoforms of tau, produced from a single gene by alternative mRNA splicing (6, 7). They differ by the presence or absence of a 29- or 58-aa insert in the N-terminal half and by the inclusion, or not, of a 31-aa repeat, encoded by exon 10 of Tau, in the C-terminal half of the protein (Fig. 1a). The exclusion of exon 10 leads to the production of three isoforms, each containing three repeats, and its inclusion leads to a further three isoforms, each containing four repeats. The repeats constitute the microtubule-binding region of tau, and similar levels of Three- And four-repeat isoforms are expressed in adult human brain. Repeat sequences homologous to those in tau are also present in the high-molecular-weight microtubule-associated proteins MAP2 and MAP4. The genomes of C. elegans and Drosophila melanogaster encode only one protein with tau-like repeats.
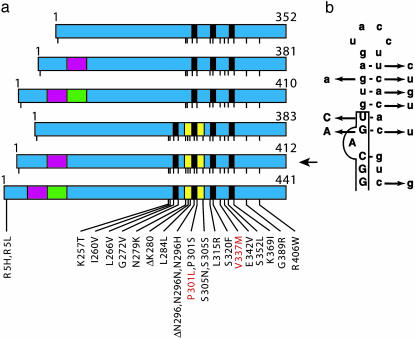
Fig. 1.
Mutations in Tau in FTDP-17. (a) Schematic diagram of the six tau isoforms (352–441 aa) expressed in adult human brain, with mutations in the coding region indicated by using the numbering of the 441-aa isoform. The six isoforms are produced by alternative mRNA splicing from a single gene and differ by the presence or absence of three inserts, shown in red (encoded by exon 2), green (encoded by exon 3), and yellow (encoded by exon 10). Exon 2 codes for the 29-aa insert, exons 2 and 3 code for the 58-aa insert, and exon 10 codes for the 31-aa repeat. Nineteen missense mutations, two deletion mutations, and three silent mutations are shown. Kraemer et al. (5) produced lines of C. elegans transgenic for the 412-aa isoform (arrow) without and with mutations P301L or V337M (indicated in red). (b) Predicted stem-loop structure in the pre-mRNA at the boundary between exon 10 and the intron that follows it, with nine mutations that destabilize the stem loop. Exon sequences are boxed and shown in uppercase letters, and intron sequences are shown in lowercase letters.
Tau mutations in FTDP-17 are either missense, deletion, or silent mutations in the coding region, or intronic mutations located close to the splice-donor site of the intron following exon 10 (1). So far, 31 different mutations have been described in >80 families with FTDP-17 (Fig. 1). Functionally, Tau mutations fall into two largely nonoverlapping categories, those that influence the alternative splicing of tau pre-mRNA and those whose primary effect is at the protein level. The intronic mutations and most coding region mutations in exon 10 increase the splicing of this exon, changing the ratio between three- and four-repeat isoforms (3, 4). Approximately half of the known mutations have their primary effect at the RNA level. They affect exon splicing enhancer and silencer sequences in exon 10 (8) or destabilize a predicted stem-loop structure located at the boundary between exon 10 and the intron that follows it (3, 4, 9) (Fig. 1b). Thus, to a significant extent, FTDP-17 is a disease of alternative mRNA splicing. The other mutations affect tau isoforms directly. In accordance with their location in the microtubule-binding region, most missense mutations and the deletion mutations lead to a reduced ability of tau to promote microtubule assembly (10, 11). A number of mutations may cause FTDP-17, at least in part, by promoting the assembly of tau into filaments (12, 13).
