Abstract
Smad proteins mediate transforming growth factor β (TGF-β)-inducible transcriptional responses. Protein inhibitor of activated signal transducer and activator of transcription (PIAS) represents a family of proteins that inhibits signal transducer and activator of transcription and also regulates other transcriptional responses. In an effort to identify Smad-interacting proteins by a yeast three-hybrid screen with Smad3 and Smad4 as baits, we identified PIASy, a member of the PIAS family. In yeast, PIASy interacts strongly with Smad4 and also with receptor-regulated Smads. In mammalian cells, PIASy binds most strongly with Smad3 and also associates with other receptor-regulated Smads and Smad4. The interaction between Smad3 and PIASy is increased in the presence of TGF-β and occurs through the C-terminal domain of Smad3. Moreover, Smad3, Smad4, and PIASy can form a ternary complex. PIASy does not inhibit Smad complex binding to DNA, but it represses Smad transcriptional activity. Interestingly, conditional overexpression of PIASy selectively inhibits a subset of endogenous TGF-β-responsive genes, which includes the cyclin-dependent kinase inhibitor p15, and the plasminogen activator inhibitor 1. We further show that PIASy can interact constitutively with histone deacetylase 1 (HDAC1) and that addition of HDAC inhibitor trichostatin A (TSA) can prevent the inhibitory function of PIASy. Taken together, our studies indicate that PIASy can inhibit TGF-β/Smad transcriptional responses through interactions with Smad proteins and HDAC.
Ttransforming growth factor β (TGF-β) exerts a wide variety of biological activities through transcriptional regulation of diverse genes (1–4). Smad2, Smad3, and Smad4 are the key intracellular mediators of TGF-β signaling. On binding of TGF-β to its transmembrane serine/threonine kinase receptors, Smad2 and Smad3 are phosphorylated by the receptor, form complexes with Smad4, and accumulate in the nucleus to regulate transcription (1–4).
Smads are usually recruited to TGF-β-responsive promoters through interactions with DNA-binding partners (1–4). The N-terminal domains of Smad3 and Smad4 also possess DNA-binding activities (5–8). The consensus-binding site for Smad3 and Smad4 is the 8-bp palindromic sequence GTCTAGAC, designated as the Smad-binding element (SBE) (5). Most of the TGF-β-responsive promoters contain one or more copies of the half-site GTCT or AGAC (1–4, 6, 7).
Smads can activate transcription by recruiting transcriptional coactivators that include p300/CBP, p/CAF, ARC105, MSG1, and SMIF1 (1–4, 9, 10). The C-terminal domains of Smad3 and Smad2 can interact with p300/CBP, which possess intrinsic histone acetyltransferase activities (refs. 11 and 12; reviewed in refs. 1–4). The Smad3 C-terminal domain can also recruit P/CAF, another histone acetyltransferase-containing coactivator (9). Smad-mediated transcriptional activation also requires the Smad activation domain in the linker region of Smad4 (13). The Smad activation domain physically interacts with p300 (13) and also plays important roles in the recruitment of Smad4 coactivators MSG1 (10) and SMIF (4).
Smads can inhibit transcription through binding of transcriptional corepressors (1, 3, 4). Three Smad corepressors have been identified, including the homeodomain protein TG-interacting factor (TGIF) and the Ski and SnoN proteins that are related to each other (refs. 14–18; reviewed in refs 1, 3, 4, and 19). TGIF can interact directly with histone deacetylase (HDAC) and also associate with the mSin3 and the C-terminal-binding protein (CtBP) corepressors (1, 3, 4, 14). Ski and SnoN can interact with NcoR and mSin3, which recruit HDACs (15, 17, 18, 20). Ski may also inhibit TGF-β-induced transcriptional responses by competing with Smad2 and Smad3 binding to Smad4 (21). The interactions between Smad2 and Smad3 with TGIF, and the interactions among Smad2, Smad3, and Smad4 with Ski/SnoN are all mediated through the conserved C-terminal domains in Smads (1, 3, 4, 14–19).
Protein inhibitor of activated STAT (PIAS; STAT is signal transducer and activator of transcription) represents a family of proteins originally identified through interaction with cytokine-induced STAT (22). In mammals, five PIAS proteins have been identified (23). PIAS1 and PIAS3 bind and inhibit STAT1 and STAT3 DNA-binding activities, respectively (22, 23). PIASy can inhibit STAT1- or androgen receptor-mediated transcriptional responses without blocking their DNA-binding activities (24, 25) and repress LEF1 activity by sequestration into nuclear bodies (26). PIASxα and PIASxβ are probably derived from alternatively spliced mRNA products of the same gene. PIASxα interacts with the androgen receptor and regulates its activity (27, 28). PIASxβ interacts with the homeodomain protein Msx2 (29). PIAS1, PIAS3, PIASxα, and PIASxβ also regulate steroid receptor-dependent transcriptional activation (30). In Drosophila, the PIAS ortholog dPIAS, initially identified as a suppressor of position effect variegation, designated Su(var)2–10, also known as Zimp (31), regulates chromosome structure and function (32), and interacts with STAT92E to regulate eye and blood cell development (33). In addition, the PIAS family has been shown to possess sumoylation ligase activity, which can promote the covalent addition of a small ubiquitin-related modifier to target proteins (26, 28, 34–41).
