Abstract
Pheromone biosynthesis-activating neuropeptide (PBAN), a peptide produced by the subesophageal ganglion, is used by a variety of moths to regulate pheromone production. PBAN acts directly on pheromone gland cells by using calcium and cAMP as second messengers. We have identified a gene encoding a G protein-coupled receptor (GPCR) from pheromone glands of the female moth Helicoverpa zea. The gene was identified based on sequence identity to a group of GPCRs from Drosophila that are homologous to neuromedin U receptors in vertebrates. The full-length PBAN receptor was subsequently cloned, expressed in Sf9 insect cells, and shown to mobilize calcium in response to PBAN. This response was dose-dependent (EC50 = 25 nM) with a maximum response at 300 nM and a minimal observable response at 10 nM. Four additional peptides produced by the PBAN-encoding gene were also tested for activity, and it was determined that three had similar activity to PBAN and the other was slightly less active. Peptides belonging to the same family as PBAN, namely pyrokinins, as well as the vertebrate neuromedin U peptide also induced a calcium response. We have identified a GPCR for the PBAN/pyrokinin family of peptides with a known function of stimulating pheromone biosynthesis in female moths. It is related to several receptors from insects (Drosophila and Anopheles) and to neuromedin U and ghrelin receptors from vertebrates.
Pheromones are a subset of semiochemicals that are used by organisms for chemical communication. Among the moths, sex pheromones are usually released by females to attract conspecific males. In the moth Helicoverpa zea, as in a variety of moths, pheromone amounts increase in the pheromone gland during scotophase when females are sexually receptive to males. The increase in pheromone biosynthesis during the scotophase is regulated by a peptide hormone produced in the subesophageal ganglion, located near the brain (1). This peptide, termed pheromone biosynthesis-activating neuropeptide (PBAN), which acts on pheromone glands to stimulate pheromone biosynthesis (2), was first identified as a 33-aa C-terminal amidated peptide (3). Subsequently, it was determined that the five C-terminal amino acids, FXPRLamide, represented the minimal sequence required for activity (4). This motif has been identified from a variety of peptides and most will stimulate pheromone biosynthesis when tested on pheromone glands isolated from moths (5).
Insects from a variety of orders (6) and even a crustacean (7) have peptides with the FXPRLamide motif, although not all insects regulate pheromone biosynthesis by using this peptide. For example, juvenile hormone regulates bark beetle pheromone biosynthesis (8). PBAN-like peptides found in other insects have other functions. In fact, the first peptide in the family was identified based on its ability to stimulate the contraction of hindgut muscles of the cockroach, Leucophaea maderae, and thus was named leucopyrokinin (9). Subsequently several other functions were identified for the family including melanization in Lepidoptera larvae (10), induction of embryonic diapause in Bombyx mori (11), and acceleration of puparium formation in several flies (12). These results indicate the ubiquity and multifunctional nature of the pyrokinin/PBAN family of peptides.
Action of peptide hormones on target cells usually requires their binding to a specific receptor with subsequent activation of second messengers. PBAN activity on pheromone gland cells causes the influx of extracellular calcium (13) that promotes the production of cAMP (14) in isolated H. zea pheromone glands. Although information is available on the mode of action of PBAN, identification of the receptors is lacking. The involvement of G proteins was determined pharmacologically, thus implicating G protein-coupled receptors (GPCRs) (14). Identification of peptide receptors in insects was enhanced with the complete sequencing and annotation of the Drosophila melanogaster genome (15). It was determined that ≈44 genes code for peptide GPCRs (16). Four of these GPCRs were similar to neuromedin U receptors in vertebrates, indicating that, if ligand and receptor were conserved, then the ligand for the Drosophila receptors may be similar to neuromedin U. The peptides identified thus far in insects with some sequence homology to neuromedin U are the PBAN-like peptides (Table 1). The fact that D. melanogaster does indeed contain peptides that belong to the PBAN/pyrokinin family (17) and that at least one receptor was characterized as binding FXPRLamide peptides (18) led us to use this information to search for a PBAN receptor in moths. A PCR-based cloning strategy using mRNA isolated from pheromone glands of H. zea identified a putative GPCR with a similar sequence to several D. melanogaster, Anopheles gambiae, and vertebrate receptors. Functional expression in insect cells characterized the receptor as an H. zea PBAN receptor (PBAN-R).
