Abstract
Iron–sulfur (Fe–S) clusters serve as cofactors in many proteins that have important redox, catalytic, and regulatory functions. In bacteria, biogenesis of Fe–S clusters is mediated by multiple gene products encoded by the isc and nif operons. In particular, genetic and biochemical studies suggest that IscU, Nfu, and IscA function as scaffold proteins for assembly and delivery of rudimentary Fe–S clusters to target proteins. Here we report the characterization of human Nfu. A combination of biochemical and spectroscopic techniques, including UV-visible absorption and 57Fe Mössbauer spectroscopies, have been used to investigate the ability of purified human Nfu to assemble Fe–S clusters. The results suggest that Nfu can assemble approximately one labile [4Fe–4S] cluster per two Nfu monomers, and support the proposal that Nfu is an alternative scaffold protein for assembly of clusters that are subsequently used for maturation of targeted Fe–S proteins. Analyses of genomic DNA, transcripts, and translation products indicate that alternative splicing of a common pre-mRNA results in synthesis of two Nfu isoforms with distinct subcellular localizations. Isoform I is localized in the mitochondria, whereas isoform II is present in the cytosol and the nucleus. These results, together with previous reports of subcellular distributions of isoforms of human IscS and IscU in mitochondria, cytosol, and nucleus suggest that the Fe–S cluster assembly machineries are compartmentalized in higher eukaryotes.
Iron–sulfur proteins are widely distributed in many organisms and have essential physiological roles in electron transport, metabolic reactions, as well as in transcriptional regulation (1). Enzymes involved in Fe–S cluster assembly were first identified in the nif (nitrogen fixation) operon in Azotobacter vinelandii (2). Genetic and biochemical studies have shown that two gene products, NifS and NifU, are essential for assembly of the Fe–S clusters in nitrogenase (3–6). Subsequent studies in a wide variety of prokaryotes resulted in the identification of the isc (iron–sulfur cluster assembly) operon (7, 8), which encodes the translational regulator IscR (9), cysteine desulfurase IscS (7), scaffold proteins IscU (10) and IscA (11, 12), chaperone proteins HscA and HscB (13), and ferredoxin Fdx. IscS is a cysteine desulfurase that generates alanine and elemental sulfur by means of the formation of a persulfide intermediate on a cysteine residue (4, 14). The sulfur is transferred directly to the scaffold protein IscU (15, 16) for the sequential assembly of [2Fe–2S]2+ and [4Fe–4S]2+ clusters (17). It has been proposed that the transient Fe–S clusters in IscU are subsequently delivered to specific target proteins (17).
In addition to the IscU proteins, recent genetic and biochemical studies (18) suggest that other proteins such as Nfu are also involved in the biosynthesis of Fe–S clusters. A. vinelandii NifU consists of three distinct domains: the N-terminal IscU-like domain, the central Fdx-like domain, and the C-terminal Nfu-like domain (19, 20). Mutant A. vinelandii strains with substitutions at the conserved cysteine residues in the IscU-like and Fdx-like domains of NifU exhibit less severe defects than the ΔnifU strain, indicating that the Nfu-like domain has an important role in Fe–S cluster assembly (20). Likewise, yeast strains that lack Nfu1p exhibit loss of Fe–S protein activities, supporting a role for NFU1 in Fe–S cluster assembly (21, 22). Furthermore, yeast NFU1 was identified in a synthetic lethal screen with SSQ1, a yeast mitochondrial chaperone gene involved in the biogenesis of Fe–S clusters (22). The cyanobacterium Synechocystis PCC6803 contains a single Nfu-like gene (syNifU) but no IscU-like genes (23). SyNifU is an essential gene for Synechocystis, suggesting that Nfu-like proteins may serve as Fe–S scaffold proteins in the absence of IscU-like proteins (18). In support of this hypothesis, Nishio and Nakai (24) reported that transfer of a [2Fe–2S]2+ cluster from SyNifU (a Synechocystis Nfu homolog) to an apoform of Synechocystis ferredoxin restores the ferredoxin electron transfer activity.
In this article, we report the characterization of human Nfu. A human Nfu homolog, named HIRIP5, was first identified in a two-hybrid screen for proteins that interact with the transcription regulator HIRA (25). However, in this article, 5′ RACE and PCR amplification experiments revealed additional 5′ sequence in the complete cDNA of human nfu. Analysis of the cDNA sequences and proteins indicated that alternative splicing leads to generation of two human Nfu isoforms that are targeted to different subcellular compartments in mammalian cells. Furthermore, biochemical studies and Mössbauer spectroscopy experiments indicate that human Nfu can bind a labile [4Fe–4S]2+ cluster that may play an important role in Fe–S cluster assembly.
Materials and Methods
Cloning and Sequence Analysis of Human Nfu Homolog. Clones containing sequences similar to yeast NFU1 were identified from the human EST database (nos. BG 718178 and NM 015700), and were further analyzed by sequencing. 5′ RACE experiments were performed by using a combination of human heart and liver Marathon-ready cDNAs (CLONTECH). 5′ RACE products were cloned into Bluescript vector pSK+ (Stratagene) and sequenced. Mapping of the intron–exon boundaries was carried out by comparison to the genomic sequence obtained from the Celera database. GST-fusion constructs were prepared by PCR cloning of the coding sequences of Nfu isoforms I and II (see Fig. 1) into pGEX-4T-1 (Amersham Pharmacia). These sequences were also cloned into pcDNA3.1(+) (Invitrogen) for in vitro translation and transient transfection experiments.
