Abstract
Ansamycins such as rifamycin, ansamitocin, and geldanamycin are an important class of polyketide natural products. Their biosynthetic pathways are especially complex because they involve the formation of 3-amino-5-hydroxybenzoic acid (AHBA) followed by backbone assembly by a hybrid nonribosomal peptide synthetase/polyketide synthase. We have reconstituted the ability to synthesize 2,6-dimethyl-3,5,7-trihydroxy-7-(3′-amino-5′-hydroxyphenyl)-2,4-heptadienoic acid (P8/1-OG), an intermediate in rifamycin biosynthesis, in an extensively manipulated strain of Escherichia coli. The parent strain, BAP1, contains the sfp phosphopantetheinyl transferase gene from Bacillus subtilis, which posttranslationally modifies polyketide synthase and nonribosomal peptide synthetase modules. AHBA biosynthesis in this host required introduction of seven genes from Amycolatopsis mediterranei, which produces rifamycin, and Actinosynnema pretiosum, which produces ansamitocin. Because the four-module RifA protein (530 kDa) from the rifamycin synthetase could not be efficiently produced in an intact form in E. coli, it was genetically split into two bimodular proteins separated by matched linker pairs to facilitate efficient inter-polypeptide transfer of a biosynthetic intermediate. A derivative of BAP1 was engineered that harbors the AHBA biosynthetic operon, the bicistronic RifA construct and the pccB and accA1 genes from Streptomyces coelicolor, which enable methylmalonyl-CoA biosynthesis. Fermentation of this strain of E. coli yielded P8/1-OG, an N-acetyl P8/1-OG analog, and AHBA. In addition to providing a fundamentally new route to shikimate and ansamycin-type compounds, this result enables further genetic manipulation of AHBA-derived polyketide natural products with unprecedented power.
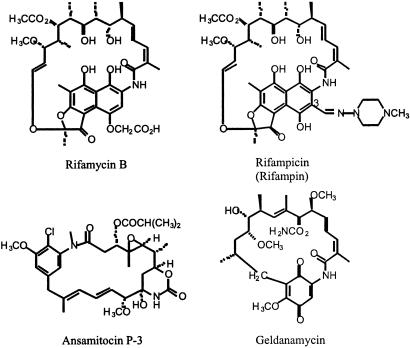
The ansamycin antibiotics (1–4) include important antibacterial agents, such as rifamycin, and anticancer agents, such as ansamitocin (5) and geldanamycin (Fig. 1). They are produced by various actinomycetes. Several chemically modified rifamycins such as rifampicin have been used since the 1960s as the first line of defense for the treatment of tuberculosis and other mycobacterial infections. As in the case of many antibiotics, the emergence of resistant pathogens has made the development of modified rifamycins an important task. Although numerous rifamycin derivatives have been synthesized from the natural products, most of the structural variations are through the modification at the C-3 position of the naphthoquinone chromophore (Fig. 1). The structural complexity of rifamycin and other ansamycins has made biosynthetic manipulation an attractive route for the generation of pharmacologically useful analogs.
Fig. 1.
Structures of natural (rifamycin B, ansamitocin P-3, and geldanamycin) and semisynthetic (rifampicin) ansamycin antibiotics.
All ansamycin antibiotics are macrocycles composed of a benzoic or naphthalenic chromophore bridged by an aliphatic polyketide chain that terminates at the chromophore with an amide linkage. The aromatic moiety is derived from a 3-amino-5-hydroxybenzoic acid (AHBA) (1) primer unit (6–9) (Fig. 2), which is activated by a nonribosomal peptide synthetase-like mechanism (10) and elaborated via addition of methylmalonyl and malonyl extender units by a multimodular polyketide synthase (11–14). The rifamycin biosynthetic gene cluster (≈90 kb) has been cloned from Amycolatopsis mediterranei S699 (12–14) and contains genes required for AHBA biosynthesis, export and resistance, regulatory genes, and a number of downstream-processing genes, in addition to a modular polyketide synthase that consists of 10 colinearly arranged modules. More recently, similar genetic analysis has also been reported for the ansamitocin gene cluster from Actinosynnema pretiosum (15) and the geldanamycin gene cluster from Streptomyces hygroscopicus (16).