Table 1. Amino acid sequences of the pyrokinin/PBAN family of peptides used in this study and the species in which they were identified.
| Species | Peptide | Peptide sequence |
|---|---|---|
| H. zea | PBAN | LSDDMPATPADQEMYRQDPEQIDSRTKYFSPRLamide |
| PGN-7 | VIFTPKLamide | |
| PGN-18 | SLAYDDKSFENVEFTPRLamide | |
| PGN-8 | TMNFSPRLamide | |
| PGN-24 | NDVKDGAASGAHSDRLGLWFGPRLamide | |
| L. maderae | Leucopyrokinin | PETSFTPRLamide |
| D. melanogaster | CAP2b-3 | TGPSASSGLWFGPRLamide |
| ETH-1 | DDSSPGFFLKITKNVPRLamide | |
| Porcine | Neuromedin U | YFLFRPRNamide |
| Aplysia californica | Myomodulin | PMSMLRLamide |
The FXPRLamide motif is shown in bold.
Materials and Methods
Materials. Synthetic H. zea PBAN, myomodulin, proctolin, FMRFamide, neuromedin U8, and leucopyrokinin were purchased from Bachem. Synthetic D. melanogaster ecdysis-triggering hormone 1 (ETH-1), CAP2b-3, and the H. zea PBAN-encoding gene neuropeptides (PGNs) 7, 8, 18, and 24 were custom-synthesized by Research Genetics (Huntsville, AL). Oxidized PBAN was prepared by adding 0.4 M H2O2 in water to 1 nmol of PBAN and incubating for 4 h at room temperature. This treatment resulted in one oxidized peptide as determined by HPLC and matrix-assisted laser desorption ionization/time-of-flight analysis (data not shown). Fluo-4/acetoxymethyl ester (AM) was purchased from Molecular Probes.
Pheromonotropic Assays. Two pheromonotropic assays were used: one that measured pheromone amounts and one that measured radiolabeled acetate incorporation into pheromone. Both were conducted as described (13, 19).
Receptor Cloning. Pheromone glands from 1- to 3-day-old female H. zea and the central nervous system of third instar larval D. melanogaster were dissected, placed on dry ice, and stored at –80°C. Poly(A)+ RNA was isolated from both tissues by using a Micro Fast Track kit (Invitrogen) and used to synthesize cDNA with a Marathon cDNA amplification kit (CLONTECH).
Synthesized cDNA from pheromone glands was amplified with a sense primer of 5′-ACNGCNTTYACNGTNGARCG-3′ and an antisense primer of 5′-GCRTGRAANGGNGCCCARCA-3′. PCR was performed under 35 cycles at 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min. The PCR product was directly sequenced and used to design further gene-specific primers to find 5′ and 3′ ends of the PBAN-R cDNA. 5′ Rapid amplification of cDNA ends (RACE) was performed by using the primers 5′-GAAGAATGATAGCGCCACTGCAACG-3′ (from nucleotide 821), 5′-CGTTCTGCCCATGATCCACATACG-3′ (from nucleotide 571), and 5′-ACACGTGCTGGTGTTTCCCAGGATG-3′ (from nucleotide 183) with a Marathon cDNA amplification kit (CLONTECH). 3′ RACE was performed by using the primers 5′-GTGTTTGCATTGTGTACCGC-3′ (from nucleotide 496), 5-AGCAATGAGAGACCTGGTCAGATGCAG-3′ (from nucleotide 739), and 5′-CGTTGCAGTGGCGCTATCATTC TTC-3′ (from nucleotide 807) with a RACE kit (Invitrogen).