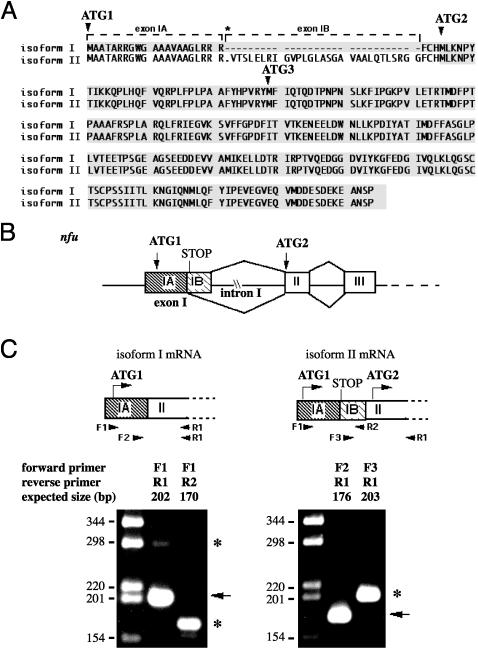
Fig. 1.
Alternative splicing of nfu pre-mRNA results in two isoforms with different translation initiation sites. (A) Predicted sequences of the two human Nfu isoforms. The boxed region indicates the coding sequences. Note that HIRIP5 was previously reported to initiate at AUG3. The asterisk indicates an in-flame stop codon present only in isoform II transcript. (B) Schematic mechanism for the use of alternative 5′ splice sites in exon I of nfu to generate two isoforms with different translational initiation sites (AUG1 or AUG2). (C) Gel electrophoretic analysis of RT-PCR DNA fragments obtained by using two different primer sets. Arrows indicate fragments amplified from isoform I, while asterisks indicate fragments amplified from isoform II.
Abs. A synthetic multiple antigenic peptide (MAP) corresponding to amino acids 239–253 of human Nfu isoform I was synthesized by Biosynthesis (Lewisville, TX), for generating rabbit polyclonal α-Nfu(MAP) 4337 (Covance Laboratories, Vienna, VA). Rabbit polyclonal Ab 4333 was generated by Covance Laboratories against purified Nfu protein generated by using the GST–Nfu isoform II construct. Abs were purified on Protein A Sepharose 4B columns (Amersham Pharmacia), or on specific MAP peptide affinity columns. Abs to human IscS and IscU were described (26, 27). Other Abs used in these studies are as follows: Mouse monoclonal α-Oct-1 (Calbiochem), and Sheep polyclonal α-manganese superoxide dismutase (α-Mn-SOD) (Calbiochem).
Cell Lysates and Subcellular Fractions. To obtain Triton lysates, human rhabdomyosarcoma (RD4) cells were washed with PBS at 4°C and lysed in 1% Triton X-100 lysis buffer (26). To obtain subcellular fractions, cell pellets were suspended in digitonin lysis buffer (0.08% digitonin/210 mM mannitol/70 mM sucrose/complete protease inhibitor mixture (Roche Molecular Biochemicals)/4 mM Hepes, pH 7.2), and were incubated for 10 min at 4°C. The digitonin lysate was centrifuged at 600 × g for 10 min at 4°C, and was separated into soluble and insoluble fractions. The insoluble fraction containing nuclei was washed, and then extracted with NER reagent (Pierce). After extraction, the sample was centrifuged at 16,000 × g for 10 min, and the supernatant constituted the nuclear fraction. The soluble fraction from the digitonin lysis was centrifuged at 7,000 × g for 9 min at 4°C, and was separated into cytosolic (supernatant) and mitochondrial fractions (pellet). The mitochondrial fraction was lysed with Triton X-100 lysis buffer.
Immunofluorescence, Immunoprecipitation, and Western Blot Analysis. RD4 and COS-7 cells (American Type Culture Collection) were grown in DMEM with 10% FCS. Transfections were carried out by using the FuGENE6 transfection reagent (Roche Molecular Biochemicals), and were analyzed 48 h after transfection. In vitro transcription and translation reactions were carried out by using the TNT coupled reticulocyte lysate system (Promega).
Transfected COS-7 cells were fixed in 4% formaldehyde for 20 min, permeabilized with methanol for 2 min, and stained with α-Nfu 4333, followed by fluorescein-conjugated donkey Abs to rabbit IgG. The samples were analyzed with a Zeiss LSM510 laser-scanning confocal microscope. Immunoprecipitation and Western blot analyses were carried out by using described procedures (27). Pulse–chase experiments were carried out first by incubating the cells at 37°C for 30 min in methionine-free DMEM (depletion medium), followed by pulse-labeling in depletion medium supplemented with 2 mCi/ml (1 Ci = 37 GBq) [35S]methionine (ICN). After labeling, cells were washed twice in PBS with 5 mM methionine at 4°C, followed by incubation in chase medium (DMEM with 5 mM methionine) at 37°C for various chase intervals. Immunoprecipitates were analyzed on SDS-polyacrylamide gels, followed by autoradiography or PhosphorImager (Molecular Dynamics) analysis.
