Abstract
The 3′ ends of RNAs associated with turnip crinkle virus (TCV), including subviral satellite (sat)C, terminate with the motif CCUGCCC-3′. Transcripts of satC with a deletion of the motif are repaired to wild type (wt) in vivo by RNA-dependent RNA polymerase (RdRp)-mediated extension of abortively synthesized oligoribonucleotide primers complementary to the 3′ end of the TCV genomic RNA. Repair of shorter deletions, however, are repaired by other mechanisms. SatC transcripts with the 3′ terminal CCC replaced by eight nonviral bases were repaired in plants by homologous recombination between the similar 3′ ends of satC and TCV. Transcripts with deletions of four or five 3′ terminal bases, in the presence or absence of nonviral bases, generated progeny with a mixture of wt and non-wt 3′ ends in vivo. In vitro, RdRp-containing extracts were able to polymerize nucleotides in a template-independent fashion before using these primers to initiate transcription at or near the 3′ end of truncated satC templates. The nontemplate additions at the 5′ ends of the nascent complementary strands were not random, with a preference for consecutive identical nucleotides. The RdRp was also able to initiate transcription opposite cytidylate, uridylate, guanylate, and possibly adenylate residues without exhibiting an obvious preference, flexibility previously unreported for viral RdRp. The unexpected existence of three different repair mechanisms for TCV suggests that 3′ end reconstruction is critical to virus survival.
Keywords: satellite RNA, RNA replication
Plus (+)-sense RNA viruses contain cis-acting elements at their 3′ ends that are important for minus (−)-strand synthesis. During the virus life cycle, the 3′ end sequences may be exposed to cellular RNases, which are widely distributed in intracellular compartments and particles such as lysosomes, vacuoles, endoplasmic reticulum, microsomes, and ribosomes (reviewed in refs. 1 and 2). Such RNases may play a role in host defense against pathogen attack in addition to general housekeeping roles such as cytoplasmic mRNA degradation (1–3). RNA viruses therefore must develop strategies to protect their 3′ ends against nuclease degradation. For example, RNA viruses may sequester replicating RNAs in virus-induced membrane structures (4–6) or covalently link amino acids or poly(A) tails to their genomic 3′ ends (7, 8). In addition, some RNA viruses have developed strategies to repair missing 3′ end nucleotides. Genomic and satellite RNAs of cymbidium ringspot tombus virus with a deletion of the 3′ terminal CCC are repaired in vivo (9, 10) as were up to seven bases deleted from the 3′ end of a cucumber mosaic virus satRNA (11). In addition, de novo polyadenylation occurs in vivo at the 3′ terminus of Sindbis virus and clover yellow vein virus RNAs lacking their normal poly(A) tails (12–14). Although possible roles in the 3′ end repair processes for the viral RNA-dependent RNA polymerase (RdRp) (15), host telomerase-like enzymes (16), terminal transferases (7), and cytoplasmic polyadenylation apparatus (17) have been suggested, the repair mechanisms remain poorly understood.
Turnip crinkle virus (TCV), a single-stranded (+)-sense RNA virus containing a genome of 4,054 bases (18, 19), has been very useful in studying 3′ end repair because of its association with a number of subviral RNAs whose mutated 3′ ends can be repaired in vivo (15, 20, 21). Satellite RNA D (satD; 194 bases), like all TCV subviral RNAs, depends on the RdRp encoded by TCV for replication (22). SatD shares little sequence similarity with TCV (references to TCV henceforth refer to the genomic RNA) beyond the 3′ terminal seven-base motif CCUGCCC-3′. Satellite RNA C (satC; 356 bases) is composed of nearly full length of satD at its 5′ end linked to two regions from TCV including 151 contiguous bases from the TCV 3′ end. SatC transcripts with deletions of 6–8 bases at the 3′ end were repaired to the wild-type (wt) sequence in vivo by abortive synthesis of oligoribonucleotides complementary to the TCV 3′ end by the RdRp and subsequent use of these abortive products to prime (−)-strand synthesis of satC (15).
In this report, we have found two additional 3′ end repair mechanisms for satC: homologous recombination between the similar 3′ ends of satC and TCV and the replacement of deleted bases in a manner that apparently does not involve the TCV terminal motif. Using an in vitro RdRp transcription system, nontemplate bases were found at the 5′ ends of nascent (−)-strands, indicating that repair occurred before initiation of transcription by the TCV RdRp. The composition of the nontemplate bases was not random but tended to consist of 2–5 consecutive identical nucleotides with a preference for multiple guanylate residues.
Materials and Methods
Construction of SatC Mutants.
PCR was used to generate 3′ end deletions in satC. The 5′ primer used in the PCR was T7C5′ (5′-GTAATACGACTCACTATAGGGATAACTAAGGG), which contains a T7 RNA polymerase promoter and bases homologous to positions 1–14 of satC. The 3′ primer was C3′Δ3 (5′-CAGGCCCCCCGTCCGAGGA), C3′Δ4 (5′-AGGCCCCCCGTCCGAGGA), or C3′Δ5 (5′-GGCCCCCCGTCCGAGGA). These three primers were complementary to the 3′ terminal sequence of satC but lacking the 3′ end three, four, and five bases, respectively. The template used for the PCR was plasmid pT7C(+), which contains a T7 RNA polymerase promoter upstream of full-length satC sequence (23). The PCR products were treated with Escherichia coli DNA polymerase I large fragment (Biolabs, Northbrook, IL) to generate blunt ends and then cloned into the SmaI site of pUC19.