Receptor Expression in Sf9 Cells. The ORF of the PBAN-R cDNA was amplified by using the sense primer of 5′-TTAAGTAAAATGACATTGTCAGCG-3′ (from –9 including NotI site) and antisense primer of 5′-GCAACGGCCTTTAAATTACGGAGC-3′ (from nucleotide 1084 including XbaI site). The restriction-site-adapted PCR product was ligated into a pIB/V5-His vector, digested with NotI and XbaI, transformed into Sf9 cells with Cellfectin reagent (Invitrogen), and selected with blasticidin by using the InsectSelect BSD system kit (Invitrogen). Likewise, the ORF of the D. melanogaster CG8795 receptor cDNA was amplified by using the sense primer of 5′-ATGCTGCCCACTAACAGTTCCG-3′ and antisense primer of 5′-TTAAAAGGCGGCCCGCTCTT-3′, and the PCR product was used to transform Sf9 cells as described for the PBAN-R.
Single-Cell Calcium Imaging. Approximately 2 × 104 cells (200 μl per well) were seeded into a 96-well cell-culture plate with black walls and a clear bottom and incubated overnight at 28°C. Fluo-4/AM in saline was made freshly by mixing 20% (wt/vol) Pluronic F-127 in DMSO with a 1 mM stock Fluo-4/AM in DMSO to produce final concentrations of 2 μM Fluo-4/AM, 0.01% Pluronic F-127, and 0.24% DMSO. The saline consisted of 21 mM KCl, 12 mM NaCl, 18 mM MgCl2, 3 mM CaCl2, 170 mM glucose, 1 mM probenecid, and 10 mM Pipes at pH 7.2. Cells were incubated in the dark with 2 μM Fluo-4/AM in saline for 20 min, washed with saline twice, and incubated in the dark at room temperature for 20–60 min before challenge with peptide ligands. Cell fluorescence measurements were taken every 10 s, and after 40 s ligand was added and fluorescent changes were measured for up to 5 min. Background fluorescence was subtracted from the measurements after challenge with ligand. After 5 min of ligand exposure, 1 μM ionomycin was added to obtain a maximum fluorescence reading. Ligand-exposure values were expressed as relative to the maximum value obtained with ionomycin, which creates ion channels in the cell membrane, thus flooding the cytosol with extracellular calcium. At least 10 cells at each ligand concentration were processed for changes in fluorescence intensity. Sf9 cells that were not transformed with the vector and cells that were transformed with the vector without the insert were also tested, and these cells did not give a response when challenged with 1 μM PBAN. Individual cell fluorescence was measured on a Prairie Technologies (Middleton, WI) scanning confocal microscope by using the blue spectrum of light. Recorded measurements were processed by using metamorph (Universal Imaging, Media, PA) and analyzed by using Microsoft's excel.
Results
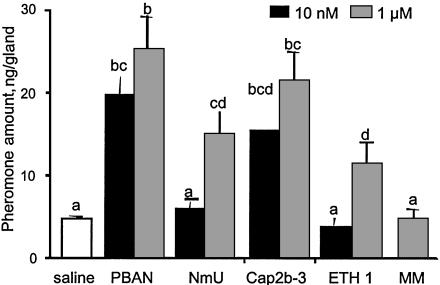
Hewes and Taghert (16) indicated that several GPCRs in Drosophila are homologous to neuromedin U receptors from vertebrates. Neuromedin U has a C-terminal motif similar to PBAN (Table 1); therefore, we reasoned that neuromedin U may stimulate pheromone biosynthesis in isolated pheromone glands of H. zea. As shown in Fig. 1, neuromedin U at 1 μM stimulated pheromone glands to produce pheromone at about the same level as 10 nM PBAN. Other peptides tested included CAP2b-3 from Drosophila, which was just as active as PBAN. The other Drosophila peptide, ETH-1, had activity similar to neuromedin U. Myomodulin did not have activity at a 1 μM concentration. Neuromedin U was tested by using isolated pheromone glands of Helicoverpa armigera and had about the same activity as when tested with H. zea and, in addition, stimulated the production of cAMP (data not shown). These results indicate that the peptide neuromedin U can stimulate pheromone biosynthesis in moths, indicating homologous receptors, which forms the basis of our search for the PBAN receptor in pheromone glands.
Fig. 1.