In Vitro Fe–S Cluster Assembly in Human Nfu Protein. Escherichia coli BL21(DE3)-expressing GST-Nfu were lysed with B-PER reagent (Pierce) in an anaerobic glove box under an N2 atmosphere. Cluster reconstitution was initiated by addition of 0.25 mM 57Fe-enriched (95% plus enrichment) ferric chloride to a reaction mixture containing cell lysate, 2 mM sodium sulfide, 2 mM cysteine, and 5 mM dithiothrietol. After 3 h, GST-Nfu protein was isolated from the reaction mixture by using Glutathione Sepharose 4B (Amersham Pharmacia). Nfu was cleaved from the GST tag with thrombin, and was further purified by using a HiTrap Q Sepharose column (Amersham Pharmacia). Fractions containing purified Nfu (as judged by UV-visible spectrum and SDS/PAGE) were pooled and concentrated in an Amicon diaflow ultrafilter equipped with a YM-10 membrane (Millipore). The concentration of purified Nfu protein was determined by the BCA protein assay (Pierce), and by using molecular mass = 26 kDa for Nfu monomer. The iron content of purified Nfu was determined by extracting the iron with2MHCl at 95°C for 20 min, and the extract was analyzed by using the ferrozine assay in the presence of ascorbate (28).
Spectroscopic Measurements. UV-visible absorption spectra were recorded on a Beckman DU640 spectrophotometer. Mössbauer spectra were recorded in either a weak-field spectrometer equipped with a Janis Research (Wilmington, MA) 8DT variable-temperature cryostat, or a strong-field spectrometer furnished with a Janis CNDT/SC SuperVaritemp cryostat that houses an 8-T superconducting magnet. Both spectrometers operate in a constant acceleration mode in transmission geometry. The zero velocity of the spectra refers to the centroid of a room temperature spectrum of a metallic iron foil.
Results
Cloning of Human Nfu Homolog. An initial search of DNA databases for human homologs of yeast Nfu1p identified HIRIP5 (GenBank accession no. NM 015700), which shares 39% sequence identity to Nfu1p (25). However, further analysis identified ≈20 human ESTs that were 90% identical to HIRIP5, but which differed in two significant ways in the 5′ region. First, an insert of 90 nt from the 5′ region of HIRIP5 was absent in these ESTs. Second, at least five of these ESTs extended the 5′ sequence to include an upstream in-frame ATG. Thus, these results suggested that there were two distinct Nfu mRNAs in human cells. To obtain the complete cDNA sequence, we carried out 5′ RACE experiments (Fig. 1A). Analysis of genomic sequence and mapping of the intron–exon boundaries supported the hypothesis that the two isoforms were generated by alternative splicing of nfu pre-mRNA (Fig. 1B). Exon I contains two potential 5′ donor splice sites that are alternatively used to generate isoform I or isoform II. As a result, isoform I contains exon IA, but not the 90-nt exon IB, whereas isoform II (HIRIP5) contains both exon IA and exon IB. Moreover, although AUG1 is present in both transcripts, initiation at AUG1 in isoform II would result in synthesis of a short upstream ORF because of termination at an in-frame stop codon present in exon IB (Fig. 1C). Thus, translation of a functional Nfu isoform II would initiate at AUG2 and generate a protein that lacks the first 24 aa of isoform I.
The presence of multiple nfu isoforms was confirmed by 5′ RACE and PCR amplification experiments. Nested PCR of the 5′ RACE products using a primer upstream of AUG1 (primer F1) and a primer at the exon II–exon III junction (primer R1) generated a 202-bp product, as expected for the isoform I sequence (Fig. 1C Left). Conversely, PCR using primer F1 and a primer within exon IB (primer R2) generated the 170-bp product, as predicted from the sequence for isoform II. These results thus confirmed the presence of both isoforms I and II. Use of a different exon IB-specific primer (primer F3) also generated PCR products of the expected size for isoform II (Fig. 1C Right). Furthermore, PCR using a primer F1 with primer R1 also generated a minor product of 292 bp (Fig. 1C Left), as expected for isoform II. These results imply that isoform I is more abundant, and are consistent with the fact that isoform I was represented by >20 ESTs in the database, whereas isoform II was reported only once (25).
Sequence analysis further suggested that the two Nfu isoforms might be targeted to different subcellular locations. Sorting programs identified a putative mitochondrial targeting sequence in isoform I, whereas isoform II lacks the putative mitochondrial targeting sequence, and is expected to reside in the cytosol (29). Isoform I mRNA encodes a putative mitochondrial precursor protein of 254 aa with a predicted molecular mass of 28.4 kDa, which is expected to be processed by mitochondrial processing peptidases to a mature form with a molecular mass of 21.8 kDa (30). Isoform II encodes a protein of 230 aa with a predicted molecular mass of 25.9 kDa.
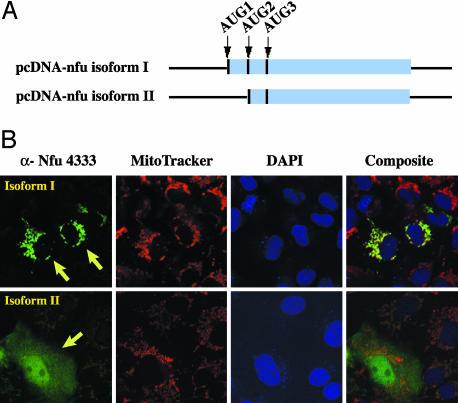
Nfu Isoforms Are Targeted to Different Subcellular Compartments. The subcellular localization of Nfu proteins was examined by immunofluorescence microscopy in COS-7 cells that were transfected with expression constructs of the two different isoforms (Fig. 2A), by using a rabbit polyclonal Ab 4333 raised against purified Nfu protein. As expected, in cells transfected with a construct encoding isoform I, staining was detected in the mitochondria, as judged by colocalization with the mitochondrial marker, MitoTracker 633. In contrast, when cells were transfected with a construct encoding isoform II, staining was detected in the cytosol and the nucleus (Fig. 2B).
