Abstract
Background
As the developmental costs of genomic tools decline, genomic approaches to non-model systems are becoming more feasible. Many of these systems may lack advanced genetic tools but are extremely valuable models in other biological fields. Here we report the development of expressed sequence tags (EST's) in an orthopteroid insect, a model for the study of neurobiology, speciation, and evolution.
Results
We report the sequencing of 14,502 EST's from clones derived from a nerve cord cDNA library, and the subsequent construction of a Gene Index from these sequences, from the Hawaiian trigonidiine cricket Laupala kohalensis. The Gene Index contains 8607 unique sequences comprised of 2575 tentative consensus (TC) sequences and 6032 singletons. For each of the unique sequences, an attempt was made to assign a provisional annotation and to categorize its function using a Gene Ontology-based classification through a sequence-based comparison to known proteins. In addition, a set of unique 70 base pair oligomers that can be used for DNA microarrays was developed. All Gene Index information is posted at the DFCI Gene Indices web page
Conclusion
Orthopterans are models used to understand the neurophysiological basis of complex motor patterns such as flight and stridulation. The sequences presented in the cricket Gene Index will provide neurophysiologists with many genetic tools that have been largely absent in this field. The cricket Gene Index is one of only two gene indices to be developed in an evolutionary model system. Species within the genus Laupala have speciated recently, rapidly, and extensively. Therefore, the genes identified in the cricket Gene Index can be used to study the genomics of speciation. Furthermore, this gene index represents a significant EST resources for basal insects. As such, this resource is a valuable comparative tool for the understanding of invertebrate molecular evolution. The sequences presented here will provide much needed genomic resources for three distinct but overlapping fields of inquiry: neurobiology, speciation, and molecular evolution.
Background
Identifying the genetic basis of interesting phenotypic variation in non-model systems is often limited by the lack of sophisticated molecular resources, such as complete genome sequences and DNA microarrys, that are available in model genetic taxa such as Drosophila [1], Anopheles [2], Caenorhabditis [3] and Apis [4]. However, the declining costs of developing genomic tools and the proliferation of accessible methods by which these tools can be generated holds promise for genomic-scale studies in organisms that offer profound insights into fundamental biological questions. Thus, there is a growing need to develop better genomic resources for these emerging systems.
The Orthoptera contain many such emerging systems. Consisting of over 25,000 species [5], the order Orthoptera is composed of two major lineages, the crickets and katydids (Ensifera) and the grasshoppers (Caelifera) [6,7] which diverged approximately 300 MYA. While well known for their economic impact on world-wide agriculture [8-13], they have been intensively studied in a wide variety of biological areas. For example, orthopterans have been used to study various aspects of neurobiology [14-17], physiology [18-21], behavior [10,22-24], development [17,25-28], sexual selection [29-35], and evolution [7,32,36-43]. However, very few genomic tools have been developed for this group of insects.
While genomic studies of many orthoptera are ongoing [44,45], large scale genomic resources have been developed for only one species in this order, Locusta migratoria (Caelifera) [45,46]. Research on Locusta has produced 12,161 unique sequences and provides a necessary counterpoint to the heavy phylogenetic bias in extant genomic resources. [47-50]. However, as described above, orthopterans are a phylogenetically diverse lineage which are being used to study a broad set of biological questions. The Gene Index presented here was developed to address three distinct but overlapping areas of orthopteran biology: neurobiology, speciation, and evolution.
For over 50 years, the Orthoptera have been used as a neurobiological model system by which the relationship between neural activity, muscular response and behavior are studied [51]. In particular, the study of orthopteran flight and song, or stridulation, have provided valuable insights into the physiological basis of behavior and the structure and function of Central Pattern Generating (CPG) circuits [52-55]. CPG circuits are responsible not only for orthopteran flight and song, but also for nearly all vital functions, such as circulation, respiration, digestion and locomotion, in both vertebrates and invertebrates. Since at least 1973, neuroethologists have called for the development of genetic tools to understand the creation, function, and diversification of the neural circuits responsible for cricket stridulation [56]. One result has been the analysis of the inheritance of species-specific songs [57,58] and a quantitative trait locus study of song (Shaw et al. in press). Yet the tools necessary to study the action and influence of individual genes remain largely absent. The EST's of this Gene Index, since they are derived from a nerve cord library, contain genes expressed in nervous system. Many of the EST's identified here may be involved in the construction of the flight and/or stridulation CPG.
Furthermore, our study organism, Laupala kohalensis, is a superb organism with which to investigate the genetic basis of CPG construction and evolution. The 38 species of Laupala have diverged within the past five million years [59]. The diversification of Laupala has been extraordinarily rapid, as Laupala contains the fastest diversifying arthropod clade recorded to date [59]. The radiation is also noteworthy for the extremely limited number of features that distinguish species. Members of this genus appear morphologically and ecologically similar and many closely related species often differ by fewer than 0.1% of nuclear gene bases [60]. However, pulse rates of male calling songs have diverged extensively in Laupala [61]. Given the diversity of pulse rate CPG's in this clade and the limited amount of genetic divergence that separates species, the release of the Laupala Gene Index will provide an extraordinary genomic tool by which CPG evolution may be studied.