Analysis of SatC Progeny Accumulating in Plants.
PCR products containing 3′ end deletions or plasmids linearized with SmaI, SacI, or EcoRI were subjected to T7 RNA polymerase transcription in vitro (24). SacI- and EcoRI-digested plasmids generated transcripts with eight (5-′GGGUACCG) and 18 (5-′GGGUACCGAGCUCGAAUU) plasmid-derived bases at the 3′ ends, respectively, whereas the PCR products and SmaI-digested plasmids produced transcripts lacking plasmid-derived bases.
To test the amplification capability of repaired satC with non-wt 3′ ends, full-length cDNAs were amplified by PCR using primers T7C5′ and either 5′-GGGCUGGGCCCCCCGUCCGA, 5′-CCCAGGCCCCCCGA, 5′-GGGCCCGCCCCCCGUCCGA, or 5′-AAACGGCCCCCCGUCCGA. Transcripts were synthesized by T7 RNA polymerase directly from the PCR products and inoculated onto six turnip plants. SatC species accumulating in uninoculated leaves at 2 weeks postinoculation were analyzed as described above.
For plant inoculations, transcripts (0.2 μg/plant) of mutant and wt satC were coinoculated with TCV transcripts (2 μg/plant) prepared from SmaI-digested plasmid pT7TCVms (24), onto four 2-week-old turnip seedlings (25). Total RNA was isolated from uninoculated leaves 2 weeks later. Gel-purified satC species were polyadenylated by using poly(A) polymerase (Amersham Pharmacia), reverse-transcribed into cDNA using primer T17-adapter (5′-CCACTCGAGTCGACATCGA[T]17), and Moloney murine leukemia virus reverse transcriptase (GIBCO/BRL) and then subjected to PCR using primer C5′ (homologous to the 5′ terminal 19 bases) and primer “adapter” (5′-CCACTCGAGTCGACATCGA), as described (21). The PCR products were cloned into pUC19 and sequenced.
In Vitro Transcription Using the TCV RdRp and Cloning of RNA Products.
In vitro transcription using partially purified TCV RdRp was carried out as described (26). To clone the products synthesized in vitro, 20 μg of satC templates with or without 3′ end deletions was subjected to the RdRp transcription reactions. After denaturing gel (5% acrylamide/8 M urea) electrophoresis, the radiolabeled products were visualized by autoradiography and purified as described (21). The purified products then were amplified by 5′ rapid amplification of cDNA ends PCR using primer C5′ for the reverse transcription reaction, as described with modifications (27). The first-strand cDNA was purified from agarose gels, and poly(A) was added to the 3′ ends by using terminal deoxynucleotidal transferase (GIBCO/BRL). To amplify the poly(A)-tailed cDNA by PCR, oligonucleotides T17-adapter was used with C5′ or C22–41 (5′-CAAUACUACGCAACUAAUGC, homologous to positions 22–41 of satC). The PCR products were cloned and sequenced as described above.
Results and Discussion
In Vivo Repair of 3′ End Deletions in SatC.
SatC shares 90% sequence similarity with TCV in their 3′ proximal regions. Both satC and TCV terminate with hairpin structures followed by the motif CCUGCCC-OH 3′. Together, these two elements comprise the minimal promoter for (−)-strand synthesis (23). SatC with a 3′ end deletion of 6 or 7 bases is infectious in vivo, and all cloned progeny regained the wt motif. The repaired sequence at the 3′ end of satC was derived from the 3′ end of TCV; abortive initiation of TCV transcripts by the RdRp resulted in the synthesis of short oligoribonucleotides complementary to the TCV 3′ end that were used as primers by the RdRp for extension on truncated satC templates (15).
To determine whether deletions of less than 6 bases are repaired in a similar fashion, turnip seedlings were inoculated with TCV along with transcripts of wt satC or satC with 3′ end deletions of 3–5 bases. Plants inoculated with wt satC transcripts generated progeny in uninoculated leaves that were nearly all wt (15 of 16 clones) (Table 1). When plants were inoculated with satC transcripts with a deletion of the 3′ terminal three bases (CΔ3) or four bases (CΔ4), 18 of 20 clones and 12 of 16 clones, respectively, had regained the wt 3′ end sequence (Table 1). In contrast, none of the clones obtained from plants inoculated with satC transcripts with a 5-base deletion (CΔ5) contained the wt 3′ end (Table 1). Twelve of 19 species recovered terminated with the motif CCGGCCC-3′, which differed by a single position from wt (CCUGCCC-3′). Species that terminated with CCCGCCC-3′, CGGGCCC-3′, CCGUUU-3′, and ΔCGGCCC-3′ also were recovered. Inoculation of plants with transcripts of CΔ5 was repeated with very similar results (data not shown). Because a deletion of 6 bases was always repaired to the wt motif (15), these results suggest that satC with 3′ end deletions of 3–5 bases can be repaired by a different mechanism, although we cannot rule out that some or all wt progeny generated from CΔ3 and CΔ4 transcripts are repaired by the previously elucidated mechanism of extension on abortive initiation products by using the 3′ end of TCV.
Table 1.