Fig. 2.
Nfu isoform I is localized to mitochondria, whereas isoform II is localized to the cytosol and nucleus. (A) Schematic diagram of the nfu constructs used for in vitro translation and COS-7 cell transfection. (B) COS-7 cells transfected with constructs containing either isoform I or isoform II were stained with rabbit polyclonal Abs to Nfu (α-NFU 4333) and were analyzed by immunofluorescence microscopy. Arrows denote transfected cells. Mitochondria were stained with MitoTracker 633, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
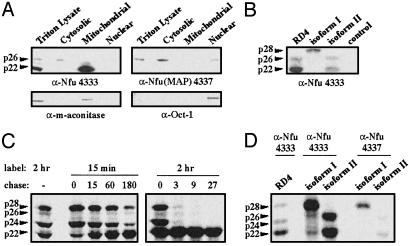
The localization of endogenous Nfu proteins was also examined by Western blot analysis and immunoprecipitation experiments. Western blot analysis of RD4 triton lysates using α-Nfu 4333 revealed a major band at ≈22 kDa (p22) and a minor band at ≈26 kDa (p26) (Fig. 3A). Subcellular fractionation indicated that p22 was present in the mitochondrial fraction, whereas p26 was present in the cytosol (Fig. 3A). The p26 protein appeared to comigrate with one of the in vitro translation products of isoform II, which was consistent with translational initiation at AUG2 to generate a cytosolic Nfu (Fig. 3B). Conversely, the presence of p22 in the mitochondrial fraction was consistent with proteolytic processing of the 28-kDa isoform I precursor protein (p28) during import into the mitochondria. To verify that p28 was processed into p22 in vivo,we carried out metabolic labeling and pulse–chase experiments. A predominant 28-kDa band (p28) appeared immediately after 15 min of metabolic labeling (Fig. 3C). With time, p28 was rapidly processed into a cleavage intermediate (p24), which was further processed into the mature mitochondrial protein p22. These results are typical of the two-step proteolytic cleavage of the leader sequence of mitochondrial proteins (31).
Fig. 3.
Endogenous Nfu proteins are present in the cytosol, mitochondria, and nucleus of RD4 cells. (A) Western blot analysis of subcellular fractions of RD4 cell lysates by using α-Nfu 4333 and α-Nfu(MAP) 4337. Analysis of a nuclear protein (Oct-1) and a mitochondrial protein (Mn-SOD) are included for comparison. (B) Western blot analysis of RD4 triton lysate (lane 1) using α-Nfu 4333. In vitro translation products of pcDNA-nfu isoform I, pcDNA-nfu isoform II, and a control construct, pPoly(A)-luciferase, were shown for comparison. (C) RD4 cells were pulse-labeled with [35S]methionine and chased at the specified times. Immunoprecipitation of the triton lysates withα-Nfu 4333 indicated a two-step processing of Nfu isoform I. (D) Immunoprecipitation of endogenous Nfu in RD4 lysates, compared with immunoprecipitation of the in vitro translation products of nfu isoform I and II. As expected, in vitro translation of isoform I gives rise to a major protein of ≈28 kDa, whereas isoform II gives rise to a protein of ≈26 kDa. The shorter product of isoform II (≈22 kDa) may be the products of in vitro translation initiated at ATG3 (Fig. 1A).
In addition, the pulse–chase experiments provided kinetic data that further supported the hypothesis that p26 is independently targeted to the cytosol. Unlike the two precursors of mitochondrial Nfu (p28 and p24), p26 was clearly detected by Western blot analysis (Fig. 3 A and B), indicating a longer half-life for p26. The pulse–chase experiments indicated that the kinetic behavior of p26 was distinct from that of p28 and p24. Immediately after 15 min of metabolic labeling (chase = 0 min), the biosynthetic ratio between p28 and p26 was ≈20:1. However, at the 180-min chase point, the biosynthetic ratio between p28 and p26 became 6:1, indicating that p26 has a longer half-life. Likewise, after 2 h of metabolic labeling, p28 decreased by >13-fold at the 3-h chase point, whereas p26 decreased by only 2.6-fold. Taken together, these results indicated that p26 was not a precursor of mitochondrial Nfu and was independently targeted to the cytosol.
The endogenous Nfu isoforms were also examined by using a second Ab α-Nfu(MAP) 4337, which was raised against an 18-aa peptide in the C terminus of Nfu. Interestingly, in contrast to α-Nfu 4333, Western blot analysis using α-Nfu(MAP) 4337 identified only p26 in the cytosolic and nuclear fraction, but did not detect p22 in the mitochondrial fraction (Fig. 3A). Western blot analysis of the recombinant GST-fusion proteins showed that α-Nfu(MAP) 4337 is fully capable of detecting both isoforms in vitro (data not shown). Likewise, both Abs 4333 and 4337 can immunoprecipitate the in vitro translation products of isoform I or isoform II constructs (Fig. 3D). Thus, the experiments carried out by using α-Nfu(MAP) 4337 not only provided further evidence of the presence of a Nfu isoform in the cytosol and the nucleus, but also suggested that posttranslational modification of the C terminus of the mitochondrial Nfu renders it undetectable by an Ab against the Nfu C terminus (Fig. 3A).