In an effort to search for new regulators of TGF-β-signaling pathways, we performed a yeast three-hybrid screen by using both Smad3 and Smad4 as baits, and identified PIASy as a Smad-interacting protein. Our studies indicate that PIASy can inhibit Smad-mediated transcriptional responses by binding to Smads and HDAC.
Materials and Methods
Yeast Three-Hybrid Screen and Two-Hybrid Assay. Smad4 and Smad3 were cloned into the EcoRI and NotI sites of the pBridge three-hybrid vector (CLONTECH), respectively. The bait was used to screen a human fetal brain cDNA library and a human liver cDNA library (CLONTECH) in the AH109 yeast strain. To examine the interactions of PIASy with various Smads in yeast, Smad fused to the LexA DNA-binding domain in the pEG202 vector, and the PIASy cDNA cloned in the pJG4–5 vector were cotransformed into the EGY48 strain and analyzed for β-galactosidase activity as described (42).
Immunoprecipitation, Immunoblot, and Ternary Complex Detection. COS or 293T cells were transfected by lipofectamine plus reagent. Immunoprecipitation, immunoblot, and detection of ternary complex were performed as described (43). For detection of endogenous Smad3 and PIASy interaction, a Smad3-specific Ab (Zymed) and a PIASy-specific Ab (24) were used.
GST Pull-Down Assay. 35S-labeled PIASy (15 μl) synthesized by in vitro translation was mixed with 15 μg of GST or GST-Smad3 beads in 1.0 ml of binding buffer (40 mM Hepes, pH 7.4/100 mM KCl/0.1% Nonidet P-40/1 mM DTT/0.1 mM PMSF) at 4°C for 2 h. The bound protein was analyzed by SDS/PAGE and was visualized by autoradiography.
Generation of Tetracycline (tet)-Controlled PIASy Stable Clones. Mouse PIASy cDNA was inserted into the pUHD10-3 hygromycin vector (44) and transfected by lipofectamine plus reagent into Mv1Lu tet-controlled transactivator (tTA) cells that harbor the bacterial tet repressor fused to the VP16 transcriptional activation domain (44). PIASy stable clones were selected in the presence of 0.5 mg/ml G418, 0.3 mg/ml hygromycin, and 2 μg/ml tet for 2–3 wk. Of 20 clones analyzed, 4 clones expressed PIASy in the absence of tet, and inhibited TGF-β induction.
RNA Isolation and Northern Blot Analysis. The parental and the PIASy Mv1Lu tTA cells were maintained in the presence of 2 μg/ml tet. At the time of splitting the cells, half of the plates were continually cultured in the presence of tet, whereas the other half of the plates had tet removed. After removal of tet (30 h), cells were treated with 500 pM TGF-β in complete medium for the indicated time. Poly(A)+ RNA were then isolated and analyzed in Northern blot as described (45).
Gel Mobility Shift Assay and DNA Affinity Precipitation. Flag-PIASy protein was purified from transfected 293T cells by anti-Flag M2 affinity gel. HaCaT cells were treated with or without 500 pM TGF-β for 1 h. Cell extracts were incubated with a 32P-labeled 39-bp probe containing two copies of the SBE (5′-CTAGGATAGCGTCTAGACATAGTCTAGACTGAGTCCTAG-3′) (17) in the absence or presence of exogenously added Flag-PIASy protein, and analyzed by gel shift as described (45). The DNA pull-down assay was carried out as described (14). HaCaT cells were treated with or without 500 pM TGF-β for 1 h. Cell lysates (1 ml) containing 1 mg of protein were incubated with 0.5 μg of biotinylated WT 2× SBE oligonucleotide or mutant 2× SBE oligonucleotide (5′-CTAGGATAGCACCTAAAAATAACCTAAAATGAGTTCTAG-3′), along with 10 μg of poly(dI-dC) in the presence of exogenously added 250 ng of Flag-PIASy protein, followed by addition of streptavidin-agarose beads. The bound proteins were then analyzed by immunoblots.
Luciferase Reporter Gene Assay. Mv1Lu/L17 cells in 60-mm dishes were transfected by DEAE-dextran and treated with or without 500 pM TGF-β for 18–24 h as described (45). Luciferase activities were normalized with cotransfected renilla luciferase control. TSA was dissolved in DMSO as 1 mg/ml stock solution. Various concentrations of TSA or the DMSO vehicle control were added at the same time as the addition of TGF-β. Results are from three or more independent transfections.
Results
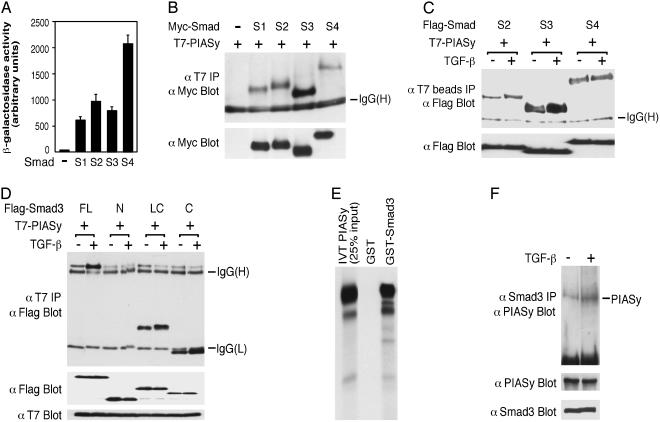
Identification of PIASy as a Smad-Interacting Protein from a Yeast Three-Hybrid Screen. To identify Smad-interacting proteins, we performed a yeast three-hybrid screen by using both Smad4 and Smad3 as baits. Full-length Smad4 and Smad3 were cloned into the pBridge vector (CLONTECH), in which Smad4 was fused to the GAL4 DNA-binding domain and Smad3 was expressed as a native protein. The construct itself does not have significant basal transcriptional activity (data not shown). A number of Smad-interacting proteins, including several known ones, such as JunB and SnoN, were identified from screening 4 × 106 transformants of a human fetal brain cDNA library and 2 × 106 transformants of a human liver library. A partial cDNA clone of PIASy was identified from the fetal brain library. Subsequent analysis indicates that in yeast PIASy interacts strongly with Smad4 and also interacts with Smad3 and Smad2 as well as Smad1 that transduces the bone morphogenetic protein signal (Fig. 1A).