New 3′ end sequences in satC progeny generated in vivo from transcripts with or without 3′ end deletions
| Construct | Plant | 3′ end sequence | Construct | Plant | 3′ end sequence |
|---|---|---|---|---|---|
| WT | [GGACGGGGGGCCUGCCC] | CΔ4 | [GGACGGGGGGCCU] | ||
| 1 & 2 | GGACGGGGGGCCUGCCC (8, 7) | 1 & 2 | GGACGGGGGGCCUGCCC (7, 5) | ||
| 2 | 3′Δ18 (1) | 1 | GGACGGGGGGCCUGCCCU (1) | ||
| GGACGGGG (1) | |||||
| WT+8b | [GGACGGGGGGCCUGCCCggguaccg] | 2 | GGACGGGGGGCCUGCCCG (1) | ||
| 1, 2, 3 | GGACGGGGGGCCUGCCC (6, 11, 4) | GGACGGGGGGCCUUU (1) | |||
| 1 & 3 | GGACGGGG (1, 2) | ||||
| 1 | GGACGGGGGGCCUU (1) | CΔ4+8b | [GGACGGGGGGCCUggguaccg] | ||
| 3 | GGACGGGGGGCCUUU (1) | 1 & 2 | GGACGGGGGGCCUGCCC (4, 3) | ||
| GGACGGGGGGCCUGC (1) | GGACGGGGGGCCUGCCCU (2, 1) | ||||
| ∗GGACGGGGGGCCUggg (1, 3) | |||||
| CΔ3 | [GGACGGGGGGCCUG] | 1 | GACGGGG (1) | ||
| 1, 2, 3 | GGACGGGGGGCCUGCCC (10, 4, 4) | 2 | GGACGGGGGGCCUUU (1) | ||
| 1 | GGACGGGGGGCCUGCCCU (1) | TCV/satC recombinants: | |||
| 3 | GGACGGGGGGCCU (1) | 2 | GCGCGGGGGGGGGGCCUGCCCG (1) | ||
| CΔ3+8b | [GGACGGGGGGCCUGggguaccg] | ||||
| 1, 2, 3 | GGACGGGGGGCCUGCCC (1, 1, 1) | CΔ5 | [GGACGGGGGGCC] | ||
| 2 | GGACGGGGGGCCUG (1) | 1 & 2 | GGACGGGGGGCCGGCCC (3, 9) | ||
| GGACGGG (1) | 1 | GGACGGGGGGCCCGCCC (1) | |||
| 3 | GGACGGGGGGCCCUGCCC (1) | ∗GGACGGGGGGCGGGCCC (1) | |||
| ∗GGACGGGGGGCCCAGCCC (1) | ∗GGACGGGGGGCCGUUU (1) | ||||
| GGACGGGG (3) | GGACGGGG (2) | ||||
| TCV/satC recombinants: | GGACGG (1) | ||||
| 1 & 2 | GCGCGGGGGGGGCCUGCCCG (1, 1) | 2 | GGACGGGGGGCGGCCC (1) | ||
| 2 & 3 | GCGCGGGGGGGGGGCCUGCCCG (3, 1) | ||||
| GCGCGGGGGGGGCCUGCCC (4, 4) | CΔ5+8b | [GGACGGGGGGCCggguaccg] | |||
| 1 | GCGCGGGGGGGGGGCCUGCCC (6) | 1 & 2 | GGACGGGGGGCCUGCCC (4, 3) | ||
| GCGCGGGGGGGGGCCUGCCCG (1) | 1 | GGACGGGGGGCCUGGG (4) | |||
| GCGCGGGGGGGGGCCUGCCC (1) | 2 | GGACGGGGGGCCUGCCCU (1) | |||
| GCGGGGGGGGGCCUGCCC (1) | GGACGGGGGGCCUG (2) | ||||
| GCGCGG (1) | GGACGGG (1) | ||||
| 2 | GCGCGGGGGGGGCCUGCCCC (1) | TCV/satC recombinants: | |||
| 3 | GCGCGGGGGGGGGGCCUGCCC (1) | 1 & 2 | GCGCGGGGGGGGGGCCUGCCCG (1, 1) | ||
| GCGCGGGGGGGGCCUGCUU (1) | 2 | GCGCGGGGGGGGCCUGCCC (1) |
Sequences from position 340 to the 3′ end of satC transcripts used for inoculations are enclosed in brackets. Sequences identical to all or portions of the 3′ terminal motif (CCUGCCC-3′) are in bold. Lowercase letters represent plasmid-derived bases. Bases in italics denote the repaired 3′ end sequences of progeny satC-like species. Number of clones of each sequence recovered from individual plants is in parentheses. All clones terminated with poly(A) added during cloning, so 3′ end nontemplate additions of adenylates are not discernible. ∗ denotes sequences reinoculated to plants (progeny described in Table 2). WT, wild-type satC.
The transcripts used to inoculate plants were generated by using T7 RNA polymerase, which is known to add nontemplate bases to the 3′ ends of nascent transcripts (28). To rule out this origin of the nontemplate sequences at the 3′ termini of some recovered satC species, constructs were generated that terminated with eight plasmid-derived bases, which would precede any bases added by T7 polymerase.
