Abstract
The ubiquitin–proteasome pathway plays a critical role in the degradation of short-lived and regulatory proteins in a variety of cellular processes. The F-box proteins are part of the ubiquitin–ligase complexes, which mediate ubiquitination and proteasome-dependent degradation of phosphorylated proteins. We previously identified NFB42, an F-box protein that is highly enriched in the nervous system, as a binding partner for the herpes simplex virus 1 UL9 protein, the viral replication-initiator protein, in a yeast two-hybrid screen. In the present work, we find that coexpression of NFB42 and UL9 genes in 293T cells leads to a significant decrease in the level of UL9 protein. Treatment with the 26S-proteasome inhibitor MG132 restores the UL9 protein to normal levels. We have observed also that the UL9 protein is polyubiquitinated in vivo and that the interaction between NFB42 and the UL9 protein is dependent upon phosphorylation of the UL9 protein. These results suggest that the interaction of the UL9 protein with NFB42 results in its polyubiquitination and subsequent degradation by the 26S proteasome. They suggest further a mechanism by which latency of herpes simplex virus 1 can be established in neuronal cells.
Herpes simplex virus 1 (HSV-1) is a clinically important pathogen that can generate a variety of infections in humans (1, 2). The primary site of HSV-1 infection is generally the dermal epithelium, where the virus replicates efficiently (3). During lytic infection, ≈75 genes are expressed from the linear 152-kbp HSV-1 genome, of which seven (UL9, UL30, UL42, UL5, UL52, UL8, and UL29) are indispensable for DNA replication (4). The product of the UL9 gene, the UL9 protein, functions to initiate HSV-1 DNA replication. It can bind to and unwind the three origins of HSV-1 DNA replication, and it thereby recruits the replication machinery to these origins (5). The UL9 protein may, therefore, be the limiting component in HSV-1 DNA replication and could be the site at which HSV-1 DNA replication is regulated.
In a yeast two-hybrid screen with the UL9 protein as bait, we identified the human neural F-box 42-kDa protein (NFB42 or FBX2) as a binding partner for the UL9 protein (6). NFB42 is highly enriched in the nervous system (cerebral cortex, midbrain, and cerebellum/brainstem), where it induces growth arrest (7). F-box proteins, so named because the 50-aa F-box motif was identified initially in cyclin F (8), are critical components of the SCF (Skp1–cullin-1–F-box protein) ubiquitin–ligase complex involved in targeting proteins for degradation via the ubiquitin–proteasome pathway (9, 10). Ubiquitin-mediated proteolysis is known to play a critical role in the degradation of short-lived and regulatory proteins (11) and requires at least three components. The 76-aa ubiquitin is activated as a thiol ester in an ATP-dependent reaction by the ubiquitin-activating enzyme (E1). After activation, ubiquitin is transferred from E1 to the one of several ubiquitin-conjugating enzymes (E2). Transfer of ubiquitin to its final protein substrate is catalyzed by the E3 ubiquitin–ligase family. The ubiquitinated proteins are then degraded by the 26S proteasome. Currently, a single E1 enzyme, >13 different E2 enzymes, and >100 E3 enzymes have been identified in humans (12).
The SCF complexes are a group of E3 enzymes composed of Skp1, cullin-1 (Cdc53 in yeast), and an F-box protein. Rbx1 (Roc1) has been identified recently as another component of these complexes (13, 14). Skp1 binds to both cullin-1 and the F-box protein. Rbx1 functions in E2 recruitment, and cullin-1 functions as a bridging molecule in the complex. F-box proteins bind to Skp1 via the F box at amino-terminal regions and also contain substrate-binding domains at their carboxyl termini. It is believed that proteins targeted for degradation are marked first by phosphorylation, which then allows for recognition by the F-box proteins (10, 15).
We report here that the phosphorylated form of the HSV-1 UL9 protein interacts with NFB42, is polyubiquitinated by the ubiquitin ligase, and is then degraded via the 26S-proteasome pathway.
Materials and Methods
Reagents. Carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (MG132) was purchased from Calbiochem. It was dissolved in dimethyl sulfoxide. The Complete protease inhibitor mixture was obtained from Roche Molecular Biochemicals. N-ethylmaleimide and other reagents were obtained from Sigma or sources noted in the text.