In addition to providing a powerful platform for comparative studies of CPG evolution, Laupala is a well-developed model system for the study of reproductive isolation and the formation of species [33,34,38,59,60,62-66]. The 38 species within this genus are believed to have diverged in part via coordinated evolution in male song and female acoustic preference [33,34,65]. While there exists an extensive body of literature on the evolution of sexual isolation and the formation of species, identifying the specific genetic basis of either process has been limited to an extremely small number of taxa for which the appropriate genetic tools have been developed. The release of this cricket Gene Index will allow researchers to build on the genetic work of Hoy and Paul [56], which demonstrated a polygenetic basis of cricket songs, and Shaw [58,66], which supported Hoy and Paul's findings and identified several chromosomal regions associated with song, by providing the tools necessary to identify specific genes involved in cricket stridulation, sexual isolation and the formation of species. Identifying the genes involved in any of these processes would represent a significant achievement.
From a comparative perspective, the publication of the Laupala Gene Index is a significant advancement in the tools available to study molecular evolution in insects. To date, major insect genome projects have focused primarily on the Diptera (e.g., fruitflies and mosquitoes; [1,2]), Hymenoptera (e.g. honeybee; [67]), and Lepidoptera (moths and butterflies; [68-70]). All of these lineages belong to a single superorder (Endopterygota) and, thus, represent only a small portion of the phylogenetic diversity encompassed by the broader class Insecta (Figure 1 &2). While the evolution of complete metamorphosis (Holometabolous, Endopterygota) was certainly one of the most significant events in the history of insect diversification [71], the heavy phylogenetic bias of previously developed genomic resources has severely limited broader inferences about the evolutionary history of insects in general. Indeed, only recently have researchers begun to address this phylogenetic bias in studies of arthropod evolution [72,73] and the genomes of an Aphid [74] and Louse [75] soon will be available. Therefore, the compilation of a basal insect genomic resource, such as the one presented here, will facilitate genomic comparisons across 350 million years of insect diversification, and will serve as a phylogenetic link to even more distant comparisons, such as crustaceans (e.g.Daphnia) and chelicerates (e.g. tick), and beyond. For example, one of the early developmental studies of arthropod body patterning genes utilized EST sequences cloned from Schistocerca (Orthoptera: Caelifera) and Tribolium (Coleotpera) to demonstrate the homology between the Drosophila hox gene zen and its' human ortholog, HOX3 [76]. Thus, the benefits of developing sophisticated genomic resources for non-model organisms are potentially much broader than typically recognized.
Figure 1.
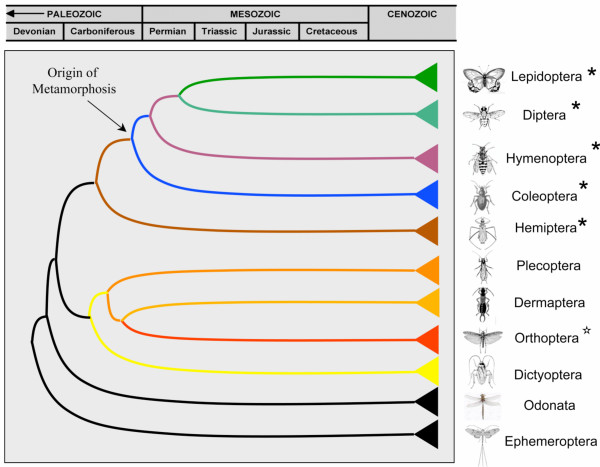
A Simplified winged-insect phylogeny showing the evolutionary origin of complete metamorphosis (adapted from Grimadi and Engel 2005; Figure 4.24, page 146.
Figure 2.
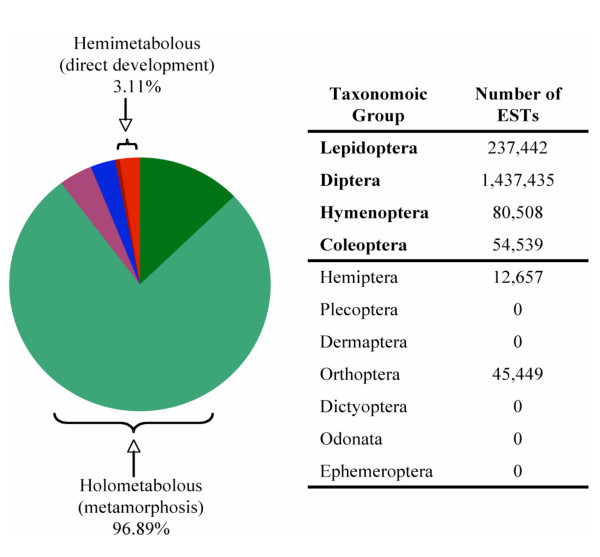
Pie chart showing the heavy phylogenetic bias towards Holometabolous insects in the total number of EST's deposited in NCBI's dbEST database [105].
The current study represents the first major initiative to develop a large genomic resource for a cricket species of the orthopteran suborder Ensifera (crickets and katydids). We present the sequences of 14,502 Expressed Sequence Tags (EST) from a Laupala kohalensis nerve cord cDNA library. We expect that the release of this Gene Index will provide much needed tools for the study of CPG construction and evolution, sexual selection and speciation, and the molecular evolution of arthropods.
Results
Two separate, normalized cDNA libraries were constructed from a single pool of RNA extracted from the nerve cord tissue of several individual crickets. A total of approximately 22,000 clones were isolated from these libraries. 388 clones were sequenced from the first library (LK01); 14114 clones were sequenced from the second library (LK04). A total of 14,502 sequences were generated. Preliminary sequence analysis revealed that 5' end sequencing of the EST's provided higher quality reads than those generated from the 3' end. As a result, the majority of our sequencing effort was directed at sequencing the 5' end of the EST's. 14,261 sequences were generated from the 5' end and 241 sequences were generated from the 3' end of the insert. Of the 14,502 sequences, 14,377 were greater than 100 bases after the vector and linker sequences were stripped. Of these 14,377 sequences, read lengths ranged from 100 bases to 1051 bases. The average read length was 704 bases. Table 1 summarizes the results of the cDNA sequencing and basic bioinformatics analysis. All 14,377 sequences were submitted to GenBank and can be accessed through the accession numbers EH628894-EH643270.