Amounts of pheromone found in isolated pheromone glands of H. zea after incubation with the indicated amounts of various peptides. NmU, porcine neuromedin U; MM, myomodulin. Isolated glands were incubated with the peptides for 30 min and then extracted, and pheromone titers were determined. Bars represent the amount of pheromone per gland + SEM (n > 4). Different letters at the top of each bar indicate statistically different treatments (one-way analysis of variance, P < 0.05).
Cloning of H. zea PBAN-R. A PCR-based cloning strategy was initiated by using degenerate primers designed against highly conserved sequences of three Drosophila receptors: CG8784, CG8795, and CG9918. By using cDNA synthesized from mRNA obtained from pheromone glands, an initial 411-nt clone was identified that had a 43% identical deduced amino acid sequence to the Drosophila CG8795 receptor. Additional primers were used to obtain 5′ and 3′ sequences. A full-length clone was amplified and sequenced to establish the complete nucleotide sequence. Fig. 2 shows the complete nucleotide sequence and deduced amino acid sequence for this clone. A 1,323-bp-long nucleotide sequence was obtained, and, by using the indicated ATG as initiation codon and the first in-frame stop codon TGA, an ORF of 1,038 nt encoding 346 aa is predicted. Hydropathy analysis indicates the presence of seven transmembrane domains, which are characteristic of GPCRs. Also present is the ERY variant of the DRY domain just downstream of transmembrane 3, which is highly conserved among GPCR and is crucial for coupling to and activation of G proteins. Two putative N-glycosylation sites are present in the N-terminal region and between transmembrane domains 4 and 5. Several serine and threonine residues are present in the C-terminal tail as possible phosphorylation signals.
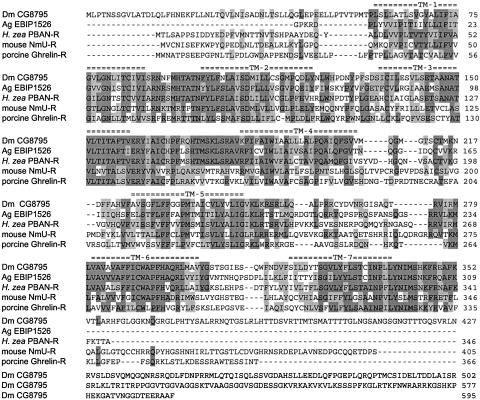
Fig. 2.
cDNA and deduced amino acid sequence of the H. zea PBAN-R. Nucleotides are numbered starting at the first ATG codon in the ORF, and the first in-frame stop codon is indicated by *. The putative polyadenylation signal is underlined twice. The deduced amino acid sequence is indicated with the predicted transmembrane (TM) domains underlined and labeled TM1–TM7. Putative extracellular N-glycosylation sites are marked with ‡. The nucleotide sequences used for primer location in obtaining the ORF of the full-length clone are underlined. The nucleotide sequences corresponding to the degenerate primers used in the first round of PCR are shown in bold and underlined.
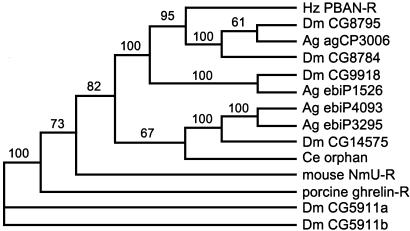
A blast search of GenBank with the deduced amino acid sequence revealed similarities with several known invertebrate and vertebrate GPCRs. The most similar insect and vertebrate receptors were chosen from the blast search and aligned by using clustalw (Fig. 3) (20). The putative GPCRs from A. gambiae are incompletely annotated, but the sequence spanning the seven transmembrane domains is present. Among these five receptors, high sequence similarities occur in the transmembrane domains, with considerable divergence occurring in the N-terminal domains. The C terminus of the PBAN-R is relatively truncated, whereas Dm CG8795 has a considerably extended C terminus. These alignments, along with several other related GPCRs, were compared phylogenetically by using paup 4.0b10 (Fig. 4). The sequences with the highest identity to the putative H. zea PBAN-R were CG8795, CG8784, CG9918, ebiP1526, and agCP3006. The three Drosophila receptors apparently bind FXPRLamide peptides (18). Another related receptor from Drosophila CG14575 binds CAP2b-1 and CAP2b-2, which are FPRVamide peptides from Drosophila (18). An orphan GPCR from the nematode Caenorhabditis elegans was also found in the blast search. The above-named sequences were the most closely related to the mouse neuromedin U receptor. Two other Drosophila receptors, CG5911a and -b, that bind ETH-1 and ETH-2 (21, 22) are more distantly related. The ghrelin receptor in vertebrates also has an ≈30% sequence similarity to the H. zea PBAN-R.