Plants inoculated with transcripts of satC containing eight plasmid-derived bases at the 3′ end (wt + 8b) accumulated mainly wt satC (21 of 27 species; Table 1). None of the species retained the plasmid-derived bases. When plants were inoculated with transcripts of satC with a 3-base 3′ terminal deletion followed by eight plasmid-derived bases (CΔ3 + 8b), only three of 37 species cloned recovered the wt 3′ end motif (Table 1). Most of the cloned species (27 of 37) had sequences consistent with recombination between satC and TCV 3′ and thus contained a repaired 3′ end derived from TCV. The TCV-derived sequence was either wt or had one or more bases deleted in a region of 10 consecutive guanylates that compose the stem of the adjacent hairpin. Such alterations after recombination probably occurred because of the reduced viability of satC with an extended hairpin at this location [data not shown; satC has only six consecutive guanylates at this position that form the stem of the (−)-strand hairpin promoter].
Although site-specific, nonhomologous recombination between the satRNAs and TCV has been reported (24, 29), this is an example of homologous recombination associated with TCV. Twenty six of 27 recombinants contained a satC-specific base at position 249 and a TCV-specific base at position 268; the crossover sites were therefore between positions 249 and 268 of satC and positions 3943–3962 of TCV (92–111 bases from the TCV 3′ end). One recombinant species contained a satC cytidylate instead of a TCV uridylate at position 268, suggesting that this species was generated by recombination downstream of position 268 in satC. No base deletions, insertions, or alterations were found in the crossover regions, indicating that the recombination event was precise. Studies using Brome mosaic virus, a tripartite RNA virus, indicate that deletions or mutations in the 3′ end noncoding regions also can be repaired by homologous recombination between the genomic RNAs (30–32). Sindbis virus RNA lacking 3′ end sequence also can be corrected by homologous recombination with a defective RNA containing the intact 3′ end (33). In addition, other RNA viruses such as poliovirus, mouse hepatitis virus, and Qβ bacteriophage can repair their genomes through homologous recombination (reviewed in refs. 34 and 35). It also should be noted that homologous recombination within the 3′ terminal 10 bases of TCV and the satC constructs used in this study could generate the wt satC derived from some of the constructs.
Plants inoculated with satC transcripts containing 4- or 5-base terminal deletions and eight plasmid-derived bases (CΔ4 + 8b and CΔ5 + 8b) accumulated few detected recombinants (four of 35 clones) (Table 1). The majority of the recovered species (21 of 35) were similar to those found in plants inoculated with CΔ4 or CΔ5 transcripts and were either wt (14 clones) or contained deletions or new nontemplate residues at their 3′ ends. Eight of the non-wt clones terminated with the novel motif CCUGGG-3′, with the guanylates possibly originating from the plasmid-derived sequence.
The finding of a large number of satC species in uninoculated leaves with different 3′ terminal sequences suggests that sequences other than the wt CCUGCCC-3′ can be used by the RdRp for replication and that satC containing such repaired 3′ sequence may be a starting point for further evolution toward the more robust wt sequence. To test this possibility, satC transcripts with four unusual 3′ terminal sequences (see * sequences in Table 1) were inoculated individually onto plants and progeny sequenced from uninoculated leaves 2 weeks later. As shown in Table 2, satC transcripts terminating in CCCAGCCC-3′ and CGGGCCC-3′ were infectious and progeny were isolated from uninoculated leaves that contained this sequence. SatC containing wt 3′ end sequences and sequences intermediate between these original and wt sequences also were recovered. SatC terminating with CCUGGG-3′ or CCGUUU-3′ were also infectious, although the original sequence was not recovered from uninoculated leaves. However, plants did contain satC with a variety of wt and close to wt 3′ terminal sequences. These results indicate that mechanisms for repairing satC 3′ terminal alterations do not need to precisely replace the deleted sequence, but rather there are a number of sequences that will permit sufficient replication to allow for subsequent modification by the error-prone RdRp.
Table 2.
3′ end sequences in satC progeny generated in vivo from transcripts with repaired, non-wt 3′ ends
| Construct | Plant | 3′ end sequence |
|---|---|---|
| 1 | [GGACGGGGGGCCCAGCCC] | |
| 1, 2, 4 | GGACGGGGGGCCAGCCC (4, 5, 6) | |
| 1 | GGACGGGGAGCCAGCCC (1) | |
| 2 | GGACGGGGGGCCCAGCCC (1) | |
| GGACGGGGGGCCAGCCU (1) | ||
| 4 | GGACGGGGGGCCUGCCC (1) | |
| 2 | [GGACGGGGGGCCUGGG] | |
| 3, 4, 5 | GGACGGGGGGCCUGCCC (2, 7, 7) | |
| 3 | GGACGGGGGGCCUGCCCG (1) | |
| 3 | [GGACGGGGGGCGGGCCC] | |
| 1, 2, 3 | GGACGGGGGGCCUGCCC (1, 5, 4) | |
| 1 & 2 | GGACGGGGGGCGUGCCC (2, 2) | |
| 1 & 3 | GGACGGGGGGCCGGCCC (1, 1) | |
| 1 | GGACGGGGGGCGGGCCC (1) | |
| GGACGGGGGGCGUGCC (1) | ||
| GGACGGGGGGCCUGCCCC (1) | ||
| 4 | [GGACGGGGGGCCGUUU] | |
| 2 & 5 | GGACGGGGGGCCUGCCC (4, 7) | |
| 2 | GGACGGGGGGGGGCCUGCCC (2) | |
| 4 | GGACGGGGGGCCCGCCC (4) | |
| GGACGGGGGGCCCGCCCUUUUUU (1) | ||
| 5 | GGACGGGGGGCCUGCC (1) |
Sequences recovered from plants inoculated with satC transcripts containing the 3′ end sequences shown in brackets. Progeny identical to the transcripts are underlined. See legend to Table 1 for further details.