Fig. 1.
Interaction of PIASy with Smad3 and other Smad proteins. (A) Yeast two-hybrid assay of PIASy interaction with Smads1–4. (B) Interaction of PIASy with Smads1–4 in 293T cells. (C) TGF-β treatment increases the interactions between PIASy with Smad3 or Smad2, but has little effect on PIASy–Smad4 interaction. TβRI (T204D) was cotransfected for TGF-β stimulation in 293T cells. T7 Ab-coupled agarose beads were used in the immunoprecipitation. (D) PIASy interacts with the C-terminal domain of Smad3. FL, full length; N, N-terminal domain; LC, linker and C-terminal domain; and C, C-terminal domain. Smad3 (FL) comigrates with a nonspecific band above the IgG (heavy chain). (E) 35S-labeled-PIASy synthesized by in vitro translation (IVT) binds to GST-Smad3. (F) Endogenous interaction between Smad3 and PIASy. HaCaT cells were treated with or without TGF-β for 1 h. Cell lysates were immunoprecipitated with a Smad3 Ab and were immunoblotted with a PIASy Ab.
PIASy Interacts with Smad3 and Other Smads in Mammalian Cells. To determine whether PIASy interacts with Smads in mammalian cells, Myc epitope-tagged Smad and T7-tagged PIASy were cotransfected into 293T cells. Cells were then subjected to immunoprecipitation by a T7 Ab followed by immunoblotting by a Myc Ab. As shown in Fig. 1 B and C, PIASy interacted most strongly with Smad3 in mammalian cells. The interaction between PIASy and Smad3 is increased in the presence of TGF-β and is mediated by the C-terminal domain of Smad3 (Fig. 1 C and D), which is necessary for transcriptional activation (1, 2). The interaction between Smad2 and PIASy is also increased in the presence of TGF-β, whereas TGF-β has little effect on the interaction between Smad4 and PIASy (Fig. 1C). PIASy also associated with Smad3 in vitro (Fig. 1E). Moreover, endogenous PIASy and Smad3 interacted with each other, and the interaction was increased by TGF-β treatment as analyzed in HaCaT keratinocytes (Fig. 1F).
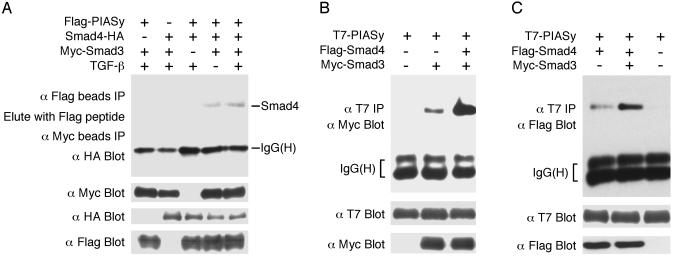
Smad3, Smad4, and PIASy Can Form a Ternary Complex. Because both Smad3 and Smad4 are essential for transcriptional activation (1–4), we determined whether Smad3, Smad4, and PIASy can form a ternary complex. Flag-PIASy, Myc-Smad3, and Smad4-HA were cotransfected into COS cells. Cell lysates were immunoprecipitated by Flag Ab coupled to agarose beads. The immunoprecipitates were eluted with Flag peptide, and the eluent was used in a second immunoprecipitation by Myc Ab agarose beads, followed by immunoblot with a HA Ab. As shown in Fig. 2A, a ternary complex was detected only when all three components were present, and TGF-β treatment increased the ternary complex formation. Moreover, cotransfection of Smad4 increased the interaction between Smad3 and PIASy (Fig. 2B). Similarly, cotransfection of Smad3 increased the association between Smad4 and PIASy (Fig. 2C). These observations suggest that Smad4 stabilizes Smad3–PIASy association and that Smad3 stabilizes Smad4–PIASy interaction.
Fig. 2.
PIASy can form a ternary complex with Smad3 and Smad4. (A) Detection of the Smad3–Smad4–PIASy ternary complex. Epitope-tagged Smad3, Smad4, and PIASy, and TβRI (T204D) for TGF-β stimulation were cotransfected into COS cells and analyzed as indicated. (B) Smad4 can increase the interaction between Smad3 and PIASy. (C) Smad3 can increase the interaction between Smad4 and PIASy. TβRI (T204D) was cotransfected for TGF-β stimulation in B and C.