Table 1.
Sequencing results of the two libraries which were examined including raw sequencing results and acceptable sequences after removing poor quality reads and contaminating sequences.
| Pooled LK libraries | Library LK01 | Library LK04 | |||||||
| EST Sequence Total Reads | all reads | 5' end reads only | 3' end reads | all reads | 5' end reads only | 3' end reads | all reads | 5' end reads only | 3' end reads |
| Number of Successful Sequences | 14502 | 14261 | 241 | 388 | 316 | 72 | 14114 | 13945 | 169 |
| Range in Length | 241–1252 | 268–1252 | 241–1128 | 758–1150 | 958–1150 | 758–1102 | 241–1252 | 268–1252 | 241–1128 |
| Mean Length | 1057 | 1058 | 1024 | 1082 | 1092 | 1041 | 1057 | 1057 | 1017 |
| High Quality EST Reads | |||||||||
| Number of Successful Sequences | 14502 | 14261 | 241 | 388 | 316 | 72 | 14114 | 13945 | 169 |
| Range in Length | 64–1096 | 64–1096 | 66–1051 | 68–1074 | 218–1074 | 68–943 | 64–1096 | 64–1096 | 66–1051 |
| Mean Length | 838 | 841 | 619 | 805 | 875 | 499 | 838 | 840 | 670 |
| EST Sequence After Vector Stripping | |||||||||
| Number of Successful Sequences | 14377 | 14158 | 219 | 354 | 295 | 59 | 14023 | 13863 | 160 |
| Range in Length | 100–1051 | 100–949 | 103–1051 | 100–926 | 100–926 | 105–916 | 100–1051 | 100–949 | 103–1051 |
| Mean Length | 704 | 705 | 657 | 486 | 473 | 553 | 710 | 710 | 695 |
A Gene Index was created from these 14,377 acceptable sequences [77]. We identified 8,607 unique sequences, representing 6,032 singletons and 2575 tentative consensus sequences (TCs). Tentative consensus sequences are composed of multiple sequencing reads with overlapping sequence alignments. The 2,575 TCs were derived from 8,345 EST's (Table 2) and ranged in length from 167 bases to 3,317 bases, with an average length of 935 bases. The number of EST's per TC ranged from 2 to 41, with a mean number of 3.24 EST's per TC. The remaining unique sequences were composed of single EST's. Singleton sequences ranged in size from 102 bases to 1019 bases, with an average length of 700 bases (Table 3).
Table 2.
Statistics of Tentative Consensus sequences (TCs)
| Number of TC | 2575 |
| Number of ESTs assembled into TC | 8345 |
| TC size range (bp) | 167–3317 |
| Mean TC length (bp) | 935 |
| Range of number of EST's in TC | 2–41 |
| Average number of EST's in TC | 3.24 |
| Number of TC with >= 20 EST's | 17 |
| Number of TC with < 5 EST's | 2205 |
Table 3.
Statistics of singletons
| Number of singletons | 6032 |
| Singleton size range (bp) | 102–1019 |
| Mean singleton length (bp) | 700 |
| Number of singletons <= 200 bp | 110 |
| Number of singletons between 200 and 500 bp | 505 |
| Number of singletons between 500 and 800 bp | 3860 |
| Number of singletons > 800 bp | 1557 |
The 8,607 unique sequences were translated into all 6 possible reading frames and compared using BLAT [78] against a comprehensive non-redundant protein database maintained by the Dana-Farber Cancer Institute. This database contains ~3 million entries collected from UniProt, SwissPro, RefSeq, GenBank resources and additional sequences from TIGR and its affiliates. The BLAT algorithm is integrated into the gene indexing bioinformatics pipeline to reduce computing times when building and annotating other large gene indices (e.g. human, [79]; mouse, [80]; and rat, [81]). In future releases, the pipeline may be modified to use additional algorithms, such as BLASTX, when working with more limited and/or phylogenetically distinct gene indices such as our cricket gene index.
5,225 of the 8,607 (60.7%) unique sequences had a significant sequence similarity match to an entry in the protein database [see Additional file 1]. 3,382 (39.3%) unique sequences returned no significant matches to entries in the database and no putative function could be assigned to them. However, 2,393 of the 3,382 (70%) sequences that did not return a significant match to a protein in the database were identified by ESTscan [82] as having putative ORF's with an average length of 295 nucleotides. This suggests that the majority of these unidentified EST's are expected to encode a protein and highlights the dearth of genomic information available for basal insect taxa.
The observed sequence similarities produced by the comparative analysis are consistent with our expectations given the tissue from which the cDNA library was constructed. While some of the unique sequences are similar to housekeeping genes, many unique sequences are similar to genes that may influence stridulation (Table 4). For example, several unique sequences are similar to genes that regulate the timing of biological events (e.g. Period and Diapause bioclock protein; Table 4), while others are involved with nervous system signal transduction (e.g. cGMP-gated cation channel protein, G-protein-coupled receptor, Shab-related delayed rectifier K+ channel, Na+/K+/2Cl-cotransporter, Nicotinic acetylcholine receptor non-alpha subunit precursor, Potassium channel tetramerisation domain-containing protein 5, Voltage-dependent anion channel, and Syntaxin 7; Table 4) and others contribute to developmental events that shape either the nervous system (e.g. Even-Skipped; Table 4) or wing development (e.g. Notch, Wnt inhibitory factor 1; Table 4). In addition to potentially influencing our primary phenotype, many of these sequences will be useful to researchers interested in insect neural function (e.g. Calmodulin, Innexin; Table 4) and insect molecular evolution (e.g. Opsin, Dyenin; Table 5).