In Vitro Synthesis of SatC by the TCV RdRp Using Templates Containing 3′ End Alterations.
The in vivo data suggest that two mechanisms exist for repairing deletions at the 3′ end of satC transcripts in addition to the previously identified repair process involving extension on primers produced by abortive initiation. These mechanisms are: (i) homologous recombination, used mainly for repairing CΔ3 + 8b transcripts; and (ii) imprecise replacement of deleted bases along with the removal (if required) of plasmid-derived bases. Assuming the involvement of the TCV RdRp in the repair process, the mechanism for replacement of the deleted bases could involve (i) RdRp-mediated polymerization of nontemplate nucleotides forming a primer used for initiation of transcription on the inoculated (+)-strand transcripts or (ii) the replaced bases could be added to the 3′ ends of nascent (+)-strands by the RdRp before terminating (+)-strand synthesis. Removal of plasmid-derived bases could occur if the RdRp recognizes satellite sequences in an internal location despite the absence of the natural 3′ terminal nucleotides. Alternatively, the RdRp may terminate nascent (+)-strand synthesis prematurely, thereby removing the plasmid-derived bases during future rounds of replication. Regardless of the repair mechanism for replacement of the deleted bases and/or removal of plasmid-derived bases, the in vivo data suggest that CΔ3 + 8b and CΔ4 + 8b/CΔ5 + 8b have different abilities to be repaired in such a fashion.
Extracts prepared from infected turnip plants that contain partially purified TCV RdRp are able to synthesize the complementary strand of added satC (+)- or (−)-strand templates (36). In vitro transcription of satC (+)-strand template with eight or 18 plasmid-derived bases at the 3′ end generated wt-length products (Fig. 1A). Similar results were obtained previously when up to 220 nonviral bases were added to the 3′ end of satC (+)-strands (37). Although efficiency of transcription was reduced when nonsatellite bases were present at the 3′ end, these results suggest that the TCV RdRp is able to initiate RNA synthesis at or near the wt 3′ end of satC when located internally in the template. Brome mosaic virus RdRp extracts also were able to initiate transcription when the natural 3′ end promoter was located in an internal position (38).
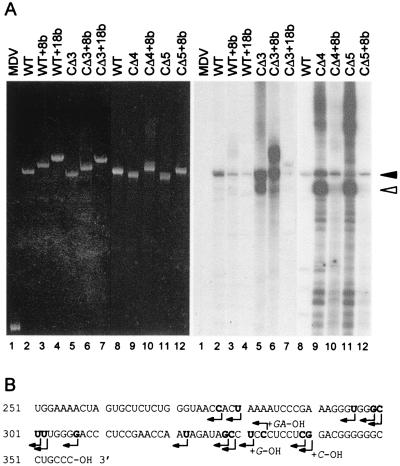
Figure 1.
Transcription of satC templates with or without 3′ end alterations using partially purified TCV RdRp. (A) The templates indicated above each lane were subjected to transcription in vitro using TCV RdRp extracts prepared from infected turnip plants. After denaturing gel electrophoresis, the gel was stained with ethidium bromide to visualize the template RNAs (Left) and subjected to autoradiography to visualize the radiolabeled products (Right). Solid arrow denotes the full-length wt product. The major faster-migrating species is denoted with an open arrow. MDV, 220-base Qβ bacteriophage midivarient RNA; WT, wild-type satC; CΔ3, CΔ4, CΔ5, satC with deletions of three, four, or five bases, respectively. Templates with additional eight or 18 plasmid-derived sequences are denoted by +8b or +18b. (B) Transcription initiation sites (denoted by arrows) of the major products synthesized from templates CΔ3, CΔ4, and CΔ5. Major products (indicated by arrows in A) were gel-purified and cloned before sequence determination (satC-sized product results are shown in Table 3). Nontemplate bases are shown in the (+)-sense orientation to the right of some initiation sites. Initiation sites to the left of adenylates are approximate because the cloning procedure does not allow for the detection of initiation sites at adenylates.
Transcription of CΔ3 and CΔ3 + 8b templates also generated species that comigrate with the wt-length product (Fig. 1A). Surprisingly, CΔ3 and CΔ3 + 8b were substantially more efficient templates than wt satC. This result was unexpected because all TCV-associated (+)- and (−)-strand RNAs, including the two subgenomic RNAs, have two or three template cytidylate residues at their transcription initiation sites (37) that are absent in CΔ3 and CΔ3 + 8b. In addition, transcription of CΔ3 and CΔ3 + 8b generated substantial amounts of products migrating faster or slower than the wt complementary strand position. High levels of similarly sized products also were obtained by using CΔ4 and CΔ5 as templates but not templates with eight plasmid-derived bases (CΔ4 + 8b and CΔ5 + 8b). It is possible that such increases in transcriptional efficiency of the templates are the result of alterations in satC structure caused by the deletion of terminal nucleotides. For example, the loss of the terminal cytidylates may result in the release of the 3′ terminus from an internal base-paired configuration.