Fig. 2.
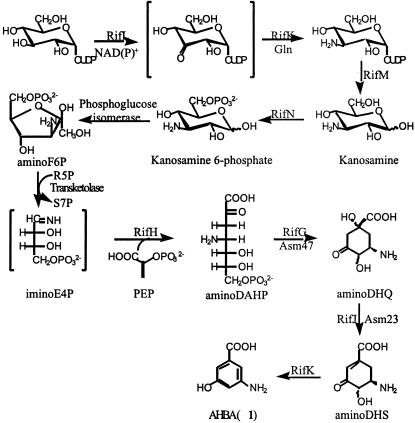
Proposed pathway for AHBA biosynthesis. AminoDHS, 5-amino analog of 3-dehydroshikimic acid; aminoDAHP, 3,4-dideoxy-4-amino-d-arabino-heptulosonic acid 7-phosphate; PEP, phosphoenolpyruvic acid; aminoDHQ, 5-deoxy-5-amino-3-dehydroquinic acid.
Of particular relevance to this study are the AHBA biosynthetic pathway and the unusually long RifA polypeptide (530 kDa). The AHBA pathway parallels the first three steps of the shikimate pathway, but is modified by introduction of glutamine-derived nitrogen at an early stage to give 3,4-dideoxy-4-amino-d-arabino-heptulosonic acid 7-phosphate (aminoDAHP) (17) instead of the normal shikimate pathway intermediate, 3-deoxy-d-arabino-heptulosonic acid 7-phosphate (DAHP). More recently, kanosamine has recently been implicated as the source of the nitrogen atom in AHBA (18). Cyclization and dehydration of 3,4-dideoxy-4-amino-d-arabino-heptulosonic acid 7-phosphate (aminoDAHP) leads to the 5-amino analog of 3-dehydroshikimic acid (aminoDHS), which is aromatized by the enzyme, AHBA synthase (17, 19, 20) to AHBA. Seven genes, rifG, -H, -J, -K, -L, -M, and -N, in the rifamycin gene cluster are believed to be necessary for the biosynthesis of AHBA (12, 21, 22). Homologs of five of these genes, asm47, -23, -44, -45, and -22 (similar to rifG,-J,-L,-M, and -N, respectively), have been found in the ansamitocin gene cluster (15).
RifA is an unusually long polypeptide (530 kDa) responsible for activating AHBA via an nonribosomal peptide synthetase-like mechanism (10), followed by elongation via three polyketide synthase modules. The tetraketide product of the RifA polypeptide, 2,6-dimethyl-3,5,7-trihydroxy-7-(3′-amino-5′-hydroxyphenyl)-2,4-heptadienoic acid (P8/1-OG), has been isolated from spontaneous (23) and engineered (24) mutants of A. mediterranei. As a step toward the goal of programming ansamycin biosynthesis in a genetically tractable heterologous host, we attempted to engineer a strain of Escherichia coli capable of producing both AHBA and P8/1-OG. E. coli was used as the host of choice due to its ease of cultivation, well established molecular biology protocols, and fast doubling times, as well as the recent demonstration that complex polyketides can be efficiently produced in this heterologous host (25, 26). The results of our studies are described here.
Materials and Methods
Chemicals, Strains, and DNA Manipulation. All chemicals were purchased from Sigma–Aldrich. DNA manipulations were performed in E. coli XL1 Blue (Stratagene) by using standard procedures. Restriction enzymes were from New England Biolabs. Polymerase chain reactions were carried out by using Pfu polymerase (Stratagene) as recommended by the manufacturer.
Constructs for AHBA Biosynthetic Genes. Seven genes from the rifamycin gene cluster (12) rifG, -H, -J, -K, -L, -M, and -N, were each cloned into the pET-28b(+) expression vector with a His6 tag at the N terminus. Plasmid pKW241 was a derivative of cosmid pHGF7515 (unpublished construct) and was designed to express rifG, -H, -J, -K, -L, -M, and -N. Plasmid pKW257 was constructed from cosmids pDDc6 and pDHc1 (15) to express asm23 and asm47. Plasmid pKW255 was constructed from pKW241 and pKW257 to express rifH, -K, -L, -M, and -N and asm23 and -47. Each of these expression plasmids were derived from individual ORF cassettes (XbaI–SpeI) that included ribosome-binding sites. These cassettes not only facilitated evaluation of expression of each gene by itself, they were also used to rapidly assemble multigene operons based on the compatibility of XbaI and SpeI sticky ends.