Fig. 3.
Alignment of H. zea PBAN-R with a PRXamide receptor from D. melanogaster (Dm CG8795), a putative GPCR from A. gambiae (Ag EBIP1526), and with two vertebrate receptors [neuromedin U receptor, GenBank accession no. NM_010341 (NmU-R), and Ghrelin receptor, GenBank accession no. U60178]. Identical amino acids are highlighted in dark gray and conserved amino acids in light gray for three or more sequences. The transmembrane domains for the H. zea PBAN-R are indicated by double dashes above the aligned sequences. Dashed lines indicate spaces to optimize alignment.
Fig. 4.
Phylogenetic tree for 14 GPCRs from insects, a nematode, and mammals that had the highest sequence similarity to H. zea PBAN-R. The tree was made with paup 4.0b10 by using the neighbor-joining distance method. Numbers indicate bootstrapping values in 1,000 replications. Dm, D. melanogaster; Ag, A. gambiae; Ce, C. elegans. Accession nos.: CG8795, AF522190; CG8784, AF522189; CG9918, AF522191; CG14575, AF522193; CG5911a, AF505863; CG5911b, AF505864; agCP3006, EAA08454; ebiP1526, EAA06972; ebiP4093, EAA03575; ebiP3295, EAA08403; NmU-R, NM_010341; ghrelin-R, U60178; Ce orphan, T15816.
Functional Identification of PBAN-R. To determine functionality, the H. zea PBAN-R was expressed by using Sf9 cells, and activity was monitored by using the fluorescent calcium chelator Fluo-4/AM and visualized by using confocal microscopy. PBAN induced a strong calcium response in a dose-dependant manner, with a calculated EC50 value of 25 nM (Fig. 5 and Table 2). The lowest concentration with an observable response was 10 nM, with a peak response at 300 nM. H. zea PBAN has two methionine residues that potentially can be oxidized; however, the EC50 of oxidized PBAN was similar to that of PBAN (Table 2). PGN-7, PGN-18, PGN-24, and leucopyrokinin had EC50 values similar to those of PBAN. The PGN peptides can be produced by the PBAN-encoding gene, and leucopyrokinin is found in the cockroach L. maderae. PGN-8 had a higher EC50 value, as did the two peptides from Drosophila (CAP2b-3 and ETH-1). Surprisingly, the porcine neuromedin U had an EC50 value of 136 nM. Several peptides, including myomodulin, were not active at 10 μM.
Fig. 5.
Dose–response profiles of the effect of synthetic H. zea PBAN on the H. zea PBAN-R- and D. melanogaster CG8795-expressing Sf9 cells. Maximum fluorescence readings were obtained by treating cells with ionomycin after challenge with peptides. The y axis indicates the peak fluorescence value relative to the ionomycin treatment. Values represent the mean ± SD of at least 10 cells per ligand concentration.
Table 2. Half-maximal effective concentrations (EC50) of the peptides tested on the expressed H. zea PBAN-R and Drosophila CG8795.
| Peptide | H. zea PBAN-R | Drosophila CG8795 |
|---|---|---|
| PBAN | 25 nM | 125 nM |
| Oxidized PBAN | 32 nM | — |
| PGN-7 | 32 nM | — |
| PGN-18 | 29 nM | — |
| PGN-8 | 200 nM | — |
| PGN-24 | 60 nM | — |
| Leucopyrokinin | 30 nM | 52 nM |
| CAP2b-3 | 375 nM | 550 nM |
| ETH-1 | 537 nM | 929 nM |
| Neuromedin U | 136 nM | >10 μM |
| Myomodulin | >10 μM | — |
| FMRFamide | >10 μM | — |
| Proctolin | >10 μM | — |
EC50 values were determined by using log-probit analysis. —, not tested.
