Abstract
The WW domains are small proteins that contain a three-stranded, antiparallel β-sheet. The 40-residue murine FBP28 WW domain rapidly formed twirling ribbon-like fibrils at physiological temperature and pH, with morphology typical of amyloid fibrils. These ribbons were unusually wide and well ordered, making them highly suitable for structural studies. Their x-ray and electron-diffraction patterns displayed the characteristic amyloid fiber 0.47-nm reflection of the cross-β diffraction signature. Both conventional and electron cryomicroscopy showed clearly that the ribbons were composed of many 2.5-nm-wide subfilaments that ran parallel to the long axis of the fiber. There was a region of lower density along the center of each filament. Lateral association of these filaments generated twisted, often interlinked, sheets up to 40 nm wide and many microns in length. The pitch of the helix varied from 60 to 320 nm, depending on the width of the ribbon. The wild-type FBP28 fibers were formed under conditions in which multiexponential folding kinetics is observed in other studies and which was attributed to a change in the mechanism of folding. It is more likely that those phases result from initial events in the off-pathway aggregation observed here.
Keywords: protein, two-state, intermediate, temperature jump, light scattering
Resolving how a protein folds and unfolds is not only a challenge in itself, but is very important for understanding protein misfolding and associated diseases, such as spongiform encephalopathy and amyloidoses (1, 2). The complex nature of protein folding requires a multidisciplinary approach, combining kinetics, structural analysis, and computer simulation. Recently, there has been increasing synergy between conclusions drawn from experimental models of fast-folding proteins and those from molecular dynamics simulation (3–6), suggesting that an accurate atomic-level description of protein folding, unfolding, and misfolding is now a realistic prospect (7).
The ultrafast-folding WW domain proteins (6, 8) are ubiquitous in eukaryotes and are involved in cellular signaling, vesicular trafficking, and have been implicated in a number of disease pathologies (9). WW domains are typically 35–45 residues long and adopt a three-stranded, antiparallel β-sheet topology (Fig. 1), with conserved tryptophan residues at the N and C termini (10–13). These small domains follow a two-state transition in both equilibrium and kinetic experiments (6, 8). Further, there is surprisingly good agreement between molecular dynamics simulations and experiments for a number of WW domain homologues, with the emerging consensus being that folding is rate limited by formation of a β-hairpin (4, 6), although it has been suggested that this mechanism can be modulated by changes in temperature (6).
Fig. 1.
The typical distorted three-stranded, antiparallel β-sheet topology of wild-type FBP28 WW (11). The conserved N-terminal (W8) and C-terminal (W30) tryptophan residues are shown in blue and red, respectively. This figure was generated by using pymol, which can be accessed at http://pymol.sourceforge.net.
A recent study (14) proposes that folding of the murine FBP28 WW domain is a multistate, unimolecular reaction with formation of a populated intermediate during folding. They suggest that changes in temperature or sequence can modulate the folding pathway from three- to two-state. These results are in contrast to our previous work (8), where we find that the folding of the murine FBP28 WW domain (and two other homologues) follows two-state transitions. In other studies, however, we had noted substantial time-dependent aggregation of the wild-type FBP28 WW domain under similar conditions and temperatures to those used by Kelly and coworkers (14). We show here that aggregation did indeed occur under those conditions and that the extended fibers that were formed had structural characteristics typical of those found in amyloidosis. These fibers displayed typical nucleation-growth kinetics, thioflavin T and Congo red binding, as well as electron and x-ray diffraction properties that were fully consistent with amyloid fiber formation. The fibrous ribbons that we observe, however, are much wider than normally found in other amyloids, because of lateral association of the subfilaments and were accordingly extremely suitable for high-resolution image analysis. The high-resolution structural data we present are consistent with these fibers containing parallel β-structure, although we cannot exclude the possibility that there is a disordered mix of parallel and anti-parallel main-chain alignment.
Materials and Methods
Reagents. All reagents were of AnalaR grade. Ultrapure guanidine hydrochloride was purchased from ICN. Thioflavin T (65% dye content) and Congo red (97% dye content) were purchased from Sigma. WW domains and mutants were prepared from recombinant DNA (10). Protein purity was determined from SDS/PAGE gels and analytical chromatography (gel filtration and reverse phase). Mass and identity of peptides were confirmed by matrix-assisted laser desorption ionization MS (MALDI-MS). Protein concentrations were determined spectrophotometrically (15). Buffers were made by using appropriate ratios of free acids and sodium salts in 18 M-Ω water, with ionic strength corrected where necessary by addition of either NaCl or Na2SO4: 10 mM sodium phosphate buffer, pH 7.0; 20 mM sodium phosphate buffer, pH 7.0 (ionic strength of 150 mM, NaCl used); 20 mM sodium 3-[N-morpholino]propanesulphonic acid (Mops), pH 7.0 (ionic strength of 150 mM, Na2SO4 used). Concentration of guanidine hydrochloride was determined by refractive index measurements (16).