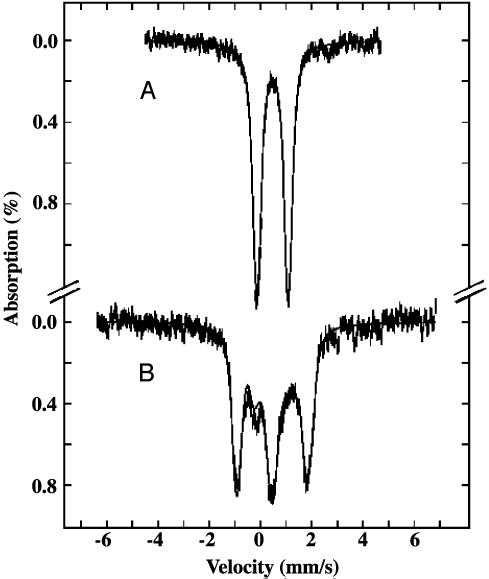
In Vitro Fe–S Cluster Assembly in Human Nfu. As purified, samples of human Nfu do not exhibit a visible chromophore (Fig. 4). On anaerobic treatment with iron and sulfide (see Materials and Methods), recombinant human Nfu isoform II acquires a broad absorption band centered at 420 nm (Fig. 4), which is indicative of a [4Fe–4S]2+ cluster (32). This Fe–S cluster is relatively stable to air over a period of 90 min, but is sensitive to the treatment with iron-chelating reagent EDTA, as demonstrated by the time-dependent decay of the UV-visible absorption bands on exposure to EDTA (data not shown). These results suggest that Nfu can protect its Fe—S cluster from oxidative degradation, even though the cluster is readily accessible to soluble chelator. Indeed, the Fe–S cluster in the purified SyNifU is also sensitive to EDTA, and is readily transferable to an apoferredoxin (24).
Fig. 4.
UV-visible absorption spectra of Nfu (43 μM) before (A) and after (B) in vitro cluster assembly.
Mössbauer spectroscopy was used to characterize the Fe–S cluster assembled on Nfu. The 4.2-K Mössbauer spectrum of the reconstituted Nfu recorded in a magnetic field of 50 mT applied parallel to the γ radiation shows a broad quadrupole doublet (Fig. 5A, hatched marks). Analysis of the spectrum indicates that it can be simulated with two unresolved doublets having parameters that are consistent with a [4Fe–4S]2+ cluster (33–38) (see Table 1 and the Fig. 5 legend). A spectrum recorded at 4.2 K, and in a strong parallel applied field of 6 T (Fig. 5B, hatched marks), further indicates that the cluster assembled on Nfu has a diamagnetic ground state, which again is consistent with the [4Fe–4S]2+ assignment. The good agreement observed between the experimental spectra and the simulations demonstrates that most, if not all, of the Fe in the Nfu sample is in a [4Fe–4S]2+ form. Protein analysis and Fe determination (see Materials and Methods) performed on the Mössbauer sample indicate that the sample contains 430 μM Nfu monomer and 840 μM Fe. These results suggest that approximately one [4Fe–4S] cluster is assembled per two monomers of Nfu.
Fig. 5.
Mössbauer spectra of human Nfu (430μM) after in vitro assembly of Fe–S cluster. The spectra (hatched marks) were recorded at 4.2 K in a field of 50 mT (A) or 6 T (B) applied parallel to the γ-radiation. The solid lines overlaid with the experimental spectra are theoretical simulations using two overlapping quadrupole doublets of the following parameters and assuming diamagnetism: δ = 0.47 mm/s, ΔEQ = 1.35 mm/s, andη = 0.5 for doublet 1, andδ = 0.47 mm/s, ΔEQ = 1.09 mm/s, and η = 0.7 for doublet 2.
Table 1. Comparison of the Mössbauer parameters of the [4Fe-4S]2+ cluster in human Nfu with those of other proteins.
| Organism | Protein | δ, mm/s | ΔEQ, mm/s | Ref. |
|---|---|---|---|---|
| Hs | Nfu | 0.47 | 1.22 | This work |
| Bs | Ferredoxin | 0.42 | 1.12 | 33 |
| Cv | Hi PIP | 0.43 | 1.14 | 34 |
| Dg | Ferredoxin I | 0.44 | 1.17 | 35 |
| Av | Ferredoxin I | 0.43 | 1.19 | 36 |
| Av | Fe protein | 0.45 | 1.12 | 37 |
The Mössbauer spectra of [4Fe-4S]2+ clusters are generally analyzed as superpositions of overlapping quadrupole doublets. The values listed here are the weighted averages. Hs, Homo sapiens; Bs, Bacillus stearothermophilus; Cv, Chromatium vinosm; Dg, Desulfovibrio giga; Av, Azotobacter vinelandii; Hi PIP, high-potential iron protein.
Discussion
We have used a combination of biochemical and spectroscopic techniques to investigate the ability of human Nfu to assemble Fe–S clusters. On incubation with iron and sulfide, human Nfu exhibits UV-visible and Mössbauer spectra characteristic of [4Fe–4S]2+ clusters. These results, together with the protein and Fe analyses, suggest the incorporation of one [4Fe–4S]2+ cluster per two monomers of Nfu. It is important to point out that, for protein-bound [4Fe–4S]2+ clusters with complete cysteinyl coordination, the isomer shifts are observed within a very narrow range of 0.42–0.45 mm/s (Table 1). The observed isomer shift of 0.47 mm/s for the Nfu [4Fe–4S]2+ cluster is relatively high, and may indicate an atypical coordination.§ It is interesting to note that the Cu(I) in the yeast copper chaperone protein Atx1 is coordinated to only two cysteine residues and an exogenous thiol group, and it was proposed that the unique coordination environment in Atx1 may facilitate formation of a bridging intermediate with its specific target protein during Cu transfer (40). It is conceivable that for human Nfu, an incomplete cysteinyl ligation of the Fe–S cluster, may also facilitate transfer of intact clusters between the scaffold and target proteins.