Constructs for Expression of pccB and accA1. A bicistronic cassette containing pccB and accA1 (24, 26) was engineered with flanking XbaI–SpeI restriction enzyme sites, with each ORF containing its own ribosome-binding site. This cassette was ligated into pKW255, yielding pKW256, which contains a complete, functional AHBA biosynthetic operon plus the pccB and accA1 genes.
Constructs for Expression of rifA. A 6.5-kbp NdeI–SpeI-restricted fragment from pSA19 (11) containing the rifLM+rifM1 gene was fused to the 6-deoxyerythronolide B synthase C-terminal linker region of eryM2 (M2C) (28–30) to yield pKW181. Similarly, a 7.9-kbp BsaBI–SpeI-restricted fragment containing rifM2+rifM3 was fused to the 6-deoxyerythronolide B synthase N-terminal linker region of eryM3 (M3N) (28–30) to yield pKW210. This cassette was cloned into pBP144 (25), resulting in a new cassette flanked with NsiI and PacI sites and yielding pKW243. The rifLM+rifM1+M2C cassette was subsequently cloned into pBP130 (25) to yield pKW242, after which the M3N+rifM2+rifM3 cassette was cloned into pKW242 to yield pKW244. Thus, pKW244 encoded the entire rifA gene product as two separate genes expressed with a 6-deoxyerythronolide B synthase-linker pair interface.
Expression of AHBA Biosynthetic Genes. E. coli BL21(DE3) (Novagen) was used as host for evaluating individual expression of AHBA biosynthetic genes. A single transformant was used to start 30 ml of LB medium cultures with kanamycin (50 μg/ml) at 37°C and 250 rpm. The starter culture was used to inoculate 5 liters of LB medium containing the same antibiotic concentration as above. This culture was grown at 250 rpm and 37°C to midlog phase (OD600 = 0.6–0.8), cooled to 4°C for 10 min, and induced with 1 mM isopropyl-β-d-thiogalactopyranoside for an additional 20 h at 13°C. The protein was purified from the cell lysate by nickel-nitrilotriacetic acid affinity chromatography (Qiagen), and analyzed on a 4–20% gradient gel (Bio-Rad) stained with Coomassie brilliant blue stain.
Purification of RifLM+RifM1+M2C and M3N+RifM2+RifM3 Proteins. Expression of RifLM+RifM1+M2C and M3N+RifM2+RifM3 proteins was achieved by using a similar procedure as described for AHBA biosynthetic gene expression above, with the exception that cell cultures were induced with 100 μM isopropyl-β-d-thiogalactopyranoside. Cell pellets were then harvested by centrifugation at 2,500 × g and resuspended in disruption buffer (200 mM sodium phosphate, pH 7.2/200 mM sodium chloride/0.2 mM DTT/1.5 mM benzamine/2 mg/liter pepstatin/2 mg/liter leupeptin/30% vol glycerol). All purification procedures were performed at 4°C or on ice. Resuspended cells were disrupted by sonication, and the lysate was clarified by centrifugation at 40,000 × g. The supernatant was brought to 40% (wt/vol) saturation with ammonium sulfate and precipitated overnight. After centrifugation at 40,000 × g, the protein pellet was redissolved in 100 mM sodium phosphate (pH 7.2) and 10% vol glycerol and exchanged into this same buffer by using PD-10 columns (Amersham Pharmacia) to remove the ammonium sulfate. This protein solution was applied onto a nickel-nitrilotriacetic acid column. After washing with 10 mM imidazole in 100 mM sodium phosphate (pH 7.2), the protein was eluted with 100 mM imidazole in the same buffer. Further purification was carried out on an anion-exchange column (HiTrap Q, Amerhsam). A gradient of 0–1 M sodium chloride in 100 mM sodium phosphate (pH 7.2), 2.5 mM DTT, and 10% vol glycerol was run at 3 ml/min for 10 column volumes. Fractions of 3 ml were collected, and those containing the desired protein (typically 0.3 M sodium chloride) were pooled and further concentrated on Amicon Ultra. Purified proteins were analyzed on a 4–20% gradient gel (Bio-Rad) stained with Coomassie brilliant blue stain. In the purification procedure described above, 20 mg/liter RifLM+RifM1+M2C and 5 mg/liter M3N+RifM2+RifM3 proteins was isolated.