Light-Scattering Experiments. Peptide stock solutions of 1–1.5 mM were freshly made from lyophilized material on the day of use to eliminate the possibility of seeding effects on aging. New disposable plastic cuvettes (1-cm path length) were routinely used to avoid cross-seeding effects from deposits that remained in reusable quartz cuvettes, and highly reproducible results were obtained. Turbidity was measured in a Varian Cary 500 Scan Spectrometer (Varian Scientific Instruments, Cary, NC).
Typically, cuvettes containing buffer only were preequilibrated for at least 30 min (with continual stirring) before manual addition of the concentrated peptide stock to the desired final concentration (between 10 and 500 μM). Light-scattering kinetics was recorded at 350, 450, and 550 nm, approximately every 60 s until 2,000–5,000 min, depending on the reaction rate. A spectral bandwidth of 1 nm was used in conjunction with an averaging time of 3–4 s. Essentially identical results were obtained from repeat experiments by using recombinant proteins from independent preparations.
Thioflavin T Assay. A slightly modified version of a standard procedure (17) was used at 37°C and performed in an Aminco-Bowman SLM series 2 luminescence spectrometer (SLM Instruments, Urbana, IL). Excitation was at 440 nm (4-nm slit width), and emission was recorded from 460 to 650 nm (8-nm slit width). Twenty microliters of 100 μM WW FBP28 samples were incubated at 37°C for 2 days in 20 mM sodium phosphate buffer, pH 7.0, ionic strength 150 mM, and added (after resuspension where necessary) to the assay buffer (2 ml of 65 μM thioflavin T in the same buffer), which was preincubated for ≈10 min with constant stirring. Samples were preincubated for 5 min before fluorescence measurement. A strong increase in emission intensity at 482 nm was diagnostic of amyloid formation.
Congo Red Assay. Suspensions of wild-type FBP28 precipitates from earlier fiber growth experiments were concentrated by a brief centrifugation in a benchtop microfuge. A small drop of the suspension was spread into streaks on a glass microscope slide and allowed to dry partially. Staining was performed by covering the precipitate with a filtered, saturated solution of Congo red (dissolved in water). The sample was overlaid with a coverslip and examined after 20-min incubation with the stain. In some areas of the sample, streaks of red staining were visible to the naked eye. The preparation was examined in a Nikon Optiphot microscope equipped with a linear polarizer, a rotating analyzer, an intense halogen source, and a strain-free ×60 NA 1.4 planapo oil-immersion objective lens. Micrographs were taken with the analyzer accurately crossed (darkest background), and with a rotation of a few degrees (gray background) with a Nikon CoolPix camera equipped with a conversion lens system and a c-mount adaptor.
Temperature-Jump (T-Jump) Fluorescence Measurements. We measured the kinetics of unfolding of wild-type FBP28 WW domain by using a modified DIA-RT capacitor-discharge T-jump fluorimeter (Dia-Log, Düsseldorf, Germany) as described (8), or a commercially available Hi-Tech PTJ-64 capacitor-discharge T-jump fluorimeter fitted with a 63-μl cell (Hi-Tech, Salisbury, U.K.). Tryptophan fluorescence was excited at 280 nm and fluorescence emission at >320 or >360 nm was measured by using cutoff filters. T-jumps of ≈3.5°C (from 6.5 to 10°C, or from 21.5 to 25°C) were obtained by using a 15-kV discharge from a 10-nF capacitor in the Hi-Tech instrument. T-jumps of ≈2°C (from 37 to 39.5°C) were obtained by using 33-kV discharges from a 10-nF capacitor in the Dia-Log instrument. Typically, 10–50 transients were averaged in each experiment. Data were fitted by using kaleidagraph (Synergy Software, Reading, PA) and, where relevant, were fitted to a two-state model (8).
Electron Microscopy. Negatively stained samples were made by absorbing onto air glow-discharged carbon-coated grids. A 3-μl drop was applied for 1 min, and the grid surface was subsequently washed with 50 μl of Tris-buffered saline, pH 7.0. Staining was performed by using 50 μl of either 2% (wt/vol) uranyl acetate or 1% (wt/vol) phosphotungstic acid (PTA) neutralized with LiOH, before blotting dry. Grids were examined with an FEI 208 electron microscope (Philips Electron Optics, Hillsbro, OR) at 80 kV. Cryoelectron microscopy (cryoem) samples were prepared on Quantifoil (Jena, Germany) holey carbon grids treated by glow discharge in an amylamine vapor. Samples were 10-fold concentrated by a short centrifugation, followed by gentle resuspension of the pellet. A 3-μl drop was blotted and immediately frozen by plunging into liquid C2H6 cryogen by standard techniques (18). Frozen grids were examined with a Gatan 626 (Pleasanton, CA) cold-stage by using an FEI Tecnai F20 200-keV field emission gun electron microscope (Philips Electron Optics) and were recorded on Kodak SO163 film.