Our observation that under anaerobic conditions a [4Fe–4S]2+ cluster can be assembled onto human Nfu appears to be inconsistent with a previous report (24) that the purified SyNifU contains a [2Fe–2S]2+ cluster. However, the state of the assembled cluster may be determined in part by the experimental conditions. For example, the E. coli transcription regulator FNR contains a [4Fe–4S]2+ cluster which, on exposure to oxygen, is converted into a [2Fe–2S]2+ cluster (41). SyNifU was purified from aerobically grown E. coli cells, a condition that could restrict the assembly of [4Fe–4S]2+ clusters, resulting in the presence of [2Fe–2S]2+ clusters in the purified protein. Interestingly, transfer of the [2Fe–2S]2+ cluster from SyNifU to an apoform of Synechocystis ferredoxin restores the ferredoxin electron transfer activity (24). In support of the relevance of the [4Fe–4S] cluster in human Nfu, incubation of IRP1 with reconstituted Nfu resulted in rapid conversion of IRP1 to active cytosolic aconitase, demonstrating the ability of Nfu to transfer its [4Fe–4S] cluster to IRP1 (W.-H.T. and T.A.R., unpublished data). Considering that the proposed function of scaffold proteins is to assemble Fe–S clusters and deliver the clusters to various target proteins, the ability to accommodate different types of Fe–S clusters may be desirable for scaffold proteins. Indeed, recent reports (6, 12, 17, 42–44) of in vitro assembly of clusters with various stoichiometries in different scaffold proteins imply that flexibility in the coordination sites may be a common feature of scaffold proteins.
By assembling and delivering Fe–S clusters to target proteins, scaffold proteins facilitate maturation of target proteins, while protecting cellular components from the chemically reactive free iron and sulfide. The ability of human Nfu to assemble a [4Fe–4S] cluster and transfer the cluster to an acceptor protein supports the proposal that Nfu is a potential scaffold protein. Previous studies (27) have shown that mammalian cells also express IscU and IscA. The presence of multiple potential scaffold proteins raises the possibility that each may have different specificities for target proteins (18). On the other hand, it is also conceivable that these putative scaffold proteins may have additional and/or different functional roles in vivo. For example, recent studies (45) have established that IscS is essential for biosynthesis of not only Fe–S clusters but also thiamine, biotin, thionucleosides in tRNA, and NAD.
In this article, we characterized two isoforms of human Nfu that are generated by alternative splicing of a common pre-mRNA. As with human IscS and IscU, the different human Nfu isoforms are targeted to the mitochondria, the cytosol, or the nucleus. Many critical metabolic pathways, including oxidative phosphorylation, heme biosynthesis, and the citric acid cycle, occur partly or entirely within the mitochondria, and many components in these pathways require Fe–S clusters for their functions. Recent yeast genetic studies (21, 22, 46–49) imply that mitochondrial iron overload may be a direct consequence of impaired Fe–S cluster assembly, and may have significant impact on cellular functions and viability. Thus, Fe–S cluster assembly/repair is an essential process in the mitochondria. However, the potential importance of isc genes in the cytosol and the nucleus is also increasingly evident. For instance, human iron regulatory protein 1 (IRP1) is a cytosolic iron sensor protein that regulates expression of proteins involved in iron metabolism, and activity of IRP1 as a translational regulator depends on the presence or absence of a [4Fe–4S]2+ cluster (50).
It should also be noted that whereas human IscS (26), IscU (27), and Nfu (this article) can clearly be detected in multiple compartments, the yeast homologs of these proteins have previously been detected only in the mitochondria (21, 22, 51), leading to the proposal that mitochondria are essential for Fe–S cluster assembly (52). Yeast use very different strategies for iron sensing, uptake, and storage, compared with human cells (53). For instance, no cytosolic IRP homologs have been identified in yeast. This divergence in iron metabolism between yeast and human may result in different requirements for Fe–S cluster assembly in yeast versus the higher eukaryotes. Although endogenous yeast Nfs1p (an IscS homolog) cannot be detected in the nucleus by Western blotting, recent nuclear transportation trap analysis and mutagenesis studies (54) clearly indicate that nuclear localization of Nfs1p is essential for cell viability, further underscoring the physiological function of extra-mitochondrial isc genes. Our studies of the mammalian isc genes have revealed that three mechanisms: alternative start codon utilization in iscS (26), alternative selection of cassette exons in iscU (27), and alternative use of 5′ donor splice sites in nfu (this article), function in mammalian cells to target isc gene products to different subcellular locations, suggesting a potentially powerful and versatile regulatory mechanism for coordinated control of these proteins in different compartments.
Acknowledgments
We thank our colleagues in our laboratories for helpful discussion and critical reading of the manuscript. This work was supported by the intramural program of the National Institute of Child Health and Human Development (T.A.R.) and National Institutes of Health Grant GM47295 (to B.H.H.) The genomic sequence data were generated through the use of the Celera Discovery System, Celera's associated databases, and public genome databases.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: MAP, multiple antigenic peptide.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY286306 and AY286307).