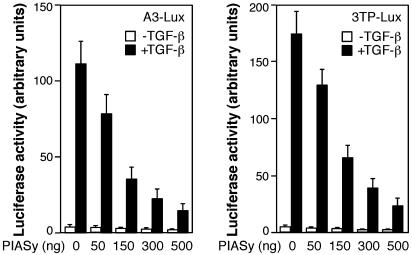
PIASy Inhibits TGF-β/Smad-Mediated Transcriptional Responses. To investigate the functional significance of PIASy–Smad interaction, we examined the effects of PIASy on TGF-β/Smad-dependent transcriptional responses. Mv1Lu/L17 mink lung epithelial cells were transiently cotransfected with PIASy and A3-Lux or 3TP-Lux, which are two well characterized TGF-β-responsive reporter genes. As shown in Fig. 3, PIASy inhibited the TGF-β-induced transcriptional activation of the two reporter genes in a dose-dependent manner. Interestingly, PIASy had little effect on Smad7 reporter gene in the same assay (data not shown), suggesting that PIASy may selectively inhibit TGF-β-responsive genes.
Fig. 3.
PIASy inhibits TGF-β/Smad transcriptional responses. Mv1Lu/L17 cells were cotransfected with the A3-Lux reporter gene, FAST-1, and various amounts of T7-PIASy plasmid (Left) or with the 3TP-Lux reporter gene and increasing doses of T7-PIASy plasmid (Right). Cells were treated with or without TGF-β and analyzed for luciferase activity.
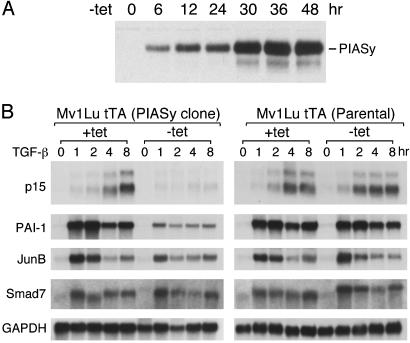
To determine whether PIASy can inhibit endogenous TGF-β-responsive genes, we generated tet-controlled PIASy stable clones, using a tet-off system. The established Mv1Lu tTA cells contain the bacterial tet repressor fused to the VP16 activation domain (44). PIASy was transfected into the Mv1Lu tTA cells. The expression of PIASy was under the control of the tet operon. Tet binding to the tet repressor-VP16 fusion inhibits activation of PIASy expression, whereas removal of tet allows activation of PIASy expression. We obtained several PIASy stable clones that expressed PIASy only in the absence of tet, but not in the presence of tet. A representative PIASy clone was analyzed for the PIASy induction time course after removal of tetracycline. As shown in Fig. 4A, PIASy expression reached ≈80–90% of maximal level after 30 h and peaked after ≈48 h.
Fig. 4.
Conditional overexpression of PIASy selectively inhibits a subset of TGF-β/Smad-responsive genes. (A) Time course of PIASy protein induction after removal of tet. A representative PIASy Mv1Lu tTA stable clone was analyzed by immunoblot with the PIASy Ab. (B) Northern blot analysis of TGF-β induction of p15, PAI-1, JunB, and Smad7 transcripts in the parental Mv1Lu tTA cells as well as in the PIASy clone with or without tet. Poly(A)+ RNA (4 μg) of each sample was used. GAPDH served as a loading control.
The PIASy clone was analyzed further for the expression of several TGF-β/Smad-responsive genes by Northern blot analysis. For comparison, the parental Mv1Lu tTA cells were also analyzed in parallel. Thirty hours after removal of tet, TGF-β was added to plates that contained tet, and to plates not containing tet. RNA was prepared from untreated and TGF-β-treated cells at different time points. The RNA samples were analyzed for the expression of p15, plasminogen activator inhibitor 1 (PAI-1), JunB, and Smad7, as well as GAPDH as a control. The PAI-1, JunB, and Smad7 are TGF-β-inducible early or immediate early responsive genes, whereas TGF-β induction of p15 starts at 2 h and peaks at 8 h in Mv1Lu cells (44). As shown in Fig. 4B, PIASy greatly inhibited the TGF-β induction of p15 and PAI-1, moderately inhibited the early response of the JunB gene, and had little effect on the expression of the Smad7 gene, which is consistent with transient transfection data. Similar results were obtained from another PIASy clone (data not shown).
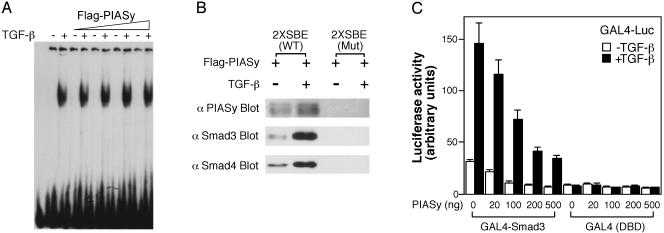
PIASy Does Not Inhibit Smad DNA-Binding Activity. We examined whether the inhibitory effect of PIASy occurred through inhibition of Smad complex binding to DNA. Flag-PIASy protein was expressed and purified from 293T cells by using the Flag Ab affinity beads. Untreated or TGF-β-treated HaCaT cell extracts were incubated with the 39-bp 2× SBE probe (17) in the absence or presence of purified PIASy protein. As shown in Fig. 5A, TGF-β treatment induced the formation of a DNA–protein complex. We and others have previously shown that this DNA–protein complex represents a Smad complex containing Smad3 and Smad4 binding to the SBE (17, 45). Exogenous Flag-PIASy protein did not inhibit the DNA-binding activity of the Smad complex (Fig. 5A). Similar results were obtained by using GST-PIASy protein or cell extracts from Smad3-, Smad4-, and PIASy-cotransfected cells (data not shown).