X-Ray Diffraction. A suspension of FBP28 fibers in water at 10 mg/ml was dried on a hydrophobic glass slide, and a piece of the resulting film mounted in an x-ray camera with the plane of the film parallel to the x-ray beam. There is a tendency for the meridional region of the x-ray diffraction pattern to be toward the plane of the protein film and the equatorial region to be perpendicular to the film. A Rikagu (The Woodlands, TX) rotating anode tube was the x-ray source, and the pattern was recorded at a distance 150 mm from the specimen by using a Mar USA (Evanston, IL) research image plate camera with a 15-min exposure time.
Results
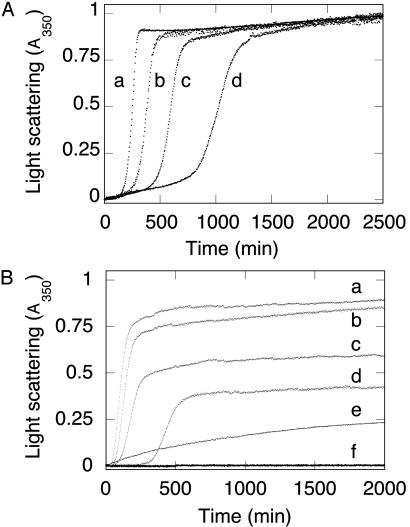
Kinetics of Light Scattering. There were three main phases in the kinetics of light scattering for wild-type FBP28 incubated at 37°C (Fig. 2A): a lag phase, a burst or growth phase, and finally a plateau. The lag phase was shortened, and the growth phase accelerated by increasing either the protein concentration (Fig. 2 A) or the ionic strength of the buffer (Fig. 2B). There was no change in the free energy of unfolding of wild-type FBP28 as a function of increasing ionic strength under the conditions where fiber growth was accelerated (data not shown). We observed significant light-scattering kinetics at peptide concentrations as low as 50 μM, with visible aggregation in the cuvette during the plateau phase. At all concentrations >50 μM, >95% of the starting material precipitated within 24–48 h. There was no visible aggregation of wild-type FBP28 at a concentration of ≈10 μM, although there was still evidence of a multiphasic increase in light scattering, and the presence of a low number of filaments by electron microscopy (data not shown). These observations were consistent with nucleated aggregation of wild-type FBP28 WW, with protein and salt concentration influencing the nucleation process, which appears to be rate determining. Similar aggregation kinetics is observed for amyloid fibrils growing in vitro (19–22).
Fig. 2.
Aggregation kinetics of wild-type FBP28 WW domain as a function of peptide concentration. (A) Light scattering of wild-type FBP28 WW versus peptide concentration at 37°C. Traces: a, 500 μM FBP28; b, 200 μM FBP28; c, 100 μM FBP28; d, 50 μM FBP28. Data were normalized to a common scale by dividing the observed absorbance at a given time by the signal at the end of the experiment. At all concentrations >10 μM, the reactions reached an apparent endpoint, with >95% of the peptide having been precipitated by centrifugation after an ≈24-h incubation. (B) Comparison of aggregation kinetics of wild-type and mutant FBP28 WW domains. Peptide (100 μM) at 37°C: Traces: a, wild-type FBP28, ionic strength = 150 mM; b, wild type, ionic strength = 100 mM; c, wild type, ionic strength = 50 mM; d, wild type, ionic strength = 16 mM; e, FBP28 W30A, ionic strength = 150 mM; f, oxidized FBP28 V5C/E35C, ionic strength = 150 mM. Under conditions where wild-type FBP28 aggregation is faster than at lower ionic strength, we saw no aggregation for FBP28 V5C/E35C. Identical results were obtained for the disulfide-reduced mutant. Lower-order association is observed for FBP28 W30A, which is consistent with the 10-nm particles seen by using electron microscopy.
As controls, we examined two mutants of FBP28 (Fig. 2B). Under identical conditions for formation of wild-type fibers, there was no visible aggregation or associated light scattering for a circularized mutant (V5C/E35C) of the FBP28 domain, which has essentially identical three-dimensional structure as the wild-type domain (N.F., T.D.S., and A.R.F., unpublished data). Under conditions where the disulfide bond was fully reduced by β-mercaptoethanol, we still did not observe an increase in light scattering. We did see some slow-light-scattering kinetics for the W30A mutant of FP28 (Fig. 2B). The kinetics observed, however, was qualitatively very distinct from that seen for the wild-type domain and was more typical of lower-order aggregation events. Further, whereas wild-type samples tested positive for amyloid fiber by using the standard thioflavin T assay, the mutants did not (data not shown). Similarly, wild-type FBP28 precipitates gave a strong apple-green birefringence under a polarizing microscope after staining with Congo red (data not shown), which is consistent with this protein forming amyloid fiber.