Footnotes
The Mössbauer parameter, isomer shift, is sensitive to the Fe coordination environment and its spin and oxidation state. In Fe–S clusters in which the Fe atoms are all high spin with tetrahedral sulfur coordination, the isomer shift is sensitive only to the oxidation state of the cluster. Furthermore, the covalent tetrahedral sulfur coordination yields an isomer shift that is characteristically smaller than values observed with the more ionic N/O ligands. An increase in isomer shift in Fe–S proteins is therefore consistent with an increase in coordination number and/or binding of N/O ligands. For example, in the C56S variant of the 2Fe-ferredoxin from Clostridium pasteurianum, binding of serine to the spin 1/2 [2Fe–2S]1+ cluster increases the overall isomer shift from 0.50 mm/s to 0.53 mm/s (39).
References
- 1.Beinert, H., Holm, R. H. & Münck, E. (1997) Science 277, 653–659. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson, M. R., Brigle, K. E., Bennett, L. T., Setterquist, R. A., Wilson, M. S., Cash, V. L., Beynon, J., Newton, W. E. & Dean, D. R. (1989) J. Bacteriol. 171, 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobson, M. R., Cash, V. L., Weiss, M. C., Laird, N. F., Newton, W. E. & Dean, D. R. (1989) Mol. Gen. Genet. 219, 49–57. [DOI] [PubMed] [Google Scholar]
- 4.Zheng, L., White, R. H., Cash, V. L., Jack, R. F. & Dean, D. R. (1993) Proc. Natl. Acad. Sci. USA 90, 2754–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu, W., Jack, R. F., Morgan, T. V., Dean, D. R. & Johnson, M. K. (1994) Biochemistry 33, 13455–13463. [DOI] [PubMed] [Google Scholar]
- 6.Yuvaniyama, P., Agar, J. N., Cash, V. L., Johnson, M. K. & Dean, D. R. (2000) Proc. Natl. Acad. Sci. USA 97, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng, L., Cash, V. L., Flint, D. H. & Dean, D. R. (1998) J. Biol. Chem. 273, 13264–13272. [DOI] [PubMed] [Google Scholar]
- 8.Tokumoto, U. & Takahashi, Y. (2001) J. Biochem. (Tokyo) 130, 63–71. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz, C. J., Giel, J. L., Patschkowski, T., Luther, C., Ruzicka, F. J., Beinert, H. & Kiley, P. J. (2001) Proc. Natl. Acad. Sci. USA 98, 14895–14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agar, J. N., Zheng, L., Cash, V. L., Dean, D. R. & Johnson, M. K. (2000) J. Am. Chem. Soc. 122, 2136–2137. [Google Scholar]
- 11.Ollagnier-de-Choudens, S., Mattioli, T., Takahashi, Y. & Fontecave, M. (2001) J. Biol. Chem. 276, 22604–22607. [DOI] [PubMed] [Google Scholar]
- 12.Krebs, C., Agar, J. N., Smith, A. D., Frazzon, J., Dean, D. R., Huynh, B. H. & Johnson, M. K. (2001) Biochemistry 40, 14069–14080. [DOI] [PubMed] [Google Scholar]
- 13.Hoff, K. G., Silberg, J. J. & Vickery, L. E. (2000) Proc. Natl. Acad. Sci. USA 97, 7790–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng, L., White, R. H., Cash, V. L. & Dean, D. R. (1994) Biochemistry 33, 4714–4720. [DOI] [PubMed] [Google Scholar]
- 15.Urbina, H. D., Silberg, J. J., Hoff, K. G. & Vickery, L. E. (2001) J. Biol. Chem. 276, 44521–44526. [DOI] [PubMed] [Google Scholar]
- 16.Smith, A. D., Agar, J. N., Johnson, K. A., Frazzon, J., Amster, I. J., Dean, D. R. & Johnson, M. K. (2001) J. Am. Chem. Soc. 123, 11103–11104. [DOI] [PubMed] [Google Scholar]
- 17.Agar, J. N., Krebs, C., Frazzon, J., Huynh, B. H., Dean, D. R. & Johnson, M. K. (2000) Biochemistry 39, 7856–7862. [DOI] [PubMed] [Google Scholar]
- 18.Nakai, M., Nishio, K., Morimoto, K., Yabe, T. & Kikuchi, S. (2002) Recent Res. Dev. Proteins 1, 1–11. [Google Scholar]
- 19.Ouzounis, C., Bork, P. & Sander, C. (1994) Trends Biochem. Sci. 19, 199–200. [DOI] [PubMed] [Google Scholar]
- 20.Agar, J. N., Yuvaniyama, P., Jack, R. F., Cash, V. L., Smith, A. D., Dean, D. R. & Johnson, M. K. (2000) J. Biol. Inorg. Chem. 5, 167–177. [DOI] [PubMed] [Google Scholar]
- 21.Garland, S. A., Hoff, K., Vickery, L. E. & Culotta, V. C. (1999) J. Mol. Biol. 294, 897–907. [DOI] [PubMed] [Google Scholar]
- 22.Schilke, B., Voisine, C., Beinert, H. & Craig, E. (1999) Proc. Natl. Acad. Sci. USA 96, 10206–10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura, Y., Kaneko, T., Hirosawa, M., Miyajima, N. & Taba, S. (1998) Nucleic Acids Res. 26, 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishio, K. & Nakai, M. (2000) J. Biol. Chem. 275, 22615–22618. [DOI] [PubMed] [Google Scholar]