Kraemer et al. (5) expressed the 412-aa isoform of human tau in nerve cells of C. elegans, either in the wild-type form or with a missense mutation of FTDP-17 (P301L or V337M, Fig. 1a). This resulted in a reduced lifespan, behavioral impairment, defective cholinergic neurotransmission, the accumulation of insoluble phosphorylated tau, and neurodegeneration, as indicated by axonal damage and nerve cell loss. Although similar changes were observed in all transgenic lines, expression of mutant tau resulted in a more severe neurodegenerative phenotype than expression of wild-type tau. Soluble, phosphorylated tau was expressed 1 day after hatching, when the nervous system looked normal. However, uncoordinated behavior was already apparent in some lines, indicating that the simple expression of mutant human tau was sufficient to cause nerve cell dysfunction. By immunohistochemistry, tau staining was observed in most nerve cells and their processes. Insoluble, phosphorylated tau was first detected 5–7 days after hatching. Its appearance was paralleled by progressive axonal degeneration and nerve cell loss, which were more severe in mutant than in wild-type tau lines. By electron microscopy, 9-day-old worms from the mutant lines showed degenerating axons throughout the ventral and dorsal nerve cords. By immunoelectron microscopy, tau staining in the axons was largely diffuse. Amorphous, tau-positive aggregates were only observed in worms transgenic for V337M tau.
How do these molecular and cellular changes compare with what has been observed in human diseases with tau pathology? Tauopathies in worms and humans share the progressive accumulation of insoluble tau and extensive neurodegeneration. However, there also some differences, the most conspicuous of which are a more modest phosphorylation and the absence of tau filaments in C. elegans. This is reminiscent of work in D. melanogaster, where the expression of wild-type and mutant human tau proteins in nerve cells led to a reduced lifespan and the loss of nerve cells, in the absence of tau filaments (14, 15). Phosphorylation of tau appeared to be more extensive in the fly than in the worm. Moreover, coexpression of human tau with the fly homologue of glycogen synthase kinase-3β, a candidate protein kinase for the hyperphosphorylation of tau, resulted in accelerated neurodegeneration and the formation of tau-immunoreactive inclusions (15). Filamentous structures were observed, but they were not shown to be decorated by anti-tau antibodies. In contrast to what has been described in FTDP-17 (16), tau-induced neurodegeneration involved programmed cell death (15). Taken together, it appears that conformationally altered, nonfilamentous human tau protein is neurotoxic in invertebrates.
Tauopathies in worms and humans share the accumulation of insoluble tau and extensive neurodegeneration.
What about vertebrate models for the human tauopathies? Expression of wild-type human tau in nerve cells of the sea lamprey has been shown to lead to the accumulation of filaments made of hyperphosphorylated tau and degenerative changes (17, 18), indicating a possible link between the formation of tau filaments and the degeneration of nerve cells. Most work has been done in the mouse (19–30), where the transgenic expression of single isoforms of wild-type human tau resulted in large numbers of nerve cells with abnormal tau-immunoreactive cell bodies and dendrites, as well as signs of axonal degeneration and muscle weakness (19–23). Abundant tau filaments were not observed, neither was substantial nerve cell loss. The level of tau phosphorylation was lower than in the human tauopathies. Overexpression of nonfilamentous tau can thus cause axonopathy and muscle weakness in the mouse. However, these transgenic lines are incomplete models for the human tauopathies.
Mouse lines expressing single tau isoforms with missense mutations of FTDP-17 in nerve cells have provided more complete models (25–30). Some of these lines exhibited the essential molecular and cellular features of human tauopathies, including the formation of abundant filaments made of hyperphosphorylated tau protein and nonapoptotic nerve cell loss (25, 29). Filamentous tau was hyperphosphorylated to the same extent as in human tauopathy brains, and most of the same sites were also phosphorylated in soluble tau, consistent with the view that hyperphosphorylation precedes filament assembly. In a recent study, an increase in the phosphorylation of soluble tau resulted in increased filament formation, suggesting that phosphorylation of tau can drive filament formation (31).