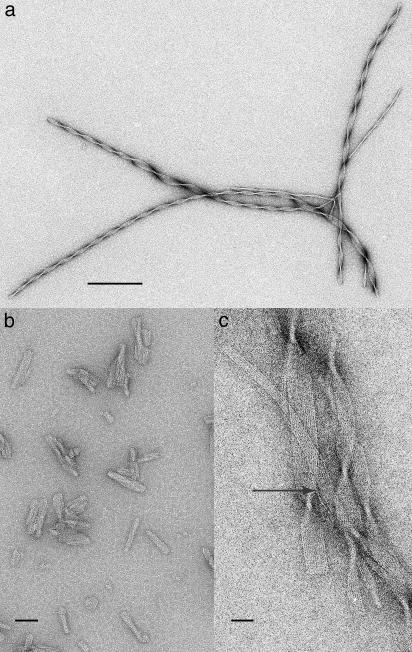
Electron Microscopy Characterization of FBP28 Aggregates. We examined the insoluble material by using electron microscopy at various stages in the aggregation reaction. Solutions containing aggregated wild-type FBP28 WW domain incubated at pH 7.0 and 37°C for 2 days contained very long (>1 μm) twirling ribbon-like fibers. Fig. 3a shows the helical nature of a group of fibers. Depending on the width of the fibers (between 5 and 40 nm), the pitch of the helical twist varied between 60 and 320 nm. Similar fibers were seen for wild-type FBP28 WW domain at all peptide concentrations from 10 to 500 μM with a morphology that was apparently independent of ionic strength between 16 and 150 mM. At higher magnification (Fig. 3c), the ribbons showed clear 2.5-nm subfilaments running along the long axis of the ribbon that were often interlinked by single and small groups of subfilaments. The existence of discrete single subfilaments indicated that the molecule was mostly localized within the 2.5-nm block. The ribbon ends were very sharp and cut at right angles, which implied that the growth of the subfilaments depended on stabilization from neighboring molecules.
Fig. 3.
Electron microscopy of negatively stained wild-type FBP28 aggregates. (a) Fibers were formed by incubation of 100 μM wild-type FBP28 in phosphate buffer, pH 7.0, at 37°C for 72 h. Negative staining was performed with uranyl acetate. The twisting filaments have morphology typical of amyloid fiber. (b) Under the same conditions as in a, wild-type FBP28 formed small protein chunks after only 15 min of incubation, which means that it may be filament nuclei. (c) Higher-magnification images of wild-type FBP28 incubated as in a. Rods (2.5 nm) of protein (white) run the length of the ribbon in groups from 3 to 18. Negatively stained with phosphotungstic acid (PTA) at pH 7.0, the ribbon separates into an individual thread (arrow). (Scale bars, 200 nm in a; 50 nm in b; and 20 nm in c.)
The species formed within the first 15 min of incubation (under conditions where the lag phase lasts ≈300 min) consisted of mixtures of aggregates of significantly smaller dimensions than the mature fibers (Fig. 3b). Many showed 2.5-nm striations and would appear to correspond to an early stage in fiber formation with the nuclei for fiber formation being a unit of ≈2.5-nm diameter, which rapidly grew into rods that can associate into twisted sheets.
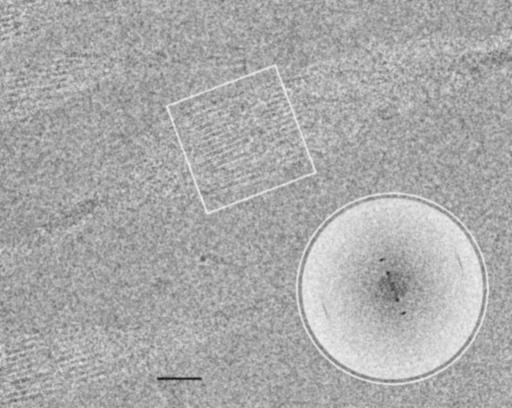
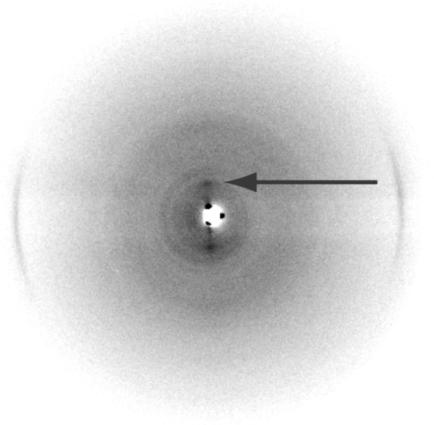
Unstained images from cryo-em (Fig. 4) showed the same morphology found by negative staining. The 37-nm-wide ribbon twists in the frozen water layer and shows 2.5-nm subfilaments with a 1.25-nm region of lower density, where the sheet is face on to the beam. The computed Fourier transform of this region (Fig. 4 Inset) showed the two orders of 2.5 nm on the equator (across the ribbon), together with a 0.47-nm spacing on the meridian (along the ribbon). Although the subfilament image is only clearly resolved in the face-on section, the meridional spacing can be found in transforms taken from small areas along the complete length and no other equatorial spacings were found (data not shown). Because the image covers an entire 180° twist of the ribbon, the subfilaments must be associated into a two-dimensional sheet, otherwise a range of different superpositions would be expected [as found for actin bundles (23)], because the image resolves detail to at least 0.47 nm. X-ray diffraction of the dried material (Fig. 5) confirmed the strong 0.47-nm meridional spacing, together with weak equatorial spacings at ≈2.8 and 1.0 nm, typical of amyloid (24) and scrapie prion fibers (25).