Fig. 5.
(A) PIASy does not inhibit Smad DNA-binding activity. Cell extracts from TGF-β-treated or untreated HaCaT cells were incubated with 2× SBE DNA probe in the absence or presence of increasing amount of purified Flag-PIASy protein (50, 100, 200, and 400 ng) and were then subjected to electrophoresis. (B) PIASy can be recruited to the SBE through interaction with Smads in DNA-affinity precipitation assay. (C) PIASy inhibits GAL4-Smad3 transcriptional activity. Mv1Lu/L17 cells were cotransfected with GAL4-Luc reporter gene, GAL4 DNA-binding domain GAL4 (DBD), or GAL4-Smad3, and increasing amount of T7-PIASy plasmid, treated with or without TGF-β, and analyzed for luciferase activity.
Because Flag-PIASy protein did not result in a convincing supershift of the Smad–DNA complex in the gel mobility shift assay, it is possible that the PIASy–Smad–DNA complex is unstable. To determine whether PIASy and Smads can interact on DNA, we performed DNA affinity precipitation experiments by using a biotinylated SBE oligonucleotide. As shown in Fig. 5B, immunoprecipitation of the biotinylated SBE oligonucleotide by streptavidin beads followed by immunoblotting of the precipitates by PIASy Ab revealed that PIASy can be recruited to the WT SBE oligonucleotide, but not to mutant SBE oligonucleotide. In addition, similar to Smad3 and Smad4, PIASy binding to SBE was increased by TGF-β treatment (Fig. 5B). Thus, PIASy can be recruited to the SBE on DNA through interaction with Smads.
PIASy Represses Smad Transcriptional Activity. Because PIASy does not inhibit Smad DNA-binding activity, we analyzed whether PIASy can directly inhibit Smad transcriptional activity. We used a GAL4 fusion assay by fusing Smad3 to the GAL4 DNA-binding domain. As shown in Fig. 5C, PIASy inhibited both the basal and TGF-β-induced transcriptional activity of GAL4-Smad3 in a dose-dependent manner, whereas it had only a modest effect on the GAL4 DNA-binding domain alone. Thus, PIASy inhibition of TGF-β/Smad-mediated transcriptional responses can occur directly by repression of Smad transcriptional activity.
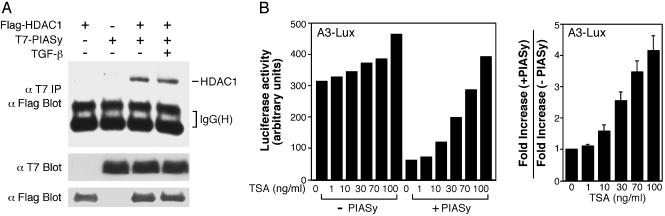
PIASy Can Interact with HDAC and Inhibit Transcription in an HDAC-Dependent Manner. To determine whether PIASy can inhibit Smad-dependent transcription by binding to HDAC, we first asked whether PIASy and HDAC1 can interact with each other. T7-PIASy and Flag-HDAC1 were cotransfected into COS cells and treated with or without TGF-β. As shown in Fig. 6A, PIASy and HDAC1 can interact with each other constitutively in an immunoprecipitation–immunoblot assay.
Fig. 6.
PIASy can inhibit Smad-mediated transcriptional activation in an HDAC-dependent manner. (A) Interaction of PIASy and HDAC1. Flag-HDAC1, T7-PIASy, and TβRI (T204D) for TGF-β stimulation were cotransfected into COS cells and analyzed as indicated. (B) TSA can disrupt the inhibitory effect of PIASy. Mv1Lu/L17 cells were cotransfected with the A3-Lux reporter gene and FAST-1 in the absence or presence of 300 ng of PIASy DNA. Cells were treated with TGF-β and increasing amounts of TSA at the same time for 18–24 h and then were analyzed for luciferase activity. (Left) One representative experiment. (Right) Summary of several experiments. For each dose of TSA, the fold increase refers to the luciferase activity in the presence of TSA divided by the luciferase activity in the absence of TSA. The fold increase in the presence of cotransfected PIASy divided by the fold increase in the absence of PIASy then was calculated and plotted against each dose of TSA. This ratio in the absence of TSA is set as 1.
The observation above prompted us to ask whether TSA, an inhibitor of HDAC, can prevent PIASy from inhibiting transcription. Mv1Lu/L17 cells were cotransfected with A3-Lux with or without PIASy and treated with TGF-β, along with increasing doses of TSA. One representative experiment is shown in Fig. 6B Left, and the results of several experiments are summarized in Fig. 6B Right. TSA enhanced the luciferase activity in a dose-dependent manner, but the effect was much more significant in the presence of cotransfected PIASy than in the absence of PIASy. Thus, PIASy can repress transcription in an HDAC-dependent manner.
Discussion
We have shown in this article that PIASy inhibits TGF-β transcriptional responses. Smad3, Smad4, and PIASy can form a ternary complex. PIASy can associate with HDAC1, and the inhibitory effect of PIASy can be disrupted by treatment with TSA. Taken together, our studies suggest that PIASy can inhibit TGF-β-mediated transcription by recruiting HDAC to Smads. Similarly, PIASxβ, another member of the PIAS family, can interact with HDAC3 (46), which is in the same class as HDAC1. PIASxα and PIASxβ have also been implicated to recruit HDAC to inhibit IL-12-mediated and STAT4-dependent gene activation (47). Thus, some PIAS proteins may repress transcription through association with HDAC.