- 25.Lorain, S., Lecluse, Y., Scamps, C., Mattei, M.-G. & Lipinsku, M. (2001) Biochim. Biophys. Acta 1517, 376–383. [DOI] [PubMed] [Google Scholar]
- 26.Land, T. & Rouault, T. A. (1998) Mol. Cell 2, 807–815. [DOI] [PubMed] [Google Scholar]
- 27.Tong, W.-H. & Rouault, T. (2000) EMBO J. 19, 5692–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stookey, L. L. (1970) Anal. Chem. 42, 779–781. [Google Scholar]
- 29.Nakai, K. & Horton, P. (1999) Trends Biochem. Sci. 24, 34–35. [DOI] [PubMed] [Google Scholar]
- 30.Gavel, Y. & von Heijne, G. (1990) Protein Eng. 4, 33–37. [DOI] [PubMed] [Google Scholar]
- 31.Pfanner, N., Craig, E. A. & Honlinger, A. (1997) Annu. Rev. Cell Dev. Biol. 13, 25–51. [DOI] [PubMed] [Google Scholar]
- 32.Orme-Johnson, W. H. & Orme-Johnson, N. R. (1982) in Iron-Sulfur Proteins, ed. Spiro, T. G. (Wiley, New York), Vol. 4, pp. 67–146. [Google Scholar]
- 33.Middleton, P., Dickson, D. P. E., Johnson, C. E. & Rush, J. D. (1978) Eur. J. Biochem. 88, 135–141. [DOI] [PubMed] [Google Scholar]
- 34.Middleton, P., Dickson, D. P. E., Johnson, C. E. & Rush, J. D. (1980) Eur. J. Biochem. 104, 289–296. [DOI] [PubMed] [Google Scholar]
- 35.Moura, J. J. G., Moura, I., Kent, T. A., Lipscomb, J. D., Huynh, B. H., LeGall, J., Xavier, A. V. & Münck, E. (1982) J. Biol. Chem. 257, 6259–6267. [PubMed] [Google Scholar]
- 36.Hu, Z., Jollie, D., Burgess, B. K., Stephens, P. J. & Münck, E. (1994) Biochemistry 33, 14475–14485. [DOI] [PubMed] [Google Scholar]
- 37.Yoo, S. J., Angove, H. C., Burgess, B. K., Hendrich, M. P. & Münck, E. (1999) J. Am. Chem. Soc. 121, 2534–2545. [Google Scholar]
- 38.Pereira, A. S., Tavares, P., Moura, I., Moura, J. J. G. & Huynh, B. H. (2001) J. Am. Chem. Soc. 123, 2771–2782. [DOI] [PubMed] [Google Scholar]
- 39.Achim, C., Bominaar, E. L., Meyer, J., Peterson, J. & Münck, E. (1999) J. Am. Chem. Soc. 121, 3704–3714. [Google Scholar]
- 40.Pufahl, R. A., Singer, C. P., Pearisco, K. L., Lin, S.-J., Schmidt, P. J., Fahrni, C. J., Cizewski-Culotta, V., Penner-Hahn, J. E. & O'Halloran, T. V. (1997) Science 278, 853–856. [DOI] [PubMed] [Google Scholar]
- 41.Popescu, C. V., Bates, D. M., Beinert, H., Munck, E. & Kiley, P. J. (1998) Proc. Natl. Acad. Sci. USA 95, 13431–13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, G., Mansy, S. S., Wu, S.-P., Surerus, K. K., Foster, M. W. & Cowan, J. A. (2002) Biochemistry 41, 5024–5032. [DOI] [PubMed] [Google Scholar]
- 43.Nuth, M., Yoon, T. & Cowan, J. A. (2002) J. Am. Chem. Soc. 124, 8774–8775. [DOI] [PubMed] [Google Scholar]
- 44.Mansy, S. S., Wu, G., Surerus, K. K. & Cowan, J. A. (2002) J. Biol. Chem. 277, 21397–21404. [DOI] [PubMed] [Google Scholar]
- 45.Mihara, H. & Esaki, N. (2002) Appl. Microbiol. Biotechnol. 60, 12–23. [DOI] [PubMed] [Google Scholar]
- 46.Strain, J., Lorenz, C. R., Bode, J., Garland, S., Smolen, G. A., Ta, D. T., Vickery, L. E. & Culotta, V. C. (1998) J. Biol. Chem. 273, 31138–31144. [DOI] [PubMed] [Google Scholar]
- 47.Knight, S. A., Sepuri, N. B., Pain, D. & Dancis, A. (1998) J. Biol. Chem. 273, 18389–18393. [DOI] [PubMed] [Google Scholar]
- 48.Lange, H., Kaut, A., Kispal, G. & Lill, R. (2000) Proc. Natl. Acad. Sci. USA 97, 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voisine, C., Cheng, Y. C., Ohlson, M., Schilke, B., Hoff, K., Beinert, H., Marszalek, J. & Craig, E. A. (2001) Proc. Natl. Acad. Sci. USA 98, 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rouault, T. A. & Kalusner, R. D. (1996) Trends Biochem. Sci. 21, 174–177. [PubMed] [Google Scholar]
- 51.Kispal, G., Csere, P., Prohl, C. & Lill, R. (1999) EMBO J. 18, 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lill, R., Diekert, K., Kaut, A., Lange, H., Pelzer, W., Prohl, C. & Kispal, G. (1999) Biol. Chem. 380, 1157–1166. [DOI] [PubMed] [Google Scholar]
- 53.Van Ho, A., Ward, D. M. & Kaplan, J. (2002) Annu. Rev. Microbiol. 56, 237–261. [DOI] [PubMed] [Google Scholar]
- 54.Nakai, Y., Nakai, M., Hayashi, H. & Kagamiyama, H. (2001) J. Biol. Chem. 276, 8314–8320. [DOI] [PubMed] [Google Scholar]