Fig. 4.
Wild-type FBP28 filaments at ≈5 mg/ml unstained and frozen in amorphous ice. Imaged by using cryoelectron microscopy (cryo-em) at 1.8 μm underfocus (protein is dark), this single twist is a 180° rotation of the ribbon's projected density. The dense crossovers are clear, but the 2.5-nm rods of density are visible only close to where the ribbon is face-on. There is a low-density region within each rod. (Scale bar, 20 nm.) (Inset) A calculated Fourier transform of the boxed area. Across the ribbon are two orders of 2.5-nm spacing, whereas there is an 0.47-nm spacing along the ribbon.
Fig. 5.
X-ray diffraction of wild-type FBP28 fibers (≈10 mg/ml) after washing into water and drying down onto film. The x-ray pattern was taken in the plane of the film and shows the 0.47-nm arcs typical of cross-β structure found in amyloid filaments. A weak equatorial spacing is also seen at ≈2.8 nm (arrow).
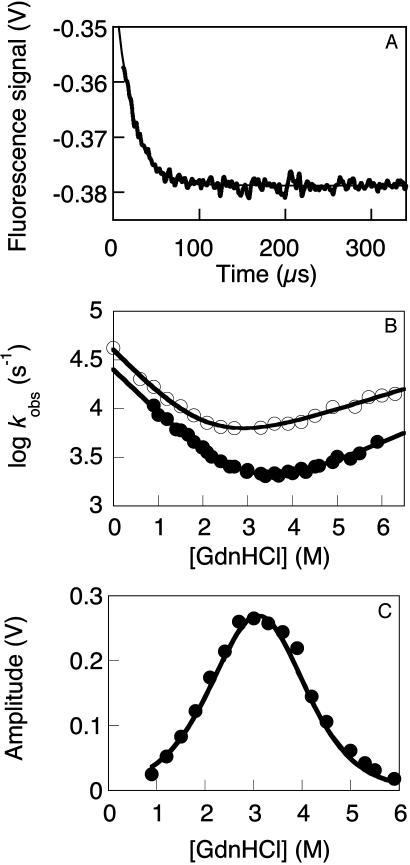
Folding Kinetics. At pH 7.0 and an ionic strength of 150 mM, we observed only the faster of the two phases reported by Nguyen et al. (14), which is consistent with our earlier observations (Fig. 6A). There was also no evidence of the slow phase at the lower ionic strength they used (data not shown). We had previously noted that aggregation was occurring at elevated temperatures, and so restricted our detailed folding studies to conditions that would not be subject to artifacts. At the lower temperatures of 10 and 25°C, we found no evidence for populated intermediates in the denaturant dependence of the observed rate constants for folding and unfolding of wild-type FBP28 (Fig. 6B), or in the measured kinetic amplitudes (Fig. 6C). This observation also extends to >20 mutants of FBP28 (N.F., M.P., and A.R.F., unpublished data). Thus, the folding kinetics of wild-type FBP28 could be quantitatively fitted to an apparent two-state transition at lower, nonphysiological temperatures, where we have little evidence for aggregation.
Fig. 6.
Two-state folding kinetics of wild-type FBP28 WW domain. (A) The T-jump-induced of unfolding at 39.5°C in 10 mM sodium phosphate buffer pH 7.0 (ionic strength of 150 mM) fits to a single exponential function, as in lower ionic strength buffers (data not shown). (B) Denaturant dependence of the observed rate for T-jump-induced unfolding of wild-type FBP28 in 20 mM sodium Mops, pH 6.5 (150 mM ionic strength). The kinetic data fitted well to a two-state transition at 10°C (•) and 25°C (○). There is no evidence of a rollover consistent with populated refolding (41), despite the broad range of denaturant concentrations, over which the experiments were measured. βT values of ≈0.7 were measured at these temperatures, which is consistent with the values observed for this and other WW domain homologues in urea (8) (βT is the ratio of the relative sensitivities of logarithms of the rate and equilibrium constants of folding, with respect to urea concentration). (C) Denaturant dependence of kinetic amplitudes for unfolding of wild-type FBP28 in 20 mM sodium Mops, pH 6.5, at 10°C. The observed unfolding amplitudes (•) show little deviation from the theoretical amplitudes calculated for a fully two-state system (dashed line), even at the lowest denaturant concentration measured. The kinetic amplitudes expected for a two-state system, as a function of the denaturant concentration, were calculated by using only the experimentally determined ΔCp (8) and mean mD-N for unfolding at equilibrium (0.86 ± 0.03 kcal/mol; M, average of 10 independent measurements where mD – N = δ(free energy of denaturation) δ[GdnHCl]). Thus, there is little evidence to support formation of a populated intermediate at 10 and 25°C. Similar two-state kinetics and amplitudes were observed for 20 other mutants of FBP28 (data not shown).