Table 4.
Genes of neurobiological interest
| Sequence ID | Gene |
| TC1375 | Calmodulin |
| 1099956307901 | Calpain B |
| 1099956293105 | cAMP-dependent protein kinase subunit R2 beta |
| 1099956429052 | cGMP-dependent protein kinase |
| TC588 | cGMP-gated cation channel protein |
| TC140 | Diapause bioclock protein |
| TC1309 | Even-Skipped |
| 1099956350726 | G-protein-coupled receptor |
| 1099817827099 | Innexin |
| 1099817862791 | Intersectin-1 |
| TC1333 | Membrane-associated ring finger |
| 1099956579253 | MscS Mechanosensitive ion channel |
| 1099956736101 | Myosin V |
| 1099956378602 | Na+/K+/2Cl-cotransporter |
| TC1855 | Nicotinic acetylcholine receptor non-alpha subunit precursor |
| TC2167 | Notch |
| 1099956498166 | Period |
| TC1283 | Potassium channel tetramerisation domain-containing protein 5 |
| 1099956317550 | Rab7 |
| TC1866 | Ras-related protein Rab-2 |
| 1099956329054 | Serpentine Receptor |
| TC1295 | Shab-related delayed-rectifier K+ channel |
| 1099956378537 | sodium and chloride-dependent high-affinity choline transporter |
| TC456 | Sparc |
| TC2021 | Stathmin |
| 1099817880653 | Swelling dependent chloride channel |
| 1099817832930 | Syntaxin 7 |
| 1099956598763 | Troponin T |
| TC2416 | Voltage-dependent anion channel |
| 1099956851891 | Wnt inhibitory factor 1 |
Table 5.
Genes of comparative interest. Uncorrected distances between Laupala and the specified taxon are shown, where possible. The mean uncorrected pairwise distance (p) between all taxa (excluding Laupala) is shown for each gene in the final column for comparison. Alignments of each gene are presented as NEXUS files in the online additional files.
| Locusta | Tribolium | Apis | Bombyx | Anopheles | Drosophila | Mean Distance (excluding Laupala) | |
| Actin | 0.0911 | 0.1752 | 0.1262 | 0.1594 | 0.1051 | 0.0911 | 0.1368 |
| Alpha-tubulin | 0.2090 | 0.2143 | 0.2288 | 0.1744 | 0.2135 | 0.1878 | 0.2115 |
| Aquaporin | 0.3164 | 0.4715 | 0.4242 | 0.4814 | 0.4400 | 0.4336 | 0.4485 |
| Dynein (Light Chain) | 0.1741 | 0.2482 | 0.1741 | 0.6043 | 0.2185 | 0.2037 | 0.2111 |
| Histone 2A | 0.3184 | 0.2720 | 0.3081 | 0.2478 | 0.2016 | 0.3218 | 0.3039 |
| HSP40 | 0.3959 | 0.4832 | 0.3592 | 0.3392 | 0.3587 | 0.4049 | 0.4287 |
| Malate Esterase | 0.3056 | 0.4032 | 0.3526 | - | 0.4140 | 0.4430 | 0.3802 |
| Myosin 2 (Light Chain) | 0.2576 | 0.3529 | 0.3132 | 0.3352 | 0.4254 | 0.3856 | 0.3652 |
| Opsin | 0.3430 | - | 0.3630 | - | 0.4173 | 0.4387 | 0.3911 |
| Polyubiquitin | 0.2046 | 0.2292 | 0.2237 | 0.2046 | 0.2846 | 0.2194 | 0.2321 |
Within our unigene set, we identified a number of genes that would be of comparative interest. To explore the Laupala unigene set as a comparative utility we compared the sequence of ten EST's from our unigene set to unigene sets available in Drosophila melanogaster, Anophelese gambiae, Bombyx mori, Apis mellifera, Tribolium casteneum, and Locusta migatoria (Table 5). The results show the evolutionary distinctiveness and phylogenetic distance between Laupala sequences and EST sequences from other genomic models. Across the ten EST's, the mean uncorrected sequence divergence (p) between Laupala and the other insect taxa surveyed was 30%. Furthermore, the mean distance between Laupala and Locusta was 89% that of the mean pairwise distance of all taxa in the analysis. Thus, despite the fact that Laupala and Locusta are both members of the insect order Orthoptera, the sequence divergence between them for this sample of EST's is close to that found among other insect orders.