Interestingly, PIASy selectively inhibits a subset of TGF-β-regulated genes. Previous studies (1, 3, 4, 14–19) have shown that Smad proteins can repress transcription by recruiting transcriptional corepressors, such as TGIF, Ski, and SnoN. It appears that PIASy, TGIF, and Ski/SnoN have distinct promoter specificities. For example, TGF-β induction of p15 is greatly inhibited by overexpression of PIASy but is not inhibited by overexpression of Ski (16). Conversely, the early responsiveness of JunB to TGF-β is only moderately inhibited by overexpression of PIASy but is significantly inhibited by overexpression of Ski (15). The mechanisms by which PIASy, TGIF, and Ski/SnoN target distinct promoters remain to be a challenging issue. One possibility is that PIASy, TGIF, and Ski/SnoN are present in different corepressor complexes that inhibit distinct sets of promoters. PIASy and TGIF also seem to have some overlapping functions. For example, overexpression of TGIF can also inhibit TGF-β induction of p15 gene (48), although it appears not as potently as by PIASy overexpression. Both TGIF and PIASy may inhibit TGF-β activated Smad complex, whereas Ski/SnoN have been proposed to function as corepressors for Smad4 to maintain TGF-β-responsive genes in a repressed state in the absence of TGF-β, and may contribute to TGF-β signal termination (18, 19). The presence of multiple regulators that share overlapping functions in the same cells confers ample flexibility under complex circumstances.
In addition, PIASy, TGIF, and Ski/SnoN are differentially expressed in various tissues (19, 25, 49, 50), which may allow them to respond differently to the TGF-β superfamily signals or other developmental cues, thus conferring cell-type-specific functions. PIASy is expressed in the mouse as early as day 7.5 of gestation (49). In situ hybridization experiments indicate that PIASy is expressed in the neuroepithelium, limb buds, and the dermal structure (49). In adult, PIASy is ubiquitously expressed, with the highest expression in testis (25) and low expression in liver and lung (49). PIASy is expressed in all three tissue culture cell lines we examined, including HaCaT, Mv1Lu, and COS cells. Whether various extracellular signals can affect PIASy levels remains to be addressed. TGIF is highly expressed in testis, ovary, and placenta, and its expression is low in skeletal muscle, heart, and brain in human adult tissues (50). Ski is expressed at high levels in hematopoietic stem cells, and at low levels in a variety of other tissues (19). The expression of SnoN is regulated by various treatments, including by TGF-β (18, 19).
Distinct from TGIF and Ski/SnoN, PIASy as well as other PIAS family members have small ubiquitin-related modifier E3 ligase activities (26, 28, 34–41). Because the ring domain mutant PIASy has a significantly lower repression activity in the reporter gene assay (data not shown), it is possible that sumoylation may play a role in PIASy-mediated transcriptional repression. HDAC1 itself may be a target for sumoylation by PIAS proteins. A previous study (51) has demonstrated that HDAC1 is sumoylated both in vivo and in vitro. Moreover, mutation of the sumoylation sites in HDAC1 reduces its transcriptional repression activity in reporter gene assays (51). Similarly, sumoylation is necessary for the proper localization and function of CtBP, and certain PIAS proteins as well as the polycomb protein Pc2 can function as small ubiquitin-related modifier E3 ligases for CtBP (39, 52). Alternatively, sumoylation of Smads may be necessary for PIASy repression of TGF-β-responsive genes.
Finally, it appears that PIASy can use different modes to inhibit transcriptional responses. PIASy can antagonize the activities of STAT1 (24), androgen receptor (25), p53 (53), LEF1 (26), and Smads. PIASy inhibits STAT1 and androgen receptor activities without interfering with their DNA-binding activities (24, 25), whereas the repression of p53 activity appears to be through inhibition of p53 binding to DNA (53). The inhibitory effect on LEF1 was shown to be through sequestration of LEF1 into nuclear bodies (26). In addition, recent studies have shown that several corepressors, such as CtBP, ligand-dependent nuclear receptor corepressor LCoR, and several repressors, such as Ikaros and Rb, can inhibit transcription in HDAC-dependent and -independent manners, based on different promoter contexts (54, 55). It will be interesting to determine whether PIASy and other PIAS members can inhibit the various responsive promoters through distinct transcriptional repression mechanisms.
In conclusion, we have identified PIASy as a regulator of TGF-β/Smad-signaling pathways. We have shown that PIASy can interact with Smads and HDAC and repress TGF-β/Smad-mediated transcription in an HDAC-dependent manner, suggesting that PIASy can inhibit transcription by recruiting HDAC to Smads. Interestingly, PIASy selectively inhibits a subset of endogenous TGF-β/Smad-responsive genes. Our findings provide insights into transcriptional regulation by Smads and PIAS proteins that play important roles in a wide variety of biological responses.