Expression Constructs. The DNA sequences corresponding to the full-length NFB42 (FBX2) and UL9 were amplified by PCR from HeLa cDNA and HSV-1 genomic DNA, respectively, with the appropriate pairs of primers and subcloned into the following vectors: pcDNA3.1/myc-His (Invitrogen) to generate pcDNA3.1-NFB42 and pcDNA3.1-UL9 for transfection into human embryonic kidney (HEK) 293T cells and for in vitro transcription/translation. The prokaryotic expression vector pFLAG-MAC (Sigma) was used to generate pFLAG-UL9 for the production of amino-terminal flag epitope-tagged UL9 protein in Escherichia coli strain DH5α. pcDNA3HA-Ub, which encodes hemagglutinin (HA)-tagged ubiquitin (HA-Ub), and pcDNA3HA-UbΔG, which encodes the HA-Ub mutant (HA-UbΔG) in which the carboxyl-terminal glycine residue is deleted, were obtained as generous gifts from Tony Eissa (Baylor College of Medicine, Houston).
Cell Culture, Transfections, and Cell Lysis. HEK293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% FBS (HyClone), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C with 5% CO2/95% air in a humidified incubator. The cells were seeded onto 60-mm plates 12–16 h before transfection. Transfection of HEK293T cells was performed by using Lipofectamine 2000 (Invitrogen) and following the manufacturer's instructions. To inhibit proteasome activity, cells were treated with MG132 at the indicated concentrations for 6 h before harvesting. To prepare a total cell extract, HEK293T cells were harvested 40 h posttransfection by centrifugation, washed twice with PBS (140 mM NaCl/2.7 mM KCl/10 mM Na2HPO4/1.8 mM KH2PO4, pH 7.4), and lysed by keeping them on ice for 30 min in 50 mM Tris·HCl, pH 7.4/150 mM NaCl/1% Triton X-100, in the presence of the Complete protease inhibitor mixture and 5 mM N-ethylmaleimide to minimize protein degradation. Protein concentrations were determined by the Bradford method (16) with BSA as a standard. Equal amounts of protein from each lysate were analyzed by SDS/PAGE and immunoblotting.
Purification of Recombinant UL9 Protein. Recombinant UL9 protein produced in Spodoptera frugiperda 21 (Sf21) insect cells by infection with baculovirus encoding the full-length UL9 was purified as described (17). For the production of flag-tagged UL9 protein in E. coli DH5α, protein induction was achieved by adding 0.3 mM isopropyl d-thiogalactopyranoside to cells in exponential phase and then incubating for 4 h at 30°C with shaking. Bacterial cells were pelleted and resuspended in extraction buffer A (50 mM Tris·HCl, pH 8.0/5 mM EDTA/0.25 mg/ml lysozyme) supplemented with Complete protease inhibitor mixture. After incubation at room temperature for 20 min, extraction buffer B (1.5 M NaCl/0.1 M CaCl2/0.1 M MgCl2/0.02 mg/ml DNase I) containing protease inhibitors was added at 1/10 vol. Cells were then treated by brief sonication, and cellular debris was removed by centrifugation at 15,000 × g for 30 min. The supernatants were mixed with an anti-flag M2 affinity gel (Sigma) preequilibrated with Tris-buffered saline (50 mM Tris·HCl, pH 7.4/150 mM NaCl). The bound proteins were eluted with 100 mM glycine (pH 3.5). The UL9 protein was >90% pure as determined by SDS/PAGE followed by staining with Coomassie brilliant blue R-250 (data not shown). Protein concentrations were determined as described above.
In Vitro Transcription and Translation of NFB42. Synthesis of full-length NFB42 by in vitro transcription and translation of pcDNA3.1-NFB42 was carried out in a 50-μl reaction volume with the TnT coupled reticulocyte lysate system (Promega) in the presence of l-[35S]methionine (New England Nuclear) according to the manufacturer's instructions. Newly synthesized protein was analyzed by SDS/PAGE.