Fig. 3.
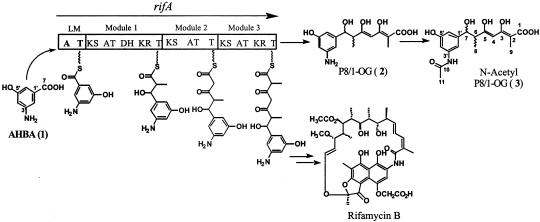
Proposed pathway for P8/1-OG and rifamycin B biosynthesis. A, adenylation domain; T, thiolation domain; KS, ketosynthase; AT, acyltransferase; DH, dehydratase; KR, ketoreductase.
Production and Analysis of AHBA. E. coli BL21(DE3) was used as host for AHBA production. A single transformant was used to start a 60-ml LB medium culture with kanamycin (50 μg/ml) at 37°C and 250 rpm. The starter culture was used to inoculate 10 liters of M9 minimal medium at the same antibiotic concentrations as above. The culture was grown to midlog phase (OD600 = 0.6–0.8), cooled on ice for 10 min, and centrifuged. The cell pellet was resuspended in 1 liter of M9 medium containing kanamycin (50 μg/ml) and 1 mM isopropyl-β-d-thiogalactopyranoside, followed by growth for an additional 48 h at 13°C. The fermentation broth was recovered via centrifugation and lyophilized. The residue was redissolved in a minimum of water, loaded onto a Sep-Pak C18 column, and eluted with methanol. The solvent was evaporated in vacuo, and the residue was purified by preparative thin layer chromatography (20% methanol and 1% acetic acid in chloroform). The isolated compound matched an authentic standard of AHBA by 1H NMR and 13C NMR.
Production and Analysis of P8/1-OG and Its N-Acetyl Analog. E. coli BAP1 (25) was used as host for P8/1-OG (2) production. A single transformant of BAP1/pKW244/pKW256 was used to inoculate 100 ml of M9 minimal medium containing carbenicillin (100 μg/ml) and kanamycin (50 μg/ml). After overnight growth at 37°C and 250 rpm, the starter culture was used to inoculate 2 liters of medium in an aerated 3 liters of Biobundle fermentor (Applikon) operated under fed-batch mode (26). Once the glucose was exhausted from the batch medium, feeding was initiated and the culture was grown to OD600 = 15. At this point the temperature was reduced to 18°C, and isopropyl-β-d-thiogalactopyranoside (200 μM) and sodium propionate (2 g/liter, every 48 h) were added.
After 120 h, the culture was centrifuged and 2 liters of the supernatant was extracted with 400 g of Dowex 2 × 8 (100–200 mesh, Cl–) to absorb acidic compounds. Neutral and alkaline compounds were washed off the column with 1.2 liters of deionized water. P8/1-OG and its N-acetyl analog were eluted off the column with 3 liters of 0.05 M HCl. Fractions containing P8/1-OG and its N-acetyl analog were neutralized with KOH (pH 7.0) and dried under reduced pressure. The residue was then stirred with 1 liter of absolute ethanol for 1 h. The insoluble KOH fraction was separated by filtration and then dried under reduced pressure. The operation was repeated twice with 250 ml of absolute ethanol. The remaining residue was then dissolved in water and polished via C18 RP-HPLC. The structure of P8/1-OG was confirmed by liquid chromatography/mass spectrometry, and the structure of the N-acetyl P8/1-OG analog was confirmed via mass spectrometry and 1H NMR spectroscopy.