Acknowledgments
We thank J. Massagué for Mv1Lu tTA cells; R. Grosschedl for T7-PIASy plasmid; R. Janknecht for Myc-tagged Smads; D. Reinberg for reagents; C. Gasper for assistance; C. Gelinas, J. Serone, D. Wotton, and N. Denissova for suggestions; and many colleagues for discussions. This work was supported by the National Foundation for Cancer Research, the Burroughs Wellcome Fund, the Emerald Foundation, the Sidney Kimmel Foundation for Cancer Research (F.L.), and the National Institutes of Health (F.L. and K.S.). F.L. is a Kimmel Scholar.
Abbreviations: TGF-β, transforming growth factor β; STAT, signal transducer and activator of transcription; PIAS, protein inhibitor of activated STAT; SBE, Smad-binding element; tet, tetracycline; tTA, tet-controlled transactivator; HDAC, histone deacetylase; TSA, trichostatin A; PAI-1, plasminogen activator inhibitor 1; TGIF, TG-interacting factor; CtBP, C-terminal-binding protein.
References
- 1.Massagué, J. & Wotton, D. (2000) EMBO J. 19, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derynck, R., Zhang, Y. & Feng, X.-H. (1998) Cell 95, 737–740. [DOI] [PubMed] [Google Scholar]
- 3.Attisano, L. & Wrana, J. L. (2000) Curr. Opin. Cell Biol. 12, 235–243. [DOI] [PubMed] [Google Scholar]
- 4.Shi, Y. & Massagué, J. (2003) Cell 113, 685–700. [DOI] [PubMed] [Google Scholar]
- 5.Zawel, L., Dai, J., Buckhaults, P., Zhou, S., Kinzler, K., Vogelstein, B. & Kern, S. (1998) Mol. Cell 1, 611–617. [DOI] [PubMed] [Google Scholar]
- 6.Yingling, J., Datto, M., Wong, C., Frederick, J., Liberati, N. & Wang, X.-F. (1997) Mol. Cell. Biol. 17, 7019–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennler, S., Itoh, S., Vivien, D., ten Dijke, P., Huet, S. & Gauthier, J.-M. (1998) EMBO J. 17, 3091–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi, Y., Wang, Y. F., Jayaraman, L., Yang, H., Massagué, J. & Pavletich, N. P. (1998) Cell 94, 585–594. [DOI] [PubMed] [Google Scholar]
- 9.Itoh, S., Ericsson, J., Nishikawa, J.-I., Heldin, C.-H. & ten Dijke, P. (2000) Nucleic Acids Res. 28, 4291–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yahata, T., de Caestecker, M. P., Lechleider, R. J., Andriole, S., Roberts, A. B., Isselbacher, K. J. & Shioda, T. (2000) J. Biol. Chem. 275, 8825–8834. [DOI] [PubMed] [Google Scholar]
- 11.Janknecht, R., Wells, N. J. & Hunter, T. (1998) Genes Dev. 12, 2114–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, X.-H., Zhang, Y., Wu, R. Y. & Derynck, R. (1998) Genes Dev. 12, 2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Caestecker, M. P., Yahata, T., Wang, D., Parks, W. T., Huang, S., Hill, C. S., Shioda, T., Roberts, A. B. & Lechleider, R. J. (2000) J. Biol. Chem. 275, 2115–2122. [DOI] [PubMed] [Google Scholar]
- 14.Wotton, D., Lo, R. S., Lee, S. & Massague, J. (1999) Cell 97, 29–39. [DOI] [PubMed] [Google Scholar]
- 15.Luo, K., Stroschein, S. L., Wang, W., Chen, D., Martens, E., Zhou, S. & Zhou, Q. (1999) Genes Dev. 13, 2196–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun, Y., Liu, X., Eaton, E., Lane, W., Lodish, H. & Weinberg, R. (1999) Mol. Cell 4, 499–509. [DOI] [PubMed] [Google Scholar]
- 17.Akiyoshi, S., Inoue, H., Hanai, J.-I., Kusanagi, K., Nemoto, N., Miyazono, K. & Kawabata, M. (1999) J. Biol. Chem. 274, 35269–35277. [DOI] [PubMed] [Google Scholar]
- 18.Stroschein, S. L., Wang, W., Zhou, S., Zhou, Q. & Luo, K. (1999) Science 286, 771–774. [DOI] [PubMed] [Google Scholar]
- 19.Liu, X., Sun, Y., Weinberg, R. A. & Lodish, H. F. (2001) Cytokine Growth Factor Rev. 12, 1–8. [DOI] [PubMed] [Google Scholar]
- 20.Nomura, T., Khan, M. M., Kaul, S. C., Dong, H.-D., Wadhwa, R., Colmenares, C., Kohno, I. & Ishii, S. (1999) Genes Dev. 13, 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, J., Krawitz, A. R., Chai, J., Li, W., Zhang, F., Luo, K. & Shi, Y. (2002) Cell 111, 357–367. [DOI] [PubMed] [Google Scholar]
- 22.Chung, C. D., Liao, J., Liu, B., Rao, X., Jay, P., Berta, P. & Shuai, K. (1997) Science 278, 1803–1805. [DOI] [PubMed] [Google Scholar]