Western Blot Analysis. Cell lysates (100 μg), immunoprecipitated proteins, or purified UL9 protein (5 μg each) was mixed with Laemmli sample buffer (60 mM Tris·HCl, pH 6.8/2% SDS/10% glycerol/100 mM DTT/0.005% bromophenol blue) and subjected to SDS/PAGE. The proteins were transferred electrophoretically onto nitrocellulose membranes (Amersham Biosciences). After blocking and extensive washing of the membranes, the proteins were probed with rabbit polyclonal anti-UL9 antibody, rabbit anti-HA antibody (Sigma), rabbit anti-phosphothreonine antibody (Research Diagnostics, Flanders, NJ), rabbit anti-phosphoserine antibody (Research Diagnostics), or monoclonal anti-β-actin (Sigma). The immunoblots were examined by enhanced chemiluminescence (Amersham Biosciences).
Immunoprecipitation. To immunoprecipitate the ubiquitin-conjugated UL9 proteins, cell lysates (500 μg) were incubated with anti-UL9 antibody in a total volume of 500 μl of immunoprecipitation buffer (10 mM Tris·HCl, pH 7.4/150 mM NaCl/1% Triton X-100/1 mM EDTA/1 mM EGTA/protease inhibitor cocktail) and allowed to rock gently at 4°C for 1 h. The immunocomplexes were recovered by incubation with protein A–agarose beads (30 μl of 25% solution; Roche Molecular Biochemicals) preequilibrated with immunoprecipitation buffer for 2 h at 4°C. After extensive washing, the immunoprecipitated proteins were eluted by heating at 95°C for 5 min in 2× Laemmli sample buffer and analyzed by SDS/PAGE and immunoblotting. To obtain the immunocomplex of 35S-labeled NFB42 and UL9 protein, the same procedure was applied, except that detection was by autoradiography.
In Vivo Ubiquitination Assays. HEK293T cells were cotransfected with 0.8 μg of pcDNA3.1-UL9, 4.0 μg of the vector, pcDNA3HA-Ub, or pcDNA3HA-UbΔG. Cells were harvested 36 h posttransfection. Total cell extracts were prepared as described above, subjected to immunoprecipitation with anti-UL9 antibody, and then analyzed by Western blotting with anti-HA or anti-UL9 antibody.
Results
The Steady-State Level of UL9 Protein Can Be Regulated by NFB42 and the 26S Proteasome. The finding that NFB42 interacts with the HSV-1 UL9 protein suggested that the HSV-1 UL9 protein can be degraded by the ubiquitin ligase–26S proteasome system. Protein degradation via the ubiquitin–proteasome pathway involves two steps. In the first step, proteins are modified by covalent attachment of a variable number of ubiquitin molecules. In the second step, the ubiquitin-tagged protein is degraded by the 26S proteasome, and the ubiquitin is recycled by the ubiquitin carboxyl-terminal hydrolases (18, 19).
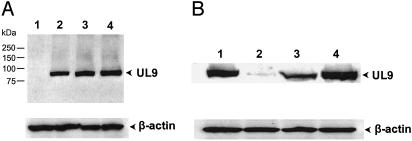
To determine whether the level of UL9 protein is affected by the 26S proteasome, HEK293T cells were transiently transfected with a DNA construct expressing the UL9 protein in the presence or absence of proteasome inhibitor MG132 (20). The cells were lysed, and the lysates were analyzed by SDS/PAGE and then immunoblotted with antibody directed against the UL9 protein. As shown in Fig. 1A, treatment of cells expressing the UL9 protein with increasing amounts of MG132 resulted in an increase in the level of UL9 protein, presumably as a consequence of inhibition of the 26S proteasome. In extracts of cells cotransfected with constructs containing the UL9 and NFB42 genes, the level of UL9 protein was reduced significantly, indicating that the steady-state level of the UL9 protein can be regulated by NFB42 (Fig. 1B, lane 2). Treatment of these cells with MG132 restored the UL9 protein to nearly normal levels (Fig. 1B, lane 3). These results suggest that interaction of the UL9 protein with NFB42 results in the ubiquitination of the UL9 protein and its subsequent degradation by the 26S proteasome.
Fig. 1.