Results and Discussion
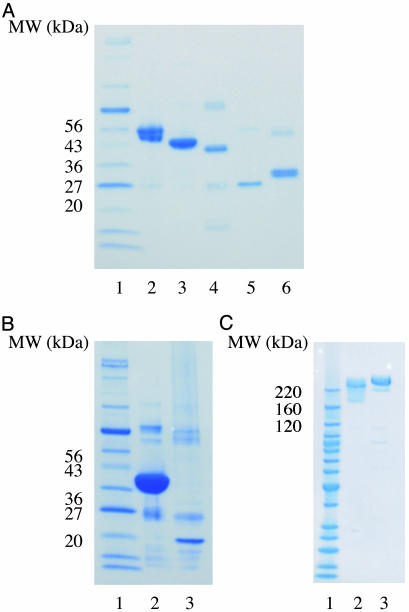
AHBA Biosynthesis. Because E. coli does not produce AHBA, we initiated our attempts toward heterologous ansamycin biosynthesis by assembly and expression of the entire AHBA biosynthetic pathway in E. coli BAP1. In vivo production of AHBA was needed to provide the primer unit for ansamycin polyketide biosynthesis. Initially we attempted expression of rifG, -H, -J, -K, -L, -M, and -N individually. Although five proteins could be expressed (Fig. 4A), two proteins, RifG and RifJ, were poorly expressed. Instead, we attempted to express their homologs from the ansamitocin biosynthetic gene cluster, asm47 and asm23. These proteins could be successfully detected and purified (Fig. 4B), allowing us to compile a multigene operon containing the rifH, rifK, rifL, rifM, rifN, asm47, and asm23 genes (Fig. 5A). The expression levels of the individual AHBA biosynthetic enzymes in this recombinant strain were in the range of 0.1–4 mg/liter culture. Alternatively, endogenous E. coli aroB and aroD could have been overexpressed and used as replacements for rifG and rifJ because it has been demonstrated that AroB and AroD can carry out the same functions as RifG and RifJ (17). AHBA production was confirmed in the resulting host by liquid chromatography/mass spectrometry. Specifically, we detected the parent [M+Na] ion of the expected compound (1) at 176 atomic mass units (amu). 1: Rf = 0.32 (silica gel, 1% acetic acid and 20% methanol in chloroform); 1H NMR (400 MHz, CD3OD) δ = 6.71 (s, 1H, H-2′), 6.66 (s, 1H, H-6′), 6.15 (s, 1H, H-4′); 13C NMR (100 MHz, CD3OD) δ = 176.0 (C7), 158.7 (C5′), 149.2 (C3′), 141.2 (C1′), 109.7 (C2′), 107.7 (C6′), and 105.7 (C4′). In the shake-flask fermentation procedure described above, the isolated yield of AHBA was 3.1 mg/liter. Parenthetically, we note that other as-yet-uncharacterized aromatic metabolites were also detected in the recombinant strain.
Fig. 4.
Purified AHBA biosynthesis and engineered RifA proteins. (A) Lane 1, protein marker; lane 2, RifH (49 kDa); lane 3, RifK (43 kDa); lane 4, RifL (40 kDa); lane 5, RifM (26 kDa); lane 6, RifN (33 kDa). (B) Lane 1, protein marker; lane 2, asm47 (38 kDa); lane 3, asm23 (19 kDa). (C) Lane 1, protein marker; lane 2, RifLM+RifM1+M2C (246 kDa); lane 3, M3N+RifM2+RifM3 (301 kDa).
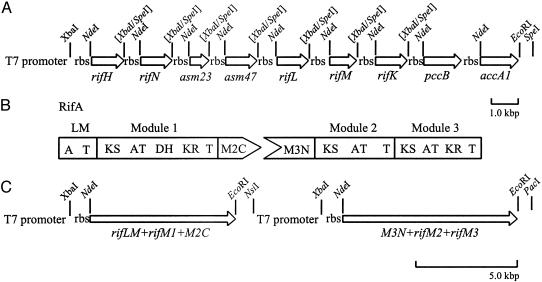
Fig. 5.
(A) Plasmid map of pKW256 for the expression of AHBA biosynthetic proteins. (B) Protein engineering of RifA. (C) Plasmid map of pKW244 for the expression of RifA as two separate proteins. A, adenylation domain; T, thiolation domain; KS, ketosynthase; AT, acyltransferase; DH, dehydratase; KR, ketoreductase.