- 23.Liu, B., Liao, J., Rao, X., Kushner, S. A., Chung, C. D., Chang, D. D. & Shuai, K. (1998) Proc. Natl. Acad. Sci. USA 95, 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, B., Gross, M., ten Hoeve, J. & Shuai, K. (2001) Proc. Natl. Acad. Sci. USA 98, 3203–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross, M., Liu, B., Tan, J., French, F., Carey, M. & Shuai, K. (2001) Oncogene 20, 3880–3887. [DOI] [PubMed] [Google Scholar]
- 26.Sachdev, S., Bruhn, L., Sieber, H., Pichler, A., Melchior, F. & Grosschedl, R. (2001) Genes Dev. 15, 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moilanen, A.-M., Karvonen, U., Poukka, H., Yan, W., Toppari, J., Janne, O. A. & Palvimo, J. J. (1999) J. Biol. Chem. 274, 3700–3704. [DOI] [PubMed] [Google Scholar]
- 28.Nishida, T. & Yasuda, H. (2002) J. Biol. Chem. 277, 41311–41317. [DOI] [PubMed] [Google Scholar]
- 29.Wu, L., Wu, H., Ma, L., Sangiorgi, F., Wu, N., Bell, J., Lyons, G. & Maxson, R. (1997) Mech. Dev. 65, 3–17. [DOI] [PubMed] [Google Scholar]
- 30.Kotaja, N., Aittomaki, S., Silvennoinen, O., Palvimo, J. J. & Janne, O. A. (2000) Mol. Endocrinol. 14, 1986–2000. [DOI] [PubMed] [Google Scholar]
- 31.Mohr, S. E. & Boswell, R. E. (1999) Gene 229, 109–116. [DOI] [PubMed] [Google Scholar]
- 32.Hari, K. L., Cook, K. R. & Karpen, G. H. (2001) Genes Dev. 15, 1334–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betz, A., Lampen, N., Martinek, S., Young, M. W. & Darnell, J. E., Jr. (2001) Proc. Natl. Acad. Sci. USA 98, 9563–9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, E. S. & Gupta, A. A. (2001) Cell 106, 735–744. [DOI] [PubMed] [Google Scholar]
- 35.Kahyo, T., Nishida, T. & Yasuda, H. (2001) Mol. Cell 8, 713–718. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt, D. & Muller, S. (2002) Proc. Natl. Acad. Sci. USA 99, 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotaja, N., Karvonen, U., Janne, O. A. & Palvimo, J. J. (2002) Mol. Cell. Biol. 22, 5222–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson, P. K. (2001) Genes Dev. 15, 3053–3058. [DOI] [PubMed] [Google Scholar]
- 39.Lin, X., Sun, B., Liang, M., Liang, Y. Y., Gast, A., Hildebrand, J., Brunicardi, F. C., Melchior, F. & Feng, X. H. (2003) Mol. Cell 11, 1389–1396. [DOI] [PubMed] [Google Scholar]
- 40.Lee, P. S., Chang, C., Liu, D. & Derynck, R. (2003) J. Biol. Chem., in press. [DOI] [PubMed]
- 41.Lin, X., Liang, M., Liang, Y. Y., Brunicardi, F. C., Melchior, F. & Feng, X. H. (2003) J. Biol. Chem. 278, 18714–18719. [DOI] [PubMed] [Google Scholar]
- 42.Zervos, A. S., Gyuris, J. & Brent, R. (1993) Cell 72, 223–232. [DOI] [PubMed] [Google Scholar]
- 43.Liu, F., Pouponnot, C. & Massagué, J. (1997) Genes Dev. 11, 3157–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynisdottir, I., Polyak, K., Iavarone, A. & Massagué, J. (1995) Genes Dev. 9, 1831–1845. [DOI] [PubMed] [Google Scholar]
- 45.Denissova, N. G., Pouponnot, C., Long, J., He, D. & Liu, F. (2000) Proc. Natl. Acad. Sci. USA 97, 6397–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tussie-Luna, M. I., Bayarsaihan, D., Seto, E., Ruddle, F. H. & Roy, A. L. (2002) Proc. Natl. Acad. Sci. USA 99, 12807–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arora, T., Liu, B., He, H., Kim, J., Murphy, T. L., Murphy, K. M., Modlin, R. L. & Shuai, K. (2003) J. Biol. Chem. 278, 21327–21330. [DOI] [PubMed] [Google Scholar]
- 48.Lo, R. S., Wotton, D. & Massagué, J. (2001) EMBO J. 20, 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sturm, S., Koch, M. & White, F. A. (2000) J. Mol. Neurosci. 14, 107–121. [DOI] [PubMed] [Google Scholar]
- 50.Bertolino, E., Reimund, B., Wildt-Perinic, D. & Clerc, R. G. (1995) J. Biol. Chem. 270, 31178–31188. [DOI] [PubMed] [Google Scholar]
- 51.David, G., Neptune, M. A. & DePinho, R. A. (2002) J. Biol. Chem. 277, 23658–23663. [DOI] [PubMed] [Google Scholar]
- 52.Kagey, M. H., Melhuish, T. A. & Wotton, D. (2003) Cell 113, 127–137. [DOI] [PubMed] [Google Scholar]
- 53.Nelson, V., Davis, G. E. & Maxwell, S. A. (2001) Apoptosis 6, 221–234. [DOI] [PubMed] [Google Scholar]
- 54.Chinnadurai, G. (2002) Mol. Cell 9, 213–224. [DOI] [PubMed] [Google Scholar]
- 55.Fernandes, I., Bastien, Y., Wai, T., Nygard, K., Lin, R., Cormier, O., Lee, H. S., Eng, F., Bertos, N. R., Pelletier, P., et al. (2003) Mol. Cell 11, 139–150. [DOI] [PubMed] [Google Scholar]