The Drosophila GPCR CG8795 was also expressed by using Sf9 cells and tested for ligand binding (Fig. 5 and Table 2). The most active peptide tested was leucopyrokinin, followed by PBAN, CAP2b-3, and ETH-1 in decreasing potency. Neuromedin U was not active at 10 μM.
Discussion
Pheromone biosynthesis in most moths is regulated by the peptide hormone PBAN, which is produced in the subesophageal ganglion and released into the hemolymph, where it stimulates pheromone biosynthesis in the pheromone gland. Synthetic PBAN and subesophageal ganglion extracts stimulate isolated pheromone glands, indicating pheromone glands as a target tissue (2). Signal transduction is accomplished through extracellular calcium influx and cAMP production. Here we report on the identification of the receptor for PBAN. It was identified by using cDNA synthesized from mRNA obtained from pheromone glands that was probed with degenerate PCR primers designed against three Drosophila genes that are homologous to neuromedin U receptors in vertebrates. The full-length clone was expressed by using Sf9 cells, and ligand binding was assessed by measuring calcium fluxes in the cytosol. Results indicate that, of the peptides tested, PBAN had the lowest EC50 value, followed by several other peptides with an FXPRLamide motif. These results provide strong evidence that the isolated cDNA encodes a protein that acts as the PBAN receptor in pheromone gland cells of H. zea.
Several peptides were tested, including those produced by the H. zea PBAN-encoding gene. The gene encoding PBAN in H. zea has posttranslational processing sites that could produce additional peptides having the FXPRLamide motif (23). One of these (PGN-24) has a high sequence similarity to the diapause hormone of B. mori (11). Of the peptides that could be produced by the PBAN-encoding gene, PBAN had the lowest EC50 value, followed by PGN-7 and PGN-18. The other two peptides, PGN-8 and PGN-24, exhibited lower potency, with PGN-8 being the least active. These results indicate that although these peptides can stimulate pheromone biosynthesis when tested on isolated pheromone glands (5), PBAN, PGN-7, and PGN-18 are most likely to stimulate pheromone biosynthesis at physiological concentrations. In addition, the peptides with similar EC50 values to PBAN are considerably shorter than PBAN, indicating that the N-terminal portion of PBAN does not necessarily confer additional binding affinity. Additional ligand-binding studies with directed amino acid substitutions in the C-terminal motif will be required to help determine sequences required for binding activity.
PBAN from H. zea has two methionine residues that could potentially become oxidized. Several reports indicate that oxidized PBAN was more effective than unoxidized PBAN when injected into female abdomens (24, 25). However, in our study oxidized PBAN was not significantly different from PBAN. The in vivo studies could have shown increased activity because of decreased degradation of PBAN in the hemolymph.
The deduced sequence of 346 aa for the H. zea PBAN-R indicates a size of 38.6 kDa. A study using a photoaffinity-biotin-labeled PBAN and pheromone glands of H. armigera demonstrated specific binding to a membrane-bound protein of ≈50 kDa (26). After accounting for the attached biotin-labeled PBAN, the apparent size of the receptor would be reduced to ≈45 kDa. The apparent differences in size could be due to several factors including unpredictable migration on SDS/PAGE due to glycosylations or other modifications and species variation. H. zea and H. armigera are closely related species that regulate pheromone production in similar ways (2) and should have a similar PBAN-R.