P8/1-OG Biosynthesis. Early attempts to overexpress rifA as a soluble polypeptide in E. coli were unsuccessful, possibly because of its large size (530 kDa). To overcome this problem, we split RifA into two separate polypeptides (RifLM+RifM1+M2C and M3N+RifM2+RifM3) (Fig. 5B). Both these truncated proteins were shown to express well in E. coli (Fig. 4C); hence the two ORFs, each containing their own ribosome-binding site and T7 promoter, were cloned on the same plasmid (Fig. 5C). A key requirement for association of these engineered polypeptides and efficient inter-polypeptide transfer of the diketide intermediate was the fusion of intermodular linker sequences from the 6-deoxyerythronolide B synthase to the C and N termini of the two proteins (Fig. 5B). Earlier studies have established the modular architecture of these linkers and have shown that they play a crucial role in facilitating selective module–module interactions (28–30). The resulting expression plasmid, pKW244, was cotransformed into E. coli BAP1 with plasmid, pKW256, carrying the AHBA biosynthetic genes and the propionyl-CoA carboxylase genes (25) (Fig. 5A). Production of the expected natural product, P8/1-OG, was confirmed by liquid chromatography/mass spectrometry; however, very little of this product was observed. We detected the parent [M+H-H2O] ions of the expected compound (2) at 292 amu. The N-acetyl P8/1-OG was the dominant product and was therefore structurally characterized via mass spectrometry and 1H NMR. Previous work with A. mediterranei mutants has shown the N-acetyl P8/1-OG product to be the dominant tetraketide (24). We detected the parent [M+Na+H-H2O] ion and [M+K+H-H2O] ion of the expected compound (3) at 357 amu and 373 amu, respectively. The chemical shifts for H-4, H-6, H-7, H-8, and H-9 of the expected compound (3) agree well with previously published 1H NMR spectra for compound 2 (23). 3: 1H NMR (400 MHz, DMSO-d6) δ = 7.09 (s, 1H, H-2′), δ = 6.14 (s, 1H, H-4′), δ = 6.96 (s, 1H, H-6′), δ = 5.98 (s, 1H, H-4), δ = 2.66 (m, J = 2.0 Hz, 1H, H-6), δ = 4.26 (d, J = 2.0 Hz, 1H, H-7), δ = 1.05 (d, J = 6.8 Hz, 3H, C8H3), δ = 1.74s (s, 3H, C9H3), and δ = 2.46 (s, 3H, C11H3); MS (ESI+) calculated for C17H21NO7 [M+Na+H-H2O] 357.3, found 357.4 and C17H21NO7 [M+K+H-H2O] 372.3, found 373.0.
In the fed-batch fermentation procedure described above, the approximate isolated yield of N-acetyl P8/1-OG was 2.5 mg/liter. However, because a considerable amount of the polyketide product was not recovered by the purification procedure, the actual product titer in the fermentation broth was probably higher. In this context it is noteworthy that P8/1-OG hydrolyzes from the acyl carrier protein domain in module 3 spontaneously even in the absence of an appropriate hydrolase. This is consistent with the observation that inactivation of the rifF encoded amide synthase in the rifamycin pathway results in spontaneous release of an assortment of acyl carrier protein-bound biosynthetic intermediates (24).
Conclusions
A hybrid AHBA pathway was assembled in E. coli BAP1 from five rifamycin genes and two ansamitocin genes. In addition to setting the stage for further analysis of this interesting pathway, installation of the AHBA biosynthetic pathway in E. coli provides a natural source of this primer unit for the biosynthesis of a variety of ansamycins as well as amine-substituted shikimate derivatives in E. coli. The significance of one-pot biosyntheses of the former class of compounds is evident from their diverse pharmacological properties (1–4); the latter family of compounds has considerable potential as specialty chemicals (31, 32). As a step in this direction, the rifA gene from A. mediterranei was expressed in the same host. High level functional expression of this 14-domain protein was facilitated by expression as two bimodular proteins, each comprising two modules. The use of matched linker pairs to facilitate inter-polypeptide chain transfer illustrates yet another use of such linkers, namely for the expression of large polyketide synthase proteins. Such large proteins are also encountered in the biosynthetic pathways of other important natural products including rapamycin (33) and epothilone (34). By reconstituting P8/1-OG biosynthesis in E. coli, it is now possible to contemplate reconstituting the entire rifamycin biosynthetic pathway in this genetically friendly host, thereby opening the door for facile generation of new rifamycin analogues.