Discussion
The most significant aspect of this study is that, under physiological conditions, a small, ultrafast-folding protein rapidly formed extended fibers that were morphologically identical to those observed in amyloidogenic proteins (2), and those fibers were highly suitable for structural characterization. Importantly, electron microscopy of the fibers isolated in the various kinetic phases provided direct structural confirmation of the kinetics. The aggregation kinetics, thioflavin T binding, and Congo red birefringence properties of the fibers formed by the wild-type FBP28 WW domain, in conjunction with the relatively high-resolution structural information, are fully consistent with this protein forming true amyloid fibers, albeit very wide ones. As the FBP28 WW domain can adopt the native conformation under conditions where there is inherent competition to form an alternative quaternary architecture, this protein is a particularly good model system for understanding detailed aspects of protein folding and misfolding, both in vitro and in silico.
Fiber Structure. There are many models for amyloid fiber structures, recently reviewed by Wetzel (26). Indeed, because of the wide variety of protein structures, it is likely that different proteins form different fibrils (27), some of which may differ radically, but others that will be variations of canonical forms, because of the constraints of packing extraneous elements. Even simple designed peptides form fibrils that are apparently built up from antiparallel β-strands (28).
The structure of insect silk from the lace-wing fly egg-stalk (29) is the archetypal cross-β structure. It has hydrogen-bonded strands that pack in a regular antiparallel meander across the fiber axis with the molecule spanning 2.5 nm. The main-chain hydrogen bonding holds the strands 0.47-nm apart, and the antiparallel nature produces a repeat between chains of the same orientation of 0.94 nm. We find only the strong 0.47-nm meridional spacing in the FBP28 fibers, however, and no evidence for regular antiparallel β-sheet organization in the diffraction pattern. Considering that FBP28 normally folds into a three-stranded, antiparallel β-sheet, the observation of what could be a parallel β-sheet fibrous assembly is, perhaps, a surprising result. However, Wetzel (26) has summarized other evidence supporting a parallel β-structure, including H/D-exchange (30) and solid-state NMR (31). Perutz et al. (32) argue that polyglutamine repeats could form a nucleus of 37–40 residues that could support further growth into a parallel β-helix with 20 residues per turn. If the main chain of FBP28 forms a helix, then most of the 40 residues would be involved, unlike disease-state amyloid fibers, where a compact core is surrounded in a coat of unstructured protein. The computed Fourier transform of the cryo-em image showed a distribution of intensity for the 0.47-nm reflection similar to that expected from successive turns in a helix, such as a β-helix, and if a first-order Bessel function of helical diffraction, suggests an angular rise of ≈5°.
It is possible to reconcile features of the Perutz β-helix model (32) with other naturally occurring β-helical folds and the FBP28 WW domain fibers. A parallel β-helix motif, initially found in soluble proteins from plants (33, 34), is also present in bacterial, insect, and yeast proteins (35). It has a similar number of residues per helical turn to the Perutz model and the extensive backbone hydrogen bonding between the parallel β-sheets. Unlike the Perutz structure, however, the plant fold has a compact core that largely excludes solvent molecules and it appears to be stabilized by tracts of stacked aromatic and aliphatic groups (34, 36), residue that interact in the β-sheet of native wild-type FBP28.
Electron microscope images show the FBP28 subfilaments to be 2.5 nm across and these rods of density also show a lower-density region in their center, giving rise to a 1.25-nm periodicity. Equatorial reflections of this spacing in x-ray diffraction have been interpreted as the side-chain packing of β-sheets (22). It is difficult to interpret our image in this way, because the main repeat of 2.5 nm is too large and the 1.25-nm periodicity is clearly its second order, not from an orthogonal view. It is currently unclear as to whether the region of low electron density corresponds to a solvent-filled channel within the fibers or a more compact core. We have observed, however, that the FBP28 fibers turn blue-black in iodine solutions, the diagnostic test for starch. It is interesting to speculate on why this color change develops when FBP28 or other amyloid deposits (37) are exposed to iodine solution. Because we know that no sugars are involved, it could be that amyloids sequester iodine in an environment similar to that in helical amylose.
The very unusual power of the 0.47-nm reflection found in electron microscope images of Alzheimer's disease peptides (38), and in FBP28 fibers is quite uncharacteristic of assemblies of biological molecules investigated in crystals (39). This power comes from the strength of the molecules to withstand the intense radiation damage of an electron beam. Crystals of aromatic compounds like anthracene have a lifetime nearly an order of magnitude longer than an aliphatic compound like paraffin (40). It could be that the six aromatic (four tyrosine and two tryptophan) residues present in the wild-type FBP28 WW domain are important to forming and stabilizing the amyloid fiber structure, which is consistent with the mutational data and resistance to radiation damage that we present here.