The Drosophila receptor CG8795 was expressed by using the same expression and assay system for comparative purposes. This GPCR has been characterized to bind FXPRLamide peptides in a study by Park et al. (18). In their study the Drosophila receptors CG8784, CG8795, CG9918, and CG14575 were expressed in Xenopus oocytes with CG8795 exhibiting EC50 values of 420 and 480 nM for CAP2b-3 and ETH-1, respectively. Two other peptides from Drosophila, PK-2 (SVPFKPRLamide) and hug γ (pELQSNGEPAYRVRTPRLamide) had the highest potency but also were found to desensitize the receptor. We found that CG8795 expressed in Sf9 cells, when challenged with CAP2b-3 and ETH-1, had somewhat higher EC50 values than those determined by Park et al. (18). However, PBAN had an EC50 value of 125 nM in our study, whereas it was active at 10 μM in the Park et al. study. It was shown recently that the Drosophila ETH receptor, CG5911b, when expressed in Chinese hamster ovary cells had an EC50 value of ≈1 nM when challenged with ETH-1 and ETH-2 (22). Interestingly, CG5911b also bound PBAN with an EC50 value of ≈3 μM, but other FXPRLamide peptides did not show activity at 10 μM (22). These results, taken together, indicate that the expression system could have differential effects when determining peptide efficacy. The results also indicate that receptors classified into families based on sequence homology have differential ligand-binding efficiencies.
As indicated by a blast search of the GenBank database, the vertebrate proteins with the closest sequence similarities to the PBAN-R were neuromedin U receptors, followed by ghrelin receptors. Both neuromedin U and ghrelin have been implicated in controlling food intake in mammals. Intracerebroventricular administration of neuromedin U into rats reduced food intake (27). On the other hand, intracerebroventricular administration of ghrelin into rats caused an increase in food intake and body weight (28). Both of these peptides are involved in the complex regulation of feeding behavior in mammals and are part of the brain/gut regulatory complex (29). The homology between the insect receptors and neuromedin U receptors is intriguing. Two forms of the neuromedin U receptor have been characterized from mammals with differential tissue localization. Receptor type 1 is found primarily in peripheral tissue, whereas receptor type 2 is found primarily in the central nervous system. Tissue localization of the Drosophila receptors CG8795, CG8784, and CG9918 is unknown but may also follow a similar pattern.
Acknowledgments
We thank Robert Harrison and Bryony Bonning at Iowa State University for helpful discussions and advice. We also thank Peter Ma at Mississippi State University for the four PGN peptides. This research was supported by United States–Israel Binational Agricultural Research and Development Fund Research Grant Award IS-2978-98R and State of Iowa funds. This is contribution no. 409/03 from the Agricultural Research Organization of the Volcani Center.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PBAN, pheromone biosynthesis-activating neuropeptide; GPCR, G protein-coupled receptor; PBAN-R, PBAN receptor; ETH, ecdysis-triggering hormone; PGN, PBAN-encoding gene neuropeptide; AM, acetoxymethyl ester.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY319852).
References
- 1.Raina, A. K. & Klun, J. A. (1984) Science 225, 531–533. [DOI] [PubMed] [Google Scholar]
- 2.Rafaeli, A. (2002) Int. Rev. Cytol. 213, 49–91. [DOI] [PubMed] [Google Scholar]
- 3.Raina, A. K., Jaffe, H., Kempe, T. G., Keim, P., Blacher, R. W., Fales, H. M., Riley, C. T., Klun, J. A., Ridgway, R. L. & Hayes, D. K. (1989) Science 244, 796–798. [DOI] [PubMed] [Google Scholar]