Acknowledgments
We thank Dr. Heinz G. Floss for providing cosmid vectors of pHGF7515, pDDc6, and pDHc1, and Dr. Allis Chien at the Stanford Mass Spectrometry Facility for providing help with mass spectrometric analysis of the products. This work was supported by National Institutes of Health Grants AI 77248 (to C.K.) and GM20011 (to C.T.W.). M.A.R. is a recipient of a Stanford graduate fellowship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AHBA, 3-amino-5-hydroxybenzoic acid; P8/1-OG, 2,6-dimethyl-3,5,7-trihydroxy-7-(3′-amino-5′-hydroxyphenyl)-2,4-heptadienoic acid; LM, nonribosomal peptide synthetase-like loading module; amu, atomic mass units; M3N, 6-deoxyerythronolide B synthase N-terminal linker region of eryM3; M2C, 6-deoxyerythronolide B synthase C-terminal linker region of eryM2.
References
- 1.Anderson, M. G., Kibby, J. J., Rickards, R. W. & Rothchild, J. M. (1980) J. Chem. Soc. Chem. Commun., 1277–1278.
- 2.Ghisalba, O. & Nuesch, J. (1981) J. Antibiot. 34, 64–71. [DOI] [PubMed] [Google Scholar]
- 3.Becker, A. M., Herlt, A. J., Hilton, G. L., Kibby, J. J. & Rickards, R. W. (1983) J. Antibiot. 36, 1323–1328. [DOI] [PubMed] [Google Scholar]
- 4.Traxler, P. & Ghisalba, O. (1982) J. Antibiot. 35, 1361–1366. [DOI] [PubMed] [Google Scholar]
- 5.Higashide, E., Asai, M., Ootsu, K., Tanida, S., Kozai, Y., Hasegawa, T., Kishi, T., Sugino, Y. & Yoneda, M. (1977) Nature 270, 721–722. [DOI] [PubMed] [Google Scholar]
- 6.Ghisalba, O. (1985) Chimia 39, 79–88. [Google Scholar]
- 7.Floss, H. G. & Beale, J. M. (1989) Angew. Chem. Int. Ed. Engl. 28, 146–177. [Google Scholar]
- 8.Floss, H. G. (1997) Nat. Prod. Rep. 14, 433–452. [DOI] [PubMed] [Google Scholar]
- 9.Hunziker, D., Yu, T. W., Hutchinson, C. R., Floss, H. G. & Khosla, C. (1998) J. Am. Chem. Soc. 120, 1092–1093. [Google Scholar]
- 10.Admiraal, S. J., Walsh, C. T. & Khosla, C. (2001) Biochemistry 40, 6116–6123. [DOI] [PubMed] [Google Scholar]
- 11.Admiraal, S. J., Khosla, C. & Walsh, C. T. (2002) Biochemistry 41, 5313–5324. [DOI] [PubMed] [Google Scholar]
- 12.August, P. R., Tang, L., Yoon, Y. J., Ning, S., Muller, R., Yu, T. W., Taylor, M., Hoffmann, D., Kim, C. G., Zhang, X., et al. (1998) Chem. Biol. 5, 69–79. [DOI] [PubMed] [Google Scholar]
- 13.Tang, L., Yoon, Y. J., Choi, C. Y. & Hutchinson, C. R. (1998) Gene 216, 255–265. [DOI] [PubMed] [Google Scholar]
- 14.Schupp, T., Toupet, C., Engel, N. & Goff, S. (1998) FEMS Microbiol. Lett. 159, 201–207. [DOI] [PubMed] [Google Scholar]
- 15.Yu, T. W., Bai, L., Clade, D., Hoffmann, D., Toelzer, S., Trinh, K. Q., Xu, J., Moss, S. J., Leistner, E. & Floss, H. G. (2002) Proc. Natl. Acad. Sci. USA 99, 7968–7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rascher, A., Hu, Z., Viswanathan, N., Schirmer, A., Reid, R., Nierman, W. C., Lewis, M. & Hutchinson, C. R. (2003) FEMS Microbiol. Lett. 218, 223–230. [DOI] [PubMed] [Google Scholar]