Effect of proteasome inhibitor MG132 and NFB42 on the level of UL9 protein. (A) HEK293T cells were transiently transfected with pcDNA3.1-UL9 expressing the UL9 protein (lanes 2–4). The cells were treated with dimethyl sulfoxide (lane 2) or MG132 (lane 3, 0.5 μM; lane 4, 1.0 μM) for 6 h, and harvested 40 h posttransfection. The cells were lysed, and the lysates were analyzed by SDS/PAGE, transferred to a nitrocellulose membrane, and blotted with anti-UL9 antibody as described in Materials and Methods. Lane 1, untreated HEK293T cells. (B) HEK293T cells were treated as follows: transfected with the UL9-containing plasmid alone (lane 1); cotransfected with constructs containing both UL9 and NFB42 (lane 2); cotransfected with constructs containing UL9, NFB42, and 1 μM MG132 for 6 h (lane 3); or cotransfected with UL9 and vector (lane 4). The level of UL9 protein was determined as described for A. Equivalence of loading was controlled with anti-β-actin antibody.
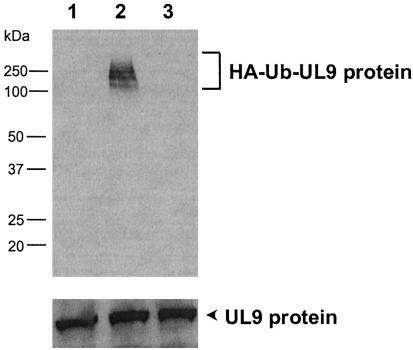
UL9 Protein Is Polyubiquitinated in Vivo. To determine whether the UL9 protein is ubiquitinated in vivo, HEK293T cells were cotransfected with plasmids expressing the UL9 protein and HA-Ub (21). In a parallel experiment, a plasmid encoding HA-Ub was replaced with either the vector DNA or with a plasmid expressing HA-UbΔG, in which the carboxyl-terminal glycine residue, which forms a thiol ester with the ubiquitin-activating enzyme, E1, is deleted (22). Thirty-six hours posttransfection, cell extracts were prepared and subjected to immunoprecipitation with an anti-UL9 antibody, followed by immunoblotting with an anti-HA antibody, to detect ubiquitin-conjugated UL9 protein. With extracts from cells expressing both the UL9 protein and HA-Ub, polyubiquitinated UL9 protein intermediates were detected as a high molecular weight ladder (Fig. 2, lane 2). In contrast, high molecular weight bands could not be detected in cells cotransfected with the mutant plasmid expressing HA-UbΔG (Fig. 2, lane 3) or with the vector (Fig. 2, lane 1).
Fig. 2.
In vivo ubiquitination of HSV-1 UL9 protein by exogenous ubiquitin. HEK293T cells were cotransfected with plasmids containing UL9 and the vector (lane 1), HA-Ub (lane 2), or HA-UbΔG (lane 3). Thirty-six hours posttransfection, the cell lysates (500 μg) were subjected to immunoprecipitation with anti-UL9 antibody, and the immunoprecipitates were analyzed by Western blotting with anti-HA antibody as described in Materials and Methods. Note that the polyubiquitinated UL9 protein intermediates are detected as a high molecular weight ladder (lane 2) and that the UL9 protein is not ubiquitinated by the mutant HA-UbΔG (lane 3). The Lower blot was generated by reprobing the Upper blot with anti-UL9 antibody and reflects the expressed UL9 protein in each lane.