Of the 5,225 sequences that matched protein entries, 408 sequences could be assigned a Gene Ontology (GO, [83,84]) term (Figures 3,4,5). 572 Biological Process GO terms were associated with predicted amino acid sequences from these 408 sequences. The 25 most frequent Biological Process GO terms are presented in Figure 3. The majority of Biological Process GO terms (488 or 85%) were assigned to five or fewer of the 408 sequences present and no Biological Process GO term was assigned to more than 45 sequences. 275 Molecular Function GO terms were associated with amino acid sequences identified in the 408 unique sequences. The 25 most frequent Molecular Function GO terms are presented in Figure 4. The majority of Molecular Function GO terms (221 or 80%) were assigned to five or fewer sequences. One Molecular Function GO term was assigned to 100 of the 408 sequences (protein binding). 212 Cellular Compartment GO terms were associated with predicted amino acid sequences identified in the 408 unique sequences. The 25 most frequent Cellular Compartment GO terms are presented in Figure 5. The 408 unique sequences contained 106 predicted nuclear proteins, and this was the most frequent Cellular Compartment GO term. Again, the majority of these GO terms, 163 (77%), were assigned to no more than five of the 408 sequences.
Figure 3.
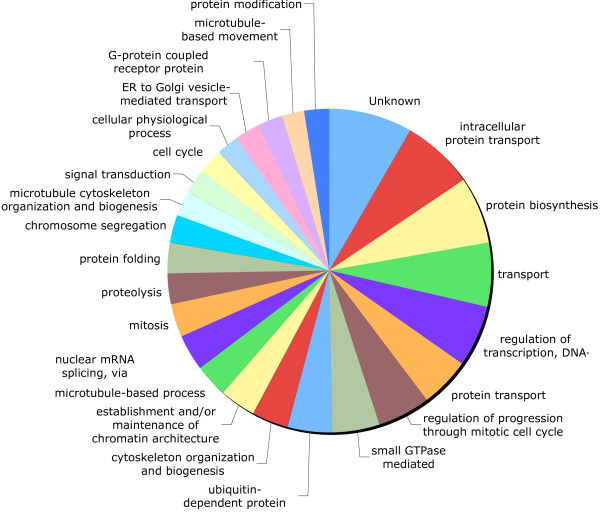
A piechart of the 25 most frequent Biological Process Gene Ontology (GO)terms.
Figure 4.
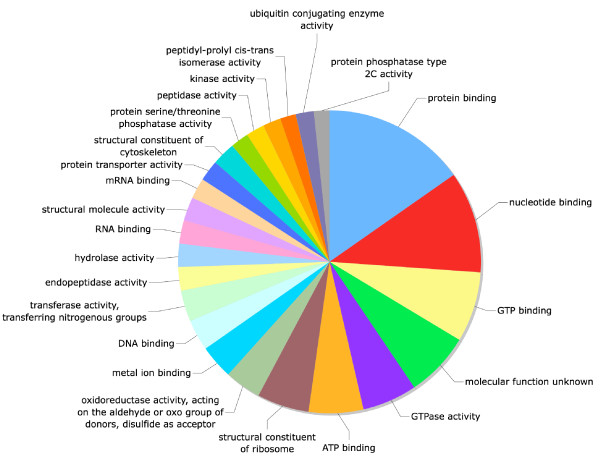
A piechart of the 25 most frequent Molecular Function Gene Ontology (GO)terms.
Figure 5.

A piechart of the 25 most frequent Cellular Compartment Gene Ontology (GO)terms.
The low redundancy of the GO terms, in addition to the large proportion of singletons in the library and the small number of EST's per TC, testify that the normalization was successful and that a large proportion of the genes expressed in the cricket developing nerve cord were identified. The putative function of the singletons and tentative consensus sequences, as inferred from the BLAT comparison and the GO term assignments, is consistent with genes expected to be expressed in a nerve cord.
Discussion
We completed an EST sequencing project to characterize genes expressed in the cricket nerve cord that underlie pulse rate of male song in L. kohalensis. By constructing a cDNA library from nymphal and adult crickets, our aim was to enhance the discovery of genes involved in the construction of the central pattern generating circuit (CPG) underlying rhythmic singing behavior. In addition, we enriched for full-length cDNA by utilizing a template-switching reverse transcriptase (SMART™ technology – BD Clontech, Mountain View, CA). Furthermore, we increased the representation of genes expressed in low-copy number by normalizing our amplified cDNA using a double-stranded nuclease (Trimmer-Direct Kit; Evrogen, Moscow). Sequencing of ~22,000 clones from this library by The Institute for Genomic Research (TIGR) produced 14,502 high quality EST's with an average length greater than 700 bases (Tables 1, 2, 3). Assembly of these EST's produced 8,607 unique sequences. We were then able to annotate 5,225 of these genes based on BLAT protein comparisons against a comprehensive non-redundant protein database maintained by the Dana-Farber Cancer Institute. Of these annotated genes, we could assign gene ontology (GO) terms to 408 genes. The diversity of our library is reflected in the large number of different GO terms assigned to these genes, including 572 Biological Process, 275 Molecular Functions, and 212 Cellular Compartment GO terms, and suggests that we were successful in our attempt to normalize cDNA representation in our library.
Cricket Gene Index
A Gene Index based on our EST sequencing project was assembled and is publicly-available at [85]. This electronic resource consists of a description of the cricket EST library, including a summary of the number of unique sequences, the distribution of tentative consensus (TC) sequences, gene annotations, GO terms, and a set of 70-mer oligonucleotide probes. The cricket Gene Index thus joins more than 30 other animal gene indices hosted by DFCI and represents the second largest EST resource for Orthoptera available online. While the cricket EST project sequenced roughly one third of that sequenced by the Locusta migratoria project (45,754 EST's, [86]) this disparity is not reflected in the total number of unique sequences identified by these two projects (L. migratoria = 12,161 unique sequences versus L. kohalensis = 8,607 unique sequences).