- 17.Kim, C.-G., Kirschning, A., Bergon, P., Zhou, P., Su, E., Sauerbrei, B., Ning, S., Ahn, Y., Breuer, M., Leistner, E. & Floss, H. G. (1996) J. Am. Chem. Soc. 118, 7486–7491. [Google Scholar]
- 18.Guo, J. & Frost, J. W. (2002) J. Am. Chem. Soc. 124, 10642–10643. [DOI] [PubMed] [Google Scholar]
- 19.Eads, J. C., Beeby, M., Scapin, G., Yu, T. W. & Floss, H. G. (1999) Biochemistry 38, 9840–9849. [DOI] [PubMed] [Google Scholar]
- 20.Kim, C. G., Yu, T. W., Fryhle, C. B., Handa, S. & Floss, H. G. (1998) J. Biol. Chem. 273, 6030–6040. [DOI] [PubMed] [Google Scholar]
- 21.Yu, T. W., Muller, R., Muller, M., Zhang, X., Draeger, G., Kim, C. G., Leistner, E. & Floss, H. G. (2001) J. Biol. Chem. 276, 12546–12555. [DOI] [PubMed] [Google Scholar]
- 22.Arakawa, K., Muller, R., Mahmud, T., Yu, T. W. & Floss, H. G. (2002) J. Am. Chem. Soc. 124, 10644–10645. [DOI] [PubMed] [Google Scholar]
- 23.Ghisalba, O., Fuhrer, H., Richter, W. J. & Moss, S. (1981) J. Antibiot. 34, 58–63. [DOI] [PubMed] [Google Scholar]
- 24.Yu, T. W., Shen, Y., Doi-Katayama, Y., Tang, L., Park, C., Moore, B. S., Hutchinson, C. R. & Floss, H. G. (1999) Proc. Natl. Acad. Sci. USA 96, 9051–9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeifer, B. A., Admiraal, S. J., Gramajo, H., Cane, D. E. & Khosla, C. (2001) Science 291, 1790–1792. [DOI] [PubMed] [Google Scholar]
- 26.Pfeifer, B., Hu, Z., Licari, P. & Khosla, C. (2002) Appl. Environ. Microbiol. 68, 3287–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez, E. & Gramajo, H. (1999) Microbiology 145, 3109–3119. [DOI] [PubMed] [Google Scholar]
- 28.Tsuji, S. Y., Cane, D. E. & Khosla, C. (2001) Biochemistry 40, 2326–2331. [DOI] [PubMed] [Google Scholar]
- 29.Wu, N., Tsuji, S. Y., Cane, D. E. & Khosla, C. (2001) J. Am. Chem. Soc. 123, 6465–6474. [DOI] [PubMed] [Google Scholar]
- 30.Wu, N., Cane, D. E. & Khosla, C. (2002) Biochemistry 41, 5056–5066. [DOI] [PubMed] [Google Scholar]
- 31.Bongaerts, J., Kramer, M., Muller, U., Raeven, L. & Wubbolts, M. (2001) Metab. Eng. 3, 289–300. [DOI] [PubMed] [Google Scholar]
- 32.Knaggs, A. R. (2001) Nat. Prod. Rep. 18, 334–355. [DOI] [PubMed] [Google Scholar]
- 33.Schwecke, T., Aparicio, J. F., Molnar, I., Konig, A., Khaw, L. E., Haydock, S. F., Oliynyk, M., Caffrey, P., Cortes, J., Lester, J. B., et al. (1995) Proc. Natl. Acad. Sci. USA 92, 7839–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang, L., Shah, S., Chung, L., Carney, J., Katz, L., Khosla, C. & Julien, B. (2000) Science 287, 640–642. [DOI] [PubMed] [Google Scholar]