Fifteen of 18 cloned products generated by using wt satC template contained either the wt 3′ end sequence, or the wt sequence with one or two additional cytidylates at the 3′ end [for ease in comparing the additional residues at the 5′ end of the (−)-strand products synthesized in vitro to the results obtained in vivo (Table 1), all additions are given in their (+)-strand configuration. The bases at the 5′ end of the (−)-strand products are guanylates] (Table 3). All clones of products synthesized by using WT + 8b were missing the nonsatellite bases. These results indicate that the TCV RdRp is able to initiate RNA synthesis internally at the wt 3′ end position of satC to generate full-length (−)-strands.
Table 3.
Addition of nontemplated bases by TCV RdRp in vitro
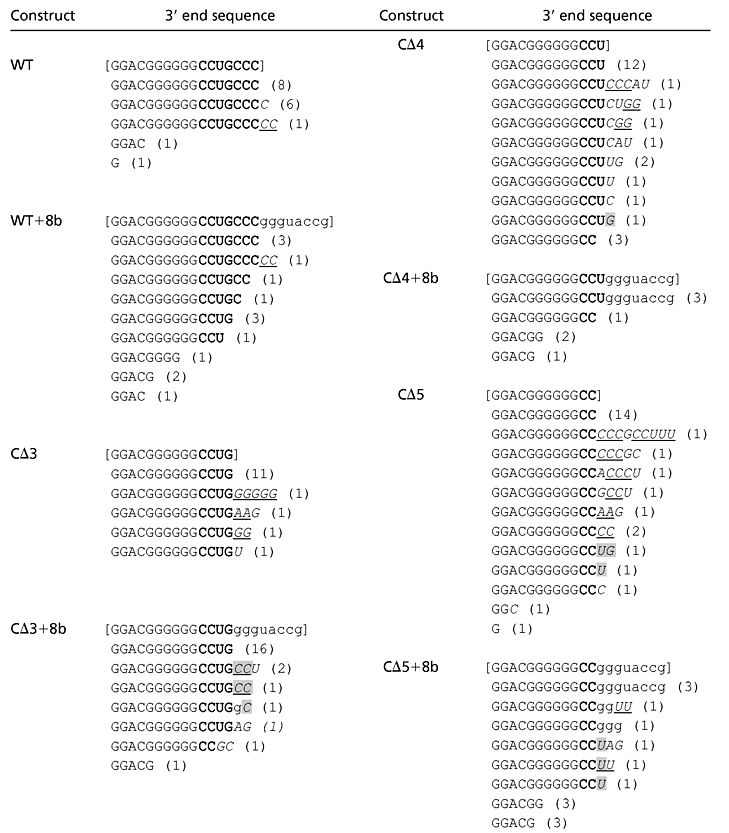 |
SatC templates (enclosed in brackets), either wt or containing 3′ end alterations, were subjected to in vitro transcription using partially purified TCV RdRp (see Fig. 1) followed by cloning of the complementary strand products. The corresponding (+)-sense orientation of the cloned products is shown. Nontemplated bases are in italics. Consecutive identical nontemplate bases are underlined. Shaded bases are identical to the wt sequence. See Table 1 for other details.
Eleven of 15 clones derived from gel-purified, wt-sized products using CΔ3 template contained the CΔ3 3′ end (Table 3). The remaining clones had 1–5 nontemplate bases preceding sequence complementary to CΔ3, indicating that polymerization of nontemplate bases occurred before initiation of transcription opposite the 3′ end of CΔ3 (Table 3). All products transcribed using CΔ3 + 8b were missing the plasmid-derived bases. Transcription initiated at the 3′-most satC residue in 88% (21 of 23) of these clones or had two to three nontemplate bases added before initiation at this position. These results indicate that the TCV RdRp was able to distinguish between satellite and nonsatellite bases and initiate transcription opposite the 3′-most satellite base using this template. Three of the clones had recovered near wt 3′ sequences (CCUGCC-3′ and CCUGCCU-3′). In contrast, when satC (−)-strands containing 5′ end deletions were used as template, the RdRp synthesized no detectable wt-length (+)-strand products in vitro (data not shown), suggesting repair does not occur after (+)-strand synthesis. The incorporation of nontemplate bases at the 5′ ends of nascent (−)-strands is likely an effective 3′ end repair mechanism of viral RNAs with short truncations. For simplicity, we will assume that the nontemplate nucleotide additions are polymerized by the TCV RdRp. However, we cannot rule out that an unidentified host enzyme in the transcription extract produces primers that are elongated by the RdRp.
The ability of the RdRp to initiate transcription of CΔ3 + 8b in the absence of a primer would reduce opportunities for correcting truncations by using nontemplate nucleotides to replace the deleted cytidylates at the 3′ end. Replacement of the deleted bases in CΔ3 + 8b therefore mainly would require a second mechanism, such as homologous recombination. In contrast, none of the clones recovered by using CΔ4 + 8b and CΔ5 + 8b templates initiated with the terminal satellite nucleotide (Table 3) and eight of 21 clones contained some or all of the plasmid-derived bases. CΔ3 + 8b and CΔ4 + 8b only differ by the presence of an additional guanylate in CΔ3 + 8b (four consecutive guanylates in CΔ3 + 8 from satellite and plasmid-derived sequence and three consecutive guanylates in CΔ4 + 8, all from the plasmid-derived sequence). However, the activity of these templates is very different in the transcription reactions (compare lanes 6 and 10 in Fig. 1A), suggesting that the two templates contain dissimilar promoter features. Homologous recombination may occur at a lower frequency than primer-mediated repair, and thus would be a less-used repair mechanism for CΔ4 + 8b and CΔ5 + 8b.