Crickets as models for behavioral genomics
Species of Orthoptera have long served as neurophysiological models of behavior. Our analysis of 14,502 EST sequences and subsequent production of 8607 singletons and tentative consensus sequences from a nerve cord derived library represents a major advance in the available genomic resources for the study of cricket neurophysiology and behavior. This resource will provide valuable tools with which to examine the underlying genetic basis of cricket stridulation, a model for the study of central pattern generation (Table 4). The resources presented here represent the first opportunity to analyze the neurophysiologic process of stridulation at the genomic scale.
Developing additional genomic resources for Laupala
We are utilizing multiple approaches in order to dissect the genetic basis of pulse rate variation in Laupala. In addition to ongoing QTL mapping efforts [64] (Shaw et al. in press), the Laupala Gene Index is a first step towards two additional genetic approaches to our study of pulse rate evolution. First, the oligonucleotide probe set developed from our Gene Index is the backbone of an oligonuclelotide micoarray being constructed to study gene expression in Laupala. These microarrays will be used to study patterns of gene expression across multiple species [87] to identify candidate genes whose expression varies with pulse rate. Second, the EST's are being screened for variation that can be used in a linkage analysis. Placing these EST's on the Laupala linkage map will facilitate comparisons between the QTL analysis and the study of gene expression. The identification of candidate genes that fall within QTL regions will strengthen the support for these candidate genes and guide our choice of which genes to use in functional studies. Furthermore, estimating the linkage relationships of EST's within Laupala and comparing them with known orthologs in model systems will allow us to identify regions of synteny across multiple species. Establishing such areas of synteny is another powerful approach to identifying strong candidate genes [88-90]. Given the now rich genomic resources available in Laupala, the extensive divergence of male song CPG and its influence on reproductive isolation, and the fairly limited genetic divergence within this genus, Laupala represents an excellent system to study the evolutionary genomics of CPG diversification.
In addition, the development of genomic resources in Laupala can be used to tackle some of the most urgent topics in evolutionary biology. Few other systems provide both the genomic tools and evolutionary power necessary to provide an understanding of how gene expression evolves in recently diverged taxa [91]. Furthermore, because male pulse rate plays a critical function in reproductive isolation in this genus, identifying the genes whose expression contributes to the construction of this phenotype will provide insight into how the evolution of gene expression contributes to reproductive isolation during the course of speciation [92].
Comparative genomics in insects
In the last 15 years, there has been a proliferation of genomic resources available for model organisms. As technology has improved, whole genome sequences have become available for a growing number of species and for the first time comparative studies of entire genomes have become possible [93-96]. However, the phylogenetic breadth of insect species in which genomic tools have been developed is extremely limited. For example, of the 37 insect genomes sequencing projects currently completed or under way, 22 (~60%) involve species of Drosophila. The remaining species are either directly related to human health (the mosquitoes Aedes aegypti and Culex pipiens, the Tsetse fly Glossina morsitans, the human louse Pediculus humanus humanus, and the Hemipteran vector of Chaga's disease Rhodnius prolixus) [97], or are of agriculture importance (the red flour beetle Tribolium casteneum, the honey bee Apis mellifera, the silkworm moth Bombyx mori, the pea aphid Acyrthosiphon pisum, and the parasitoid wasp Nasonia vitripennis). The only species with significant genomic tools that is not of biomedical or agricultural importance is the African butterfly (Bicyclus anyana), an evo-devo model for wing pattern development [98]. The vast majority of these insects are holometabolous and possess relatively small genomes [99,100]. However, this severe phylogenetic and genome-size bias limits comparative studies of insect and arthropod evolution (Figure 1 &2). The cricket Gene Index presented here represents a significant contribution to the genomic resources available for comparative molecular studies of basal insect lineages (Table 5). Based on our preliminary comparative analysis, Laupala, a representative of the Orthopteran suborder Ensifera, is as distinct from Locusta, a representative of the Califeran suborder of the Orthoptera, as it is from other insect orders.
Conclusion
We document the sequencing of 14,502 EST's derived from a Laupala kohalensis nerve cord cDNA library. From these 14,502 sequences, 8,607 unique sequences were identified. Just over 60% of the unique sequences, 5,225, had a predicted protein sequence significantly similar to a sequence in a non-redundant protein database. Of these, Gene Ontology terms could be assigned to 408 of the putative proteins. This resource was developed to address fundamental questions of biological interest. Our interests lie in identifying genes that contribute to the diversification of male song pulse rate and, by extension, speciation within the Hawaiian cricket genus Laupala. The release of this resource, however, has a much broader impact than that prescribed by our interests. Neuroethologists studying the construction and function of CPG neural circuits in insects have lamented the lack of available genetic tools necessary to study these vital neurobiological phenotypes. The release of the Laupala Gene Index contributes to meeting this need. Likewise, evolutionary biologists have lacked diverse systems with which fundamental evolutionary processes might be addressed at the genomic scale. Empirical data can be collected using the Laupala resource to examine the evolution of gene expression during the speciation process. Finally, the release of this Gene Index begins to rectify an extreme phylogenetic bias in the availability of genomic resources in insects and will facilitate comparative studies of molecular evolution across 350 MY of arthropod evolution.
Methods
Cricket rearing and RNA isolation
Laupala kohalensis were raised from laboratory-reared parents under identical and constant light (12:12) and temperature (20°C) conditions. Crickets were fed Cricket Chow (Purina) twice weekly. Groups of crickets were reared in quart-sized, glass jars outfitted with moistened Kimwipes (Kimberly-Clark) from hatching. As individuals matured to approximately the 5th post-embryonic instar, 2–4 individuals per group were moved into individual specimen cups and maintained under conditions identical to the jars.