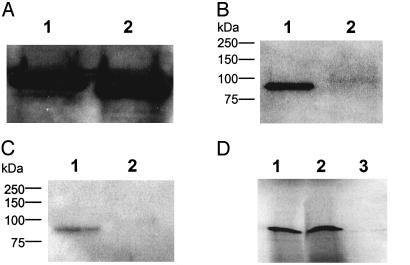
Phosphorylation of UL9 Protein Is Required for Interaction with NFB42. F-box proteins of the ubiquitin ligase–SCF complex target phosphorylated proteins for degradation (10, 23, 24). We performed a computer analysis of potential phosphorylation sites in the UL9 protein and found that it can be phosphorylated at many sites by a variety of protein kinases. To determine whether binding of NFB42 to the UL9 protein requires that the UL9 protein be phosphorylated, as is the case for other SCF substrates such as the pRB-related p130 protein (24) and p27kip1 (25), the UL9 protein expressed in E. coli was compared with that expressed in Sf21 insect cells. Insect cells can perform a variety of posttranslational modifications, including phosphorylation, glycosylation, acylation, amidation, isoprenylation, and carboxymethylation (26, 27). In contrast, proteins expressed in bacteria have few, if any, posttranslational modifications (28). As shown in Fig. 3 B and C, lanes 2, the UL9 protein purified from baculovirus-infected insect cells was phosphorylated at both serine and threonine residues. Only very slight phosphorylation was observed, however, with the E. coli-expressed UL9 protein. The UL9 proteins expressed in E. coli and Sf21 cells showed very similar reactivity with anti-UL9 antibody as judged by Western blotting (Fig. 3A). The Sf21- and E. coli-expressed UL9 proteins were incubated with 35S-labeled in vitro transcribed/translated NFB42, and the mixtures were immunoprecipitated with anti-UL9 antibody. The immunocomplexes were then resolved by SDS/PAGE and visualized by autoradiography. As shown in Fig. 3D, lane 1, interaction between the UL9 protein and NFB42 was observed only in the case of the phosphorylated UL9 protein.
Fig. 3.
Phosphorylation-dependent interaction of UL9 protein with NFB42. (A) UL9 protein (5 μg) purified from baculovirus-infected Sf21 insect cells (lane 1) and E. coli (lane 2) was subjected to SDS/PAGE and blotted with anti-UL9 antibody, as described in Materials and Methods. (B and C) The UL9 proteins purified from Sf21 insect cells and from E. coli were probed with anti-phosphoserine antibody (B, lanes 1 and 2, respectively) and anti-phosphothreonine antibody (C, lanes 1 and 2, respectively). (D) Purified UL9 protein (5 μg) from Sf21 insect cells (lane 1) and E. coli (lane 3) was mixed with 35S-labeled in vitro transcribed/translated NFB42 and immunoprecipitated with anti-UL9 antibody. The immunocomplexes were resolved by SDS/PAGE and visualized by autoradiography as described in Materials and Methods. Lane 2 shows the input 35S-labeled NFB42 added to form the immunocomplexes.
Discussion
The ubiquitin–proteasome pathway has been shown to play a crucial role in a number of cellular processes [including control of the cell cycle, regulation of gene expression, differentiation, apoptosis, DNA repair, and DNA replication (18, 29, 30)] as well as in a number of viral infections. A functionally active viral ubiquitin-conjugating enzyme was first discovered in African swine fever virus (31). The human papillomavirus E6 protein promotes the ubiquitination and degradation of p53 and thereby prevents apoptosis (32). The herpes virus-associated ubiquitin-specific protease contributes to the stabilization of p53 by the deubiquitination of p53 (33). The stimulation of the onset of HSV-1 lytic infection mediated by the RING (zinc-binding) finger protein Vmw110 (ICP0), which is the product of the HSV-1 immediate-early gene 1 and is known to have E3 ubiquitin-ligase activity (34, 35), is inhibited by inactivation of the ubiquitin–proteasome pathway (36). This finding demonstrates that the transcriptional activity of a viral genome can be regulated by the ubiquitin–proteasome pathway.
By using the yeast two-hybrid screen, we observed that the neural F-box protein NFB42 can interact with the HSV-1-encoded UL9 protein (6). The interaction between NFB42 and the UL9 protein was confirmed by in vitro coimmunoprecipitation (6). The UL9 protein is a multifunctional nuclear protein whose properties include origin-specific DNA binding and DNA-dependent ATPase and helicase activities; the UL9 protein is likely the site at which HSV-1 DNA replication is regulated.
Our findings that (i) there is an increase in the level of UL9 protein in cells treated with a specific proteasome inhibitor, MG132, and that (ii) cotransfection of HEK293T cells by constructs containing genes for the UL9 protein and NFB42 results in a decrease in the level of the UL9 protein indicate that the HSV-1 UL9 protein can be degraded by the ubiquitin– proteasome pathway after interaction with NFB42. Our finding that the UL9 protein is ubiquitinated in vivo supports this idea further. The pattern of ubiquitin conjugation to UL9 protein resembles that of the bovine papillomavirus E1 (which also has origin-specific DNA-binding and ATP-dependent DNA helicase activity; ref. 37), the human papillomavirus type 18 E2 (which is required for viral DNA replication), the E1 viral helicase (38), and the p53 tumor suppressor (32).