The native-like mutant FBP28 V5C/E35C, which has the N and C termini circularized by a disulfide linkage or its reduced form did not form fibers, indicating that the native fold did not directly form fibers, and that either or both the N and C termini were involved in the stabilization of filamentous structure. The mutant W30A also did not form fibers. Interestingly, W30A inhibited the formation of wild-type fibers under some conditions (data not shown), indicating that hybrids of wild-type and mutant strands formed destabilized structures. These data are all consistent with much of the length of the protein being involved in fibril interactions. Increasing the buffer ionic strength facilitates both fiber nucleation and growth without affecting the free energy of denaturation of the native protein. It may be that charge neutralization in the denatured state is responsible for the accelerated fiber formation. The existence of flat ends to the ribbons is strongly indicative of lateral interactions being necessary for growth.
Do Artifacts Give Rise to Multistate Kinetics? Biphasic exponential reaction kinetics is observed for unfolding of wild-type FBP28 WW domain at 39.5°C and in 10 mM sodium phosphate buffer, pH 7.0, when fluorescence lifetime is used as the spectroscopic probe (14), which is highly sensitive to aggregation state. The slower kinetic phase is observed only at final temperatures in the range of 33–58°C [after T-jumps of 8–13°C (14)], conditions that overlap those where we have found fibril formation. Nguyen et al. (14) also note that truncation of the C-terminal region of FBP28 WW or mutation of W30 to phenylalanine or alanine also eliminates the slower phase. (We find that the W30A mutation abrogates fiber formation.) In contrast, the changes in intrinsic fluorescence in our T-jump experiments were presumably not sensitive to the aggregation state of FBP28, and so were consistent with our earlier work (8) that shows two-state kinetics for the actual folding-refolding transition. We also found that wild-type FBP28 WW domain folds by a two-state transition at 10 and 25°C in urea (8) and guanidine hydrochloride. Nguyen et al. (14) do report that the kinetics of the additional (slow) phase does not depend on protein concentration. The concentration range they examined, however, overlaps that where we found protein aggregation and were able to isolate fibrils and their putative nuclei. Perhaps their measurements reflect some initiation event that is effectively independent of concentration in this range. Given that aggregation definitely occurs within the temperature range of their studies, and that the occurrence of multiexponential kinetics correlates with the conditions where aggregation is high, it is most likely that the additional phase relates to early phases in an off-pathway aggregation process and not to a switch of folding mechanism.
Acknowledgments
This article is dedicated to the memory of Max Perutz for his inspirational studies. We thank Professors Chris Dobson and Hartmut Oschkinat, Dr. Alexey Murzin, and Sandra Tremmel for helpful discussions; and Drs. Brad Amos, Stefanie Reichelt, and Heather Peto for help with the Congo red birefringence assay. We acknowledge Dr. C. M. Johnson's extensive technical expertise and input. This work was supported by the Agouron Institute (Grant AL-SA1-99.3) and the Medical Research Council (Cambridge, U.K.).
Abbreviations: T-jump, temperature jump; GdnHCl, guanidine hydrochloride.
References
- 1.Dobson, C. M. (2001) Philos. Trans. R. Soc. London B 356, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sunde, M. & Blake, C. C. F. (1998) Q. Rev. Biophys. 31, 1–39. [DOI] [PubMed] [Google Scholar]
- 3.Mayor, U., Johnson, C. M., Daggett, V. & Fersht, A. R. (2000) Proc. Natl. Acad. Sci. USA 97, 13518–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson, N., Pires, J. R., Toepert, F., Johnson, C. M., Pan, Y. P., Daggett, V., Oschkinat, H. & Fersht, A. R. (2001) Proc. Natl. Acad. Sci. USA 98, 13008–13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayor, U., Guydosh, N. R., Johnson, C. M., Grossman, J. G., Sato, S., Jas, G. S., Freund, S. M., Alonso, D. O, Daggett, V., and Fersht, A. R. (2003) Nature 421, 863–867. [DOI] [PubMed] [Google Scholar]
- 6.Jager, M., Nguyen, H., Crane, J. C., Kelly, J. W. & Gruebele, M. (2001) J. Mol. Biol. 311, 373–393. [DOI] [PubMed] [Google Scholar]