Between the hours of 08:00 and 12:00, groups of crickets were anaesthetized with carbon dioxide, and individuals were digitally imaged using a Leica MZ8 compound microscope mounted with a JVC TK-1280U camera connected to a Power Macintosh 7500/100 Apple computer via the program NIH Image. Individuals were transferred to Corning 1 ml cryovials and snap frozen through the emersion of the cryovials into liquid nitrogen and immediately moved to -70°C. All crickets were sacrificed at 12:00.
The individuals included in this study spanned the putative critical developmental period (instars 5–8) during which the neural circuit responsible for orthopteran stridulation is established [2]. 17 crickets were individually thawed under RNAlater (Ambion) and dissected to remove the nerve cord. Based on the width of the pronotum, individuals were assigned to one of 8 post-embryonic developmental stages [27]. Of the 17, 8 and 6 were sacrificed at instars 5 and 6, respectively. At these stages, neither wing buds nor ovipositors are apparent; therefore the gender could not be determined for these individuals. In addition, two males at instar 7, and one female at instar 8 were included in the study.
RNA was extracted from the pooled, dissected nerve cord using an RNAeasy mini (Qiagen) kit in combination with a QiaShredder column (Qiagen). The quality and quantity of RNA was assessed via spectrometry at 260 nm and 280 nm.
cDNA synthesis
Double-stranded cDNA was synthesized from total RNA isolated from nerve cord tissue of L. kohalensis using the Creator™ SMART™ system developed by Clontech BD Bioscience (Mountain View, CA). This method combines long-distance PCR with a proofreading polymerase and a template switching reverse transcriptase to preferentially amplify full-length cDNA's. During the first-strand synthesis, short universal priming sites with asymmetrical SfiI digestion sites are incorporated to both the 5' and 3' ends of each cDNA fragment. A second round of amplification is then performed via primer extension [101] to generate double-stranded cDNA that can then be digested and directionally cloned into an appropriate vector.
Reaction conditions for the first-strand synthesis were as follows: 2 μl of total RNA from either Laupala nerve cord tissue (~0.8 μg/μl) or control Human placenta (1.0 μg/μl), 1 μl of RNAse-free water (Ambion), 1 μl of the 5' SMART IV™ primer (BD Clontech), and 1 μl of a 3'oligo d(T) primer with a modified adaptor (CDS-3M – Evrogen, Moscow) were incubated at 72°C for 2 minutes and then placed on ice for an additional 2 minutes. To this reaction, 2 μl of 5× 1st strand buffer, 1 μl of DTT (20 mM), 1 μl dNTPs (10 mM), and 1 μl of PowerScript™ reverse transcriptase were added and the mixture was incubated at 42°C for 90 minutes. 2 μl of the first-strand template was used in the second-strand reaction in 100 μl total volume under the following cycling conditions: an initial 95°C incubation for 1 minute, 16 cycles of (95°C for 30 s, 66°C for 30 s, and 72°C for 4 minutes), and a final 72°C incubation. 5 μl of this PCR product were then visualized on a 1.0% agarose gel to assess the quality of the amplification.
cDNA normalization
We normalized our library using a Trimmer-Direct cDNA normalization kit (Evrogen, Moscow) to reduce the abundance of high copy number cDNA and to increase the probability of cloning and sequencing low copy number cDNA's. Briefly, purified cDNA (~1000 ng) was denatured at 95°C and then incubated at 68°C in hybridization buffer for 5 hours. Following this incubation, cDNA was exposed to a double-stranded nuclease enzyme (DSN, Evrogen) at three different concentrations (1,1/2, and 1/4) for 25 minutes at 68°C. This reaction was stopped by a 5 minute incubation on ice. The normalized cDNA was then amplified using primers complementary to the adaptors incorporated during the second-strand reaction. Initial amplification consisted of 7 cycles of 95°C for 30 s, 66°C for 30 s, and 72°C for 4 minutes. The reactions were the placed at 4°C while non-normalized controls were cycled for an additional 6 cycles. Aliquots of these controls were removed at 9, 11, and 13 cycles. These products were visualized to determine the optimal number of cycles, and based on these results the normalized cDNA amplifications were placed back in the theromcycler for an additional 13 cycles (total # of cycles = 20).
5 μl aliquots of the amplified, normalized cDNA from each of the 3 different DSN enzyme treatments were run out on an agarose gel along side un-normalized control (Human placenta) and experimental (Laupala nerve cord) cDNA PCR products. Visualization indicated that the 1/2 DSN and 1/4 DSN enzyme concentrations both normalized the cDNA well. Treatment with the full strength enzyme had over-degraded the samples. Therefore, we combined the normalized cDNA PCR products for the two diluted DSN treatments. This template was then used for a final round of amplification (12 cycles: 95°C, 64°C, and 72°C for 30 s) before cloning the normalized cDNA into pDNR-lib vector (BD Clontech).