Recognition of substrates for ubiquitination by the E3 ubiquitin ligases is a key step in the control of protein stability. F-box proteins have been shown to play a role in targeting the substrates for ubiquitination in a phosphorylation-dependent reaction (39). More than 50 mammalian cDNAs that encode F-box proteins have been found (40). We found that NFB42 binds specifically to the phosphorylated form of UL9 protein, which is similar to the binding of Cln 1 and 2 by SCFGrr1 (41), Sic1 by SCFcdc4 (42), and IκB-β-catenine by SCFβ-TrCP/E3RS (43). It has been found also that NFB42 recognizes N-glycosylated proteins (44). Posttranslational modification of proteins can affect their stability and function profoundly, typically, by altering their enzymatic properties or their capacity to interact with other proteins. A number of the proteins encoded by the HSV-1 genome are phosphorylated during virus infection. These proteins include the tegument proteins (VP 1/2, VP 13/14, VP16, and VP24) and the immediate-early regulatory proteins (ICP0, ICP4, ICP22, and ICP27). Recently, Isler and Schaffer demonstrated that the UL9 protein is phosphorylated during HSV-1 infection by metabolic labeling (45).
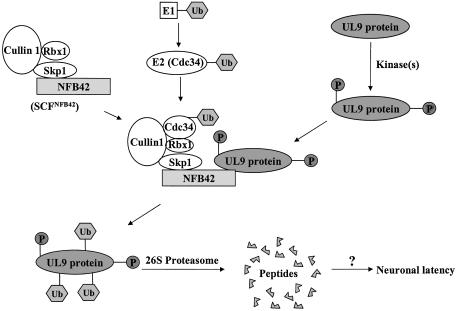
An important feature of the HSV-1 life cycle is its ability to establish a life-long latency within sensory neurons, primarily in the trigeminal and dorsal root ganglia. After primary infection at the periphery, HSV-1 gains access to sensory nerve termini. Within the neurons, the virus can either enter a productive cycle, resulting in the release of progeny virions, or it can establish latency, which allows the virus to evade the host immune system. The latent genomes can reactivate periodically, leading to recurrent episodes of disease. During the latent state only one set of RNAs (the latency-associated transcripts) have been detected (46). Despite the obvious significance of the regulatory mechanisms that govern the establishment of and reactivation from latency, these processes are poorly understood. It has been suggested that the HSV-1-encoded Vmw110 protein plays a role in the balance between the latent and lytic states of the virus (47). Based on our findings that (i) the HSV-1 UL9 protein is polyubiquitinated in vivo, (ii) the steady-state level of the protein is regulated by NFB42, and (iii) interaction with NFB42 is dependent on the phosphorylation of UL9 protein (and on a previous report demonstrating a high level of NFB42 mRNA and protein in the nervous system, but not in other tissues; ref. 7), we suggest that in neurons, UL9 protein, phosphorylated by cellular kinase(s), is recognized by the neuron-specific F-box protein, NFB42, and then degraded via the ubiquitin–proteasome pathway. We suggest that this degradation leads to the inhibition of initiation of HSV-1 DNA replication and possibly to the establishment of neuronal latency (Fig. 4).
Fig. 4.
Model depicting the role of NFB42 in the recognition and ubiquitination of HSV-1 UL9 protein. The UL9 protein is phosphorylated in HSV-1-infected cells by cellular kinase(s). In neurons, the phosphorylated UL9 protein can be recognized by NFB42, a neuron-specific F-box protein. NFB42 is linked to the SCF complex by interaction between the F box within NFB42 and Skp1, which results in the formation of SCFNFB42. Skp1 also binds to cullin-1; Cdc34, an E2 ubiquitin-conjugating enzyme, binds to the SCFNFB42 and ubiquitinates the UL9 protein bound to NFB42. Rbx1 functions in E2 recruitment. Cullin-1 functions as a bridging molecule in the SCF complex. The polyubiquitinated UL9 proteins are degraded by the 26S proteasome, leading to the inhibition of initiation of HSV-1 DNA replication and, ultimately, to neuronal HSV-1 latency.