The composition of the RdRp-generated nontemplate primers was not random. Twenty of 27 clones with two or more nontemplate bases contained multiple consecutive nucleotides (underlined in Table 3), the most common being GG1–2 (12 clones, corresponding to the underlined CC1–2 in Table 3). The preference for synthesis of multiple guanylates at the 5′ terminus of the (−)-strand product likely provides an efficient mechanism for repairing deletion of terminal cytidylates from the (+)-strand 3′ end.
Only 14 of the 41 clones with nontemplate additions generated in vitro contained nonsatellite bases that were identical in sequence to the terminal nucleotide of their satellite-specific sequence, suggesting that a polymerase slippage mechanism is not responsible for all added sequences. Such a mechanism has been proposed to explain the 5′ end heterogeneity of RNA transcripts produced by the DNA-dependent RNA polymerases of E. coli and T7 bacteriophage (39–41). However, the templates in those studies contained homooligomeric nucleotides not found on the satC templates truncated by 3 or 4 bases. Helm et al. (42) showed that T7 RNA polymerase also can add nontemplate bases when initiating transcription from templates without homooligomeric nucleotides. Template-free RNA also is reported to have been synthesized by the RdRp of Qβ bacteriophage (43), although this result is still controversial (44, 45), and the poliovirus RdRp contains terminal adenylyl transferase activity (46). These findings suggest that polymerization of nucleotides independent of the template may be an intrinsic property of RdRp.
Because the TCV RdRp is able to efficiently initiate transcription of satC templates missing the three terminal cytidylates in vitro, what function might these cytidylates have in replication in vivo? Sequence analysis of the major species produced in vitro from templates CΔ3, CΔ4, and CΔ5, which migrated slightly faster than wt-sized RNA (indicated by the open arrow in Fig. 1A), revealed that initiation sites were clustered at positions 296–307 and 322–340 (Fig. 1B). These results also indicated that the RdRp initiated transcription opposite cytidylate, uridylate, guanylate, and possibly adenylate residues without exhibiting an obvious preference (the cloning procedure obviates the detection of terminal adenylates). Such flexibility has not been reported for other viral RdRp, which usually initiate transcription with a specific nucleotide residue (47, 48). The substantial amount of incorrectly initiated transcription by the TCV RdRp using CΔ3, CΔ4, and CΔ5 templates suggests that the three terminal cytidylates assist this RdRp in correctly initiating transcription opposite a 3′ terminal nucleotide.
The question also arises why different repair mechanisms exist for satC deletions of 3–5 bases compared with 6 bases. Previous results with satD also indicated that there are different mechanisms for repair of 5- versus 6-base deletions (20). Because the uridylate in the terminal motif (CCUGCCC-3′) of satC can be substituted with any of the other three nucleotides and still permit replication in vivo (ref. 15; Table 1), repair of 4- and 5-base deletions by synthesis of nonrandom primers may occur with similar probabilities. Repair of a 6-base deletion to a sequence that is amenable to further rounds of replication by using primers synthesized independently of a template must occur at a lower frequency than extension of primers derived from abortive cycling. The ability of the TCV RdRp to repair 3′ end deletions of varying lengths by different mechanisms, combined with reports of 3′ end deletion repair in unrelated viruses, suggests that some RdRp have needed to acquire several mechanisms to cope with a hostile cellular environment.
Acknowledgments
This work was supported by National Science Foundation Grants MCB-9728277 and MCB-9630191 to A.E.S.
Abbreviations
- TCV
turnip crinkle virus
- satC
satellite RNA C
- wt
wild type
- RdRp
RNA-dependent RNA polymerase
References
- 1.Green P J. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:421–445. [Google Scholar]
- 2.Irie M. Pharmacol Ther. 1999;81:77–89. doi: 10.1016/s0163-7258(98)00035-7. [DOI] [PubMed] [Google Scholar]
- 3.Egesten A, Dyer K D, Batten D, Domachowske J B, Rosenberg H F. Biochim Biophys Acta. 1997;1358:255–260. doi: 10.1016/s0167-4889(97)00081-5. [DOI] [PubMed] [Google Scholar]
- 4.Garnier M, Candresse T, Bove J M. Virology. 1986;151:100–109. doi: 10.1016/0042-6822(86)90107-8. [DOI] [PubMed] [Google Scholar]
- 5.Bienz K, Egger D, Pfister T, Troxler M. J Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackenzie J M, Khromykh A A, Jones M K, Westaway E G. Virology. 1998;245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 7.Neufeld K L, Galarza J M, Richards O C, Summers D F, Ehrenfeld E. J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skuzeski J M, Bozarth C S, Dreher T W. J Virol. 1996;70:2107–2115. doi: 10.1128/jvi.70.4.2107-2115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalmay T, Russo M, Burgyan J. Virology. 1993;192:551–555. doi: 10.1006/viro.1993.1071. [DOI] [PubMed] [Google Scholar]