- 4.Raina, A. & Kempe, T. (1990) Insect Biochem. 20, 849–851. [Google Scholar]
- 5.Ma, P. W. K., Roelofs, W. L. & Jurenka, R. A. (1996) J. Insect Physiol. 42, 257–266. [Google Scholar]
- 6.Gäde, G. (1997) Prog. Chem. Org. Nat. Prod. 71, 1–128. [Google Scholar]
- 7.Torfs, P., Nieto, J., Cerstiaens, A., Boon, D., Baggerman, G., Poulos, C., Waelkens, E., Derua, R., Calderon, J., De Loof, A., et al. (2001) Eur. J. Biochem. 268, 149–154. [DOI] [PubMed] [Google Scholar]
- 8.Tillman, J. A., Seybold, S. J., Jurenka, R. A. & Blomquist, G. J. (1999) Insect Biochem. Mol. Biol. 29, 481–514. [DOI] [PubMed] [Google Scholar]
- 9.Holman, G. M., Cook, B. J. & Nachman, R. J. (1986) Comp. Biochem. Physiol. C Comp. Pharmacol. 85, 219–224. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto, S., Kitamura, A., Nagasawa, H., Kataoka, H., Orikasa, C., Mitsui, T. & Suzuki, A. (1990) J. Insect Physiol. 36, 427–432. [Google Scholar]
- 11.Imai, K., Konno, T., Nakazawa, Y., Komiya, T., Isobe, M., Koga, K., Goto, T., Yaginuma, T., Sakakibara, K., Hasegawa, K., et al. (1991) Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 67, 98–101. [Google Scholar]
- 12.Zdarek, J., Nachman, R. J. & Hayes, T. K. (1997) Ann. N.Y. Acad. Sci. 814, 67–72. [DOI] [PubMed] [Google Scholar]
- 13.Jurenka, R. A., Jacquin, E. & Roelofs, W. L. (1991) Proc. Natl. Acad. Sci. USA 88, 8621–8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rafaeli, A. & Gileadi, C. (1996) Insect Biochem. Mol. Biol. 26, 797–807. [Google Scholar]
- 15.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 16.Hewes, R. S. & Taghert, P. H. (2001) Genome Res. 11, 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi, M.-Y., Rafaeli, A. & Jurenka, R. (2001) Cell Tissue Res. 306, 459–465. [DOI] [PubMed] [Google Scholar]
- 18.Park, Y., Kim, Y.-J. & Adams, M. E. (2002) Proc. Natl. Acad. Sci. USA 99, 11423–11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soroker, V. & Rafaeli, A. (1989) Insect Biochem. 19, 1–5. [Google Scholar]
- 20.Wu, C. H., Huang, H., Arminski, L., Castro-Alvear, J., Chen, Y., Hu, Z.-Z., Ledley, R. S., Lewis, K. C., Mewes, H.-W., Orcutt, B. C., et al. (2002) Nucleic Acids Res. 30, 35–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iversen, A., Cazzamali, G., Williamson, M., Hauser, F. & Grimmelikhuijzen, C. J. P. (2002) Biochem. Biophys. Res. Commun. 299, 924–931. [DOI] [PubMed] [Google Scholar]
- 22.Park, Y., Kim, Y.-J., Dupriez, V. & Adams, M. E. (2003) J. Biol. Chem. 278, 17710–17715. [DOI] [PubMed] [Google Scholar]
- 23.Ma, P. W. K., Knipple, D. C. & Roelofs, W. L. (1994) Proc. Natl. Acad. Sci. USA 91, 6506–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura, A., Nagasawa, H., Kataoka, H., Inoue, T., Matsumoto, S., Ando, T. & Suzuki, A. (1989) Biochem. Biophys. Res. Commun. 163, 520–526. [DOI] [PubMed] [Google Scholar]
- 25.Raina, A. K., Kempe, T. G. & Jaffe, H. (1991) in Insect Neuropeptides: Chemistry, Biology and Action, eds. Menn, J. J., Kelly, T. J. & Masler, E. P. (Am. Chem. Soc., Washington DC), Vol. 453, pp. 100–109. [Google Scholar]
- 26.Rafaeli, A., Zakharova, T., Lapsker, Z. & Jurenka, R. A. (2003) Insect Biochem. Mol. Biol. 33, 371–380. [DOI] [PubMed] [Google Scholar]
- 27.Howard, A. D., Wang, R., Pong, S. S., Mellin, T. N., Strack, A., Guan, X. M., Zeng, Z., Williams, D. L., Jr., Feighner, S. D., Nunes, C. N., et al. (2000) Nature 406, 70–74. [DOI] [PubMed] [Google Scholar]
- 28.Tschop, M., Smiley, D. & Heiman, M. (2000) Nature 407, 908–913. [DOI] [PubMed] [Google Scholar]
- 29.Kalra, S. P., Bagnasco, M., Otukonyong, E. E., Dube, M. G. & Kalra, P. S. (2003) Regul. Pept. 111, 1–11. [DOI] [PubMed] [Google Scholar]