Work on mouse models has shown a correlation between the formation of tau filaments and neurodegeneration, but it is not known whether the two phenomena are causally related. The findings in C. elegans and D. melanogaster suggest that this may not be the case, because neurodegeneration was effected by nonfilamentous tau of reduced solubility. Alternatively, the mechanisms of neurodegeneration in worms and flies may differ from those in mice and humans.
Disease models in C. elegans offer some advantages over mouse models, in particular with regard to the speed and relative ease with which genetic modifiers of disease phenotype can be discovered and pharmacological modifiers can be screened. Once identified, their relevance for mammalian model systems can be tested. In the future, this combined approach is likely to lead to a better understanding of the detailed molecular mechanisms by which the dysfunction of tau protein can cause neurodegeneration.
See companion article on page 9980.
References
- 1.Lee, V. M.-Y., Goedert, M. & Trojanowski, J. Q. (2001) Annu. Rev. Neurosci. 24, 1121–1159. [DOI] [PubMed] [Google Scholar]
- 2.Poorkaj, P., Bird, T. D., Wijsman, E., Nemens, E., Garruto, R. M., Anderson, L., Andreadis, A., Wiederholt, W. C., Raskind, M. & Schellenberg, G. D. (1998) Ann. Neurol. 43, 815–825. [DOI] [PubMed] [Google Scholar]
- 3.Hutton, M., Lendon, C. L., Rizzu, P., Baker, M., Foelich, S., Houlden, H., Pickering-Brown, S., Chakraverty, S., Isaacs, A., Grover, A., et al. (1998) Nature 393, 702–705. [DOI] [PubMed] [Google Scholar]
- 4.Spillantini, M. G., Murrell, J. R., Goedert, M., Farlow, M. R., Klug, A. & Ghetti, B. (1998) Proc. Natl. Acad. Sci. USA 95, 7737–7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraemer, B. C., Zhang, B., Leverenz, J. B., Thomas, J. H., Trojanowski, J. Q. & Schellenberg, G. D. (2003) Proc. Natl. Acad. Sci. USA 100, 9980–9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goedert, M., Spillantini, M. G., Jakes, R., Rutherford, D. & Crowther, R. A. (1989) Neuron 3, 519–526. [DOI] [PubMed] [Google Scholar]
- 7.Andreadis, A., Brown, M. W. & Kosik, K. S. (1992) Biochemistry 31, 10626–10633. [DOI] [PubMed] [Google Scholar]
- 8.D'Souza, I., Poorkaj, P., Hong, M., Nochlin, D., Lee, V. M.-Y., Bird, T. D. & Schellenberg, G. D. (1999) Proc. Natl. Acad. Sci. USA 96, 5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varani, L., Hasegawa, M., Spillantini, M. G., Smith, M. J., Murrell, J. R., Ghetti, B., Klug, A., Goedert, M. & Varani, G. (1999) Proc. Natl. Acad. Sci. USA 96, 8229–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasegawa, M., Smith, M. J. & Goedert, M. (1998) FEBS Lett. 437, 207–210. [DOI] [PubMed] [Google Scholar]
- 11.Hong, M., Zhukareva, V., Vogelsberg-Ragaglia, V., Wszolek, Z., Reed, L., Miller, B. L., Geschwind, D. H., Bird, T. D., McKeel, D., Goate, A., et al. (1998) Science 282, 1914–1917. [DOI] [PubMed] [Google Scholar]
- 12.Nacharaju, P., Lewis, J., Easson, C., Yen, S., Hackett, J., Hutton, M. & Yen, S. H. (1999) FEBS Lett. 447, 195–199. [DOI] [PubMed] [Google Scholar]
- 13.Goedert, M., Jakes, R. & Crowther, R. A. (1999) FEBS Lett. 450, 306–311. [DOI] [PubMed] [Google Scholar]