- 10.Dalmay T, Rubino L. Virology. 1995;206:1092–1098. doi: 10.1006/viro.1995.1032. [DOI] [PubMed] [Google Scholar]
- 11.Burgyan J, Garcia-Arenal F. J Virol. 1998;72:5061–5066. doi: 10.1128/jvi.72.6.5061-5066.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill K R, Hajjou M, Hu J Y, Raju R. J Virol. 1997;71:2693–2704. doi: 10.1128/jvi.71.4.2693-2704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raju R, Hajjou M, Hill K R, Botta V, Botta S. J Virol. 1999;73:2410–2419. doi: 10.1128/jvi.73.3.2410-2419.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tacahashi Y, Uyeda I. Virology. 1999;265:147–152. doi: 10.1006/viro.1999.0027. [DOI] [PubMed] [Google Scholar]
- 15.Nagy P D, Carpenter C D, Simon A E. Proc Natl Acad Sci USA. 1997;94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao A L, Dreher T W, Marsh L E, Hall T C. Proc Natl Acad Sci USA. 1989;86:5335–5339. doi: 10.1073/pnas.86.14.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richter J D. Microbiol Mol Biol Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrington J C, Heaton L A, Zuidema D, Hillman B I, Morris T J. Virology. 1989;170:219–226. doi: 10.1016/0042-6822(89)90369-3. [DOI] [PubMed] [Google Scholar]
- 19.Oh J W, Kong Q, Song C, Carpenter C D, Simon A E. Mol Plant-Microbe Interact. 1995;8:979–987. doi: 10.1094/mpmi-8-0979. [DOI] [PubMed] [Google Scholar]
- 20.Carpenter C D, Simon A E. Virology. 1996;226:153–160. doi: 10.1006/viro.1996.0641. [DOI] [PubMed] [Google Scholar]
- 21.Carpenter C D, Simon A E. J Virol. 1996;70:478–486. doi: 10.1128/jvi.70.1.478-486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon A E, Howell S H. EMBO J. 1986;5:3423–3428. doi: 10.1002/j.1460-2075.1986.tb04664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song C, Simon A E. J Mol Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- 24.Carpenter C D, Oh J W, Zhang C, Simon A E. J Mol Biol. 1995;245:608–622. doi: 10.1006/jmbi.1994.0050. [DOI] [PubMed] [Google Scholar]
- 25.Li X H, Heaton L A, Morris T J, Simon A E. Proc Natl Acad Sci USA. 1989;86:9173–9177. doi: 10.1073/pnas.86.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan H, Song C, Simon A E. RNA. 1997;3:1401–1412. [PMC free article] [PubMed] [Google Scholar]
- 27.Guan H, Carpenter C D, Simon A E. Virology. 2000;268:345–354. doi: 10.1006/viro.1999.0153. [DOI] [PubMed] [Google Scholar]
- 28.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C X, Cascone P J, Simon A E. Virology. 1991;184:791–794. doi: 10.1016/0042-6822(91)90454-j. [DOI] [PubMed] [Google Scholar]
- 30.Bujarski J J, Kaesberg P. Nature (London) 1986;321:528–531. doi: 10.1038/321528a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao A L, Hall T C. J Virol. 1993;67:969–979. doi: 10.1128/jvi.67.2.969-979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagy P D, Bujarski J J. J Virol. 1995;69:131–140. doi: 10.1128/jvi.69.1.131-140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raju R, Subramaniam S V, Hajjou M. J Virol. 1995;69:7391–7401. doi: 10.1128/jvi.69.12.7391-7401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agol V I. Semin Virol. 1997;8:77–84. [Google Scholar]
- 35.Worobey M, Holmes E C. J Gen Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- 36.Song C, Simon A E. Proc Natl Acad Sci USA. 1994;91:8792–8796. doi: 10.1073/pnas.91.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon A E. Curr Top Microbiol Immunol. 1999;239:19–36. doi: 10.1007/978-3-662-09796-0_2. [DOI] [PubMed] [Google Scholar]
- 38.Miller W A, Bujarski J J, Dreher T W, Hall T C. J Mol Biol. 1986;187:537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- 39.Guo H C, Roberts J W. Biochemistry. 1990;29:10702–10709. doi: 10.1021/bi00499a019. [DOI] [PubMed] [Google Scholar]
- 40.Xiong X F, Reznikoff W S. J Mol Biol. 1993;231:569–580. doi: 10.1006/jmbi.1993.1310. [DOI] [PubMed] [Google Scholar]
- 41.Pleiss J A, Derrick M L, Uhlenbeck O C. RNA. 1998;4:1313–1317. doi: 10.1017/s135583829800106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helm M, Brule H, Giege R, Florentz C. RNA. 1999;5:618–621. doi: 10.1017/s1355838299982328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biebricher C K, Eigen M, Luce R. Nature (London) 1986;321:89–91. doi: 10.1038/321089a0. [DOI] [PubMed] [Google Scholar]
- 44.Hill D, Blumenthal T. Nature (London) 1983;301:350–352. doi: 10.1038/301350a0. [DOI] [PubMed] [Google Scholar]
- 45.Chetrerin A B, Chetverina H V, Munishkin A V. J Mol Biol. 1991;222:3–9. doi: 10.1016/0022-2836(91)90729-p. [DOI] [PubMed] [Google Scholar]
- 46.Neufeld K L, Galarza J M, Richards O C, Summers D F, Ehrenfeld E. J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh R N, Dreher T W. RNA. 1998;4:1083–1095. doi: 10.1017/s1355838298980694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sivakumaran K, Kao C C. J Virol. 1999;73:6415–6423. doi: 10.1128/jvi.73.8.6415-6423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]