Acknowledgments
We thank Dr. Tony Eissa (Baylor College of Medicine) for providing the cDNA encoding HA-Ub and HA-UbΔG, and Dr. Gennady Ilyin for valuable discussions. This work was supported by National Institutes of Health Research Grant AI26538 (to I.R.L.).
Abbreviations: HA, hemagglutinin; HA-Ub, HA-tagged ubiquitin; HA-UbΔG, HA-Ub mutant; HEK, human embryonic kidney; HSV-1, herpes simplex virus 1; SCF, Skp1–cullin-1–F-box protein; Sf21, Spodoptera frugiperda 21.
References
- 1.Croen, K. D., Ostrove, J. M., Dragovic, L. J., Smialek, J. E. & Straus, S. E. (1987) N. Engl. J. Med. 317, 1427–1432. [DOI] [PubMed] [Google Scholar]
- 2.Nahmias, A. J. & Roizman, B. (1973) N. Engl. J. Med. 289, 781–789. [DOI] [PubMed] [Google Scholar]
- 3.Yeung-Yue, K. A., Brentjens, M. H., Lee, P. C. & Tyring, S. K. (2002) Dermatol. Clin. 20, 249–266. [DOI] [PubMed] [Google Scholar]
- 4.Boehmer, P. E. & Lehman, I. R. (1997) Annu. Rev. Biochem. 66, 347–384. [DOI] [PubMed] [Google Scholar]
- 5.Lee, S. S. & Lehman, I. R. (1997) Proc. Natl. Acad. Sci. USA 94, 2838–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eom, C. Y. & Lehman, I. R. (2002) Proc. Natl. Acad. Sci. USA 99, 1894–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erhardt, J. A., Hynicka, W., DiBenedetto, A., Shen, N., Stone, N., Paulson, H. & Pittman, R. N. (1998) J. Biol. Chem. 273, 35222–35227. [DOI] [PubMed] [Google Scholar]
- 8.Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W. & Elledge, S. J. (1996) Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- 9.Feldman, R. M., Correll, C. C., Kaplan, K. B. & Deshaies, R. J. (1997) Cell 91, 221–230. [DOI] [PubMed] [Google Scholar]
- 10.Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J. & Harper, J. W. (1997) Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz, A. L. & Ciechanover, A. (1999) Annu. Rev. Med. 50, 57–74. [DOI] [PubMed] [Google Scholar]
- 12.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- 13.Kamura, T., Koepp, D. M., Conrad, M. N., Skowyra, D., Moreland, R. J., Iliopoulos, O., Lane, W. S., Kaelin, W. G., Jr., Elledge, S. J., Conaway, R. C., et al. (1999) Science 284, 657–661. [DOI] [PubMed] [Google Scholar]
- 14.Tan, P., Fuchs, S. Y., Chen, A., Wu, K., Gomez, C., Ronai, Z. & Pan, Z. Q. (1999) Mol. Cell 3, 527–533. [DOI] [PubMed] [Google Scholar]
- 15.Verma, R., Annan, R. S., Huddleston, M. J., Carr, S. A., Reynard, G. & Deshaies, R. J. (1997) Science 278, 455–460. [DOI] [PubMed] [Google Scholar]
- 16.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S. S., Dong, Q., Wang, T. S. & Lehman, I. R. (1995) Proc. Natl. Acad. Sci. USA 92, 7882–7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornitzer, D. & Ciechanover, A. (2000) J. Cell Physiol. 182, 1–11. [DOI] [PubMed] [Google Scholar]
- 19.Voges, D., Zwickl, P. & Baumeister, W. (1999) Annu. Rev. Biochem. 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- 20.Lee, D. H. & Goldberg, A. L. (1998) Trends Cell Biol. 8, 397–403. [DOI] [PubMed] [Google Scholar]
- 21.Ellison, M. J. & Hochstrasser, M. (1991) J. Biol. Chem. 266, 21150–21157. [PubMed] [Google Scholar]