- 14.Wittmann, C. W., Wszolek, M. F., Shulman, J. M., Salvaterra, P. M., Lewis, J., Hutton, M. & Feany, M. B. (2001) Science 293, 711–714. [DOI] [PubMed] [Google Scholar]
- 15.Jackson, G. R., Wiedau-Pazos, M., Sang, T.-K., Wagle, N., Brown, C. A., Massachi, S. & Geschwind, D. H. (2002) Neuron 34, 509–519. [DOI] [PubMed] [Google Scholar]
- 16.Atzori, C., Ghetti, B., Piva, R., Srinivasan, A. N., Zolo, P., Delisle, M. B., Mirra, S. S. & Migheli, A. (2001) J. Neuropathol. Exp. Neurol. 60, 1190–1197. [DOI] [PubMed] [Google Scholar]
- 17.Hall, G. F., Yao, J. & Lee, G. (1997) Proc. Natl. Acad. Sci. USA 94, 4733–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, G. F., Lee, V. M.-Y., Lee, G. & Yao, J. (2001) Am. J. Pathol. 158, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Götz, J., Probst, A., Spillantini, M. G., Schäfer, T., Jakes, R., Bürki, K. & Goedert, M. (1995) EMBO J. 14, 1304–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brion, J. P., Tremp, G. & Octave, J. N. (1999) Am. J. Pathol. 154, 255–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishihara, T., Hong, M., Zhang, B., Nakagawa, Y., Lee, M. K., Trojanowski, J. Q. & Lee, V. M.-Y. (1999) Neuron 24, 751–762. [DOI] [PubMed] [Google Scholar]
- 22.Spittaels, K., van den Haute, C., van Dorpe, J., Bruynseels, K., Vandezande, K., Laenen, I., Geerts, H., Mercken, M., Sciot, R., van Lommel, A., et al. (1999) Am. J. Pathol. 155, 2153–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Probst, A., Götz, J., Wiederhold, K. H., Tolnay, M., Mistl, C., Jaton, A. L., Hong, M., Ishihara, T., Lee, V. M.-Y., Trojanowski, J. Q., et al. (2000) Acta Neuropathol. 99, 469–481. [DOI] [PubMed] [Google Scholar]
- 24.Higuchi, M., Ishihara, T., Zhang, B., Hong, M., Andreadis, A., Trojanowski, J. Q. & Lee, V. M.-Y. (2002) Neuron 35, 433–446. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, J., McGowan, E., Rockwood, J., Melrose, H., Nacharaju, P., van Slegtenhorst, M., Gwinn-Hardy, K., Murphy, M., Baker, M., Yu, X., et al. (2000) Nature Genet. 25, 402–405. [DOI] [PubMed] [Google Scholar]
- 26.Götz, J., Chen, F., Barmettler, R. & Nitsch, R. (2001) J. Biol. Chem. 276, 529–534. [DOI] [PubMed] [Google Scholar]
- 27.Lim, F., Hernandez, F., Lucas, J. J., Gomez-Ramos, P., Moran, M. A. & Avila, J. (2001) Mol. Cell. Neurosci. 18, 702–714. [DOI] [PubMed] [Google Scholar]
- 28.Tanemura, K., Murayama, M., Akagi, T., Hashikawa, T., Tominaga, T., Ishikawa, M., Yamaguchi, H. & Takashima, A. (2002) J. Neurosci. 22, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen, B., Ingram, E., Takao, M., Smith, M. J., Jakes, R., Virdee, K., Yoshida, H., Holzer, M., Craxton, M., Emson, P. C., et al. (2002) J. Neurosci. 22, 9340–9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatebayashi, Y., Miyasaka, T., Chui, D.-H., Akagi, T., Mishima, K.-I., Iwasaki, K., Fujiwara, M., Tanemura, K., Murayama, M., Ishiguro, K., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 13896–13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noble, W., Olm, V., Takata, K., Casey, E., O, M., Meyerson, J., Gaynor, K., LaFrancois, J., Wang, L., Kondo, T., et al. (2003) Neuron 38, 555–565. [DOI] [PubMed] [Google Scholar]