- 7.Fersht, A. R. & Daggett, V. (2002) Cell 108, 573–582. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson, N., Johnson, C. M., Macias, M., Oschkinat, H. & Fersht, A. R. (2001) Proc. Natl. Acad. Sci USA 98, 13002–13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudol, M. (1996) Exp. Mol. Med. 28, 65–69. [Google Scholar]
- 10.Macias, M. J., Hyvonen, M., Baraldi, E., Schultz, J., Sudol, M., Saraste, M. & Oschkinat, H. (1996) Nature 382, 646–649. [DOI] [PubMed] [Google Scholar]
- 11.Macias, M. J., Gervais, V., Civera, C. & Oschkinat, H. (2000) Nat. Struct. Biol. 7, 375–379. [DOI] [PubMed] [Google Scholar]
- 12.Verdecia, M. A., Bowman, M. E., Lu, K. P., Hunter, T. & Noel, J. P. (2000) Nat. Struct. Biol. 7, 639–643. [DOI] [PubMed] [Google Scholar]
- 13.Wiesner, S., Stier, G., Sattler, M. & Macias, M. J. (2002) J. Mol. Biol. 324, 807–822. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen, H., Jager, M., Moretto, A., Gruebele, M. & Kelly, J. W. (2003) Proc. Natl. Acad. Sci. USA 100, 3948–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill, S. C. & von Hippell, P. H. (1989) Anal. Biochem. 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 16.Pace, C. N. (1986) Methods Enzymol. 131, 3286–3299. [DOI] [PubMed] [Google Scholar]
- 17.Levine, H. (1993) Protein Sci. 2, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubochet, J., Adrian, M., Chang, J. J., Homo, J. C., Lepault, J., McDowall, A. W. & Schultz, P. (1988) Q. Rev. Biophys. 21, 129–228. [DOI] [PubMed] [Google Scholar]
- 19.Perutz, M. F. & Windle, A. H. (2001) Nature 412, 143–144. [DOI] [PubMed] [Google Scholar]
- 20.Lomakin, A., Chung, D. S., Benedek, G. B., Kirschner, D. A. & Teplow, D. B. (1996) Proc. Natl. Acad. Sci. USA 93, 1125–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarrett, J. T. & Lansbury, P. T. (1993) Cell 73, 1055–1058. [DOI] [PubMed] [Google Scholar]
- 22.Fandrich, M. & Dobson, C. M. (2002) EMBO J. 21, 5682–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmid, M. F., Matsudaira, P., Jeng, T.-W., Jakana, J., Towns-Andrews, E. & Chiu, W. (1991) J. Mol. Biol. 221, 711–725. [DOI] [PubMed] [Google Scholar]
- 24.Perutz, M. F., Johnson, T., Suzuki, M. & Finch, J. T. (1994) Proc. Natl. Acad. Sci. USA 91, 5355–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inouye, H. & Kirschner, D. A. (1998) J. Struct. Biol. 122, 247–255. [DOI] [PubMed] [Google Scholar]
- 26.Wetzel, R. (2002) Structure (London) 10, 1031–1036. [DOI] [PubMed] [Google Scholar]
- 27.Jimenez, J. L., Nettleton, E. J., Bouchard, M., Robinson, C. V., Dobson, C. M. & Saibil, H. R. (2002) Proc. Natl. Acad. Sci. USA 99, 9196–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de la Paz, M. L., Goldie, K., Zurdo, J., Lacroix, E., Dobson, C. M., Hoenger, A. & Serrano, L. (2002) Proc. Natl. Acad. Sci. USA 99, 16052–16057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geddes, A. J., Parker, K. D., Atkins, E. D. T. & Beighton, E. (1967) J. Mol. Biol. 32, 343–358. [DOI] [PubMed] [Google Scholar]
- 30.Kheterpal, I., Zhou, S., Cook, K. D. & Wetzel, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13597–13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tycko, R. (2000) Curr. Opin. Chem. Biol. 4, 500–506. [DOI] [PubMed] [Google Scholar]
- 32.Perutz, M. F., Finch, J. T., Berriman, J. & Lesk, A. (2002) Proc. Natl. Acad. Sci. USA 99, 5591–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoder, M. D., Lietzke, S. E. & Jurnak, F. (1993) Structure (London) 1, 241–251. [DOI] [PubMed] [Google Scholar]
- 34.Yoder, M. D., Keen, N. T. & Jurnak, F. (1993) Science 260, 1503–1507. [DOI] [PubMed] [Google Scholar]
- 35.Jenkins, J. & Pickersgill, R. (2001) Prog. Biophys. Mol. Biol. 77, 111–175. [DOI] [PubMed] [Google Scholar]
- 36.Lietzke, S. E., Scavetta, R. D., Yoder, M. D. & Jurnak, F. (1996) Plant Physiol. 111, 73–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Virchow, R. (1854) Virchows Arch. Pathol. Anat. Physiol. Klin. Med. 6, 135–138. [Google Scholar]
- 38.Serpell, L. C. & Smith, J. M. (2000) J. Mol. Biol. 299, 225–231. [DOI] [PubMed] [Google Scholar]
- 39.Unwin, P. N. T. & Henderson, R. (1975) J. Mol. Biol. 94, 425–440. [DOI] [PubMed] [Google Scholar]
- 40.Joy, R. T. (1973) in Advances in Optical and Electron Microscopy, eds. Barer, R. & Cosslett, V. E. (Academic, London), Vol. 5, p. 297. [Google Scholar]
- 41.Matouschek, A., Kellis, J. T., Serrano, L., Bycroft, M. & Fersht, A. R. (1990) Nature 346, 440–445. [DOI] [PubMed] [Google Scholar]