- 22.Kolodziejski, P. J., Musial, A., Koo, J. S. & Eissa, N. T. (2002) Proc. Natl. Acad. Sci. USA 99, 12315–12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koepp, D. M., Schaefer, L. K., Ye, X., Keyomarsi, K., Chu, C., Harper, J. W. & Elledge, S. J. (2001) Science 294, 173–177. [DOI] [PubMed] [Google Scholar]
- 24.Tedesco, D., Lukas, J. & Reed, S. I. (2002) Genes Dev. 16, 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrano, A. C., Eytan, E., Hershko, A. & Pagano, M. (1999) Nat. Cell Biol. 1, 193–199. [DOI] [PubMed] [Google Scholar]
- 26.Hoss, A., Moarefi, I., Scheidtmann, K. H., Cisek, L. J., Corden, J. L., Dornreiter, I., Arthur, A. K. & Fanning, E. (1990) J. Virol. 64, 4799–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloc, M., Reddy, B., Crawford, S. & Etkin, L. D. (1991) J. Biol. Chem. 266, 8206–8212. [PubMed] [Google Scholar]
- 28.Marston, F. A. (1986) Biochem. J. 240, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hershko, A., Ciechanover, A. & Varshavsky, A. (2000) Nat. Med. 6, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 30.Laney, J. D. & Hochstrasser, M. (1999) Cell 97, 427–430. [DOI] [PubMed] [Google Scholar]
- 31.Hingamp, P. M., Arnold, J. E., Mayer, R. J. & Dixon, L. K. (1992) EMBO J. 11, 361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheffner, M., Huibregtse, J. M., Vierstra, R. D. & Howley, P. M. (1993) Cell 75, 495–505. [DOI] [PubMed] [Google Scholar]
- 33.Li, M., Chen, D., Shiloh, A., Luo, J., Nikolaev, A. Y., Qin, J. & Gu, W. (2002) Nature 416, 648–653. [DOI] [PubMed] [Google Scholar]
- 34.Boutell, C., Sadis, S. & Everett, R. D. (2002) J. Virol. 76, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagglund, R. & Roizman, B. (2002) Proc. Natl. Acad. Sci. USA 99, 7889–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Everett, R. D., Orr, A. & Preston, C. M. (1998) EMBO J. 17, 7161–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malcles, M. H., Cueille, N., Mechali, F., Coux, O. & Bonne-Andrea, C. (2002) J. Virol. 76, 11350–11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellanger, S., Demeret, C., Goyat, S. & Thierry, F. (2001) J. Virol. 75, 7244–7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koepp, D. M., Harper, J. W. & Elledge, S. J. (1999) Cell 97, 431–434. [DOI] [PubMed] [Google Scholar]
- 40.Ilyin, G., Serandour, A., Pigeon, C., Rialland, M., Glaise, D. & Guguen-Guillouzo, C. (2002) Gene 296, 11–20. [DOI] [PubMed] [Google Scholar]
- 41.Skowyra, D., Koepp, D. M., Kamura, T., Conrad, M. N., Conaway, R. C., Conaway, J. W., Elledge, S. J. & Harper, J. W. (1999) Science 284, 662–665. [DOI] [PubMed] [Google Scholar]
- 42.Orlicky, S., Tang, X., Willems, A., Tyers, M. & Sicheri, F. (2003) Cell 112, 243–256. [DOI] [PubMed] [Google Scholar]
- 43.Deshaies, R. J. (1999) Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida, Y., Chiba, T., Tokunaga, F., Kawasaki, H., Iwai, K., Suzuki, T., Ito, Y., Matsuoka, K., Yoshida, M., Tanaka, K. & Tai, T. (2002) Nature 418, 438–442. [DOI] [PubMed] [Google Scholar]
- 45.Isler, J. A. & Schaffer, P. A. (2001) J. Virol. 75, 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones, C. (1998) Adv. Virus Res. 51, 81–133. [DOI] [PubMed] [Google Scholar]
- 47.Everett, R. D., Meredith, M., Orr, A., Cross, A., Kathoria, M. & Parkinson, J. (1997) EMBO J. 16, 1519–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]