Size-fractionation, directional cloning, and transformation of normalized cDNA
The amplified cDNA was digested with SfiI (79 μl of normalized cDNA, 10 μl of NEB buffer 2, 10 μl restriction enzyme, and 1 μl ob BSA) for 2 hours at 50°C, and then the cDNA was ethanol precipitated and resuspended in 10 μl of RNAse-free water. SfiI digestion results in asymmetrical sticky-ends on all of the cDNA fragments and permits directional cloning. We combined several separate digestion aliquots to concentrate the cDNA. Cleaned, digested fragments were allowed to run out on a 1% agarose gel for 6 hours at low voltage to ensure good size separation. We size-fractionated the library to enrich for fragments between 1.5 kb and 4 kb. The cDNA was gel-purified and resuspended in RNAse-free water. We ligated the normalized cDNA into pDNR-lib, a plasmid vector specifically designed for cDNA library construction, and incubated these reactions at 16°C overnight. The ligations were ethanol-precipitated and resuspended in 10 μl of RNAse-free water. 2 μl (~800 ng) of the ligated vector was used to transform electro-competent cells (ElectroTen-Blue. Stratagene, La Jolla, CA) which were then grown for an hour in LB media. A serial titration was used to titer the library and to determine the number of positive transformants. Average insert size was estimated by amplifying 96 randomly chosen clones.
EST sequencing
Each library was spread on LB-Agar plates containing 100 ug/ml of chloramphenicol. Positive transformants were identified and isolated using a Q-Pix automated colony picker. Isolated clones were grown overnight in LB at 37° at 900 RPM. Plasmid DNA was isolated using a modified alkali lysis method and was used as a template in a sequencing reaction. Either M13 forward or M13 reverse was used to prime the sequencing reaction. Randomly selected clones from the two libraries were sequenced using dye-terminator chemistry (Applied Biosystems) with ABI 3730 automated sequencers. Individual nucleotides were called using TraceTuner 2.0 (Paracel), and sequence reads with quality score >20 were used to construct a cricket Gene Index.
Cricket Gene Index assembly and annotation
The cricket Gene Index database was assembled at Dana-Farber Cancer Institute as described elsewhere [102]. Cricket EST reads of sufficient quality were first subjected to a vigorous screening procedure to identify and remove the contaminating vector and adaptor sequences, poly-A/T tails, and bacterial sequences. EST's shorter than 100 bases after trimming were discarded, and the remaining 14,377 cleaned sequences were compared pair-wise using a modified version of the MegaBLAST program [103] that eliminates the generation of the final alignment lay-out to speed up the process. Following this initial pair-wise search, sequences sharing greater than 95% identity over at least 40 bases and with less than 20 bases unmatched sequence at either end were grouped into clusters, leaving unclustered sequences as singletons. Components of each cluster were then assembled using the Paracel Transcript Assembler (PTA), a modified version of CAP3 assembly program [104] to produce Tentative Consensus (TC) sequences. These virtual cDNA's with assigned TC numbers together comprise the cricket Gene Index. Following assembly, TCs and singleton EST's were searched against a non-redundant protein database using the BLAT program [78], and assigned a provisional function if they had hits exceeding a threshold BLAT score of 30 and a 30% similarity cutoff. cDNA's with high-scoring hits were also annotated with Gene Ontology (GO) terms and Enzyme Commission (EC) numbers and Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathway information using a SwissProt to GO translation table provided by the GO consortium.
Comparative analysis
To demonstrate the phylogenetic distinctiveness of these data, ten L. kohalensis unigenes were chosen based on their annotation results for a comparative analysis of sequence evolution. These 10 unigenes were translated in all 6 possible reading frames and compared using BLAT to a database containing the 6 possible reading frame translations of the unigene sets from the following organisms: Drosophila melanogaster, Anophelese gambiae,Bombyx mori, Apis mellifera, Tribolium casteneum, and Locusta migratoria. The unigene with the highest BLAT score from each of the species in the database, when one could be identified, was selected.
EST's that returned a significant BLAT hit to the Laupala sequences were aligned using a weighted CLUSTAL algorithm and default alignment parameters in the program MegAlign (DNASTAR, Inc, Madison, WI). Aligned datasets were then exported as NEXUS files [see Additional file 2, see Additional file 3, see Additional file 4, see Additional file 5, see Additional file 6, see Additional file 7, see Additional file 8, see Additional file 9, see Additional file 10, see Additional file 11, see Additional file 12] and analyzed further in PAUP * 4.0b10 (Swofford 2000). Uncorrected distances (p-distances) were calculated for all pairwise comparisons. Gene regions compared included only those with representation from all organisms; other regions were excluded from analyses. Regions with substantial gaps in alignment were also excluded.
Authors' contributions
PDD participated in the conception of the project, the design of the study, the creation of the cDNA library and the drafting of the manuscript. SPM participated the design of the study, the creation of the cDNA library and the drafting of the manuscript. JQ and FL participated in the construction of the cricket Gene Index from EST sequences and making the resources accessible online. VN participated in establishing the collaboration and DNA sequencing. KLS participated in the conception of the project, the design of the study, establishing the collaboration and the drafting of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
BLAT best hits results. This is a text file that lists the top BLAT matches for each of the 5,225 unique sequences with significant sequence similarity to known proteins.
Characters used in the gene alignments for comparative analysis. This file identifies the characters used in the comparative analysis of the 10 unigenes presented in Table 5.
NEXUS file of Actin alignment. This file presents the alignment of the six actin sequences used for comparative analysis.
NEXUS file of alpha-tubulin alignment. This file presents the alignment of the six alpha-tubulin sequences used for comparative analysis.
NEXUS file of alpha-tubulin alignment. This file presents the alignment of the six alpha-tubulin sequences used for comparative analysis.
NEXUS file of dynein (light chain) alignment. This file presents the alignment of the six dynein (light chain) sequences used for comparative analysis.
NEXUS file of histone 2a alignment. This file presents the alignment of the six histone 2a sequences used for comparative analysis.
NEXUS file of HSP40 alignment. This file presents the alignment of the six HSP40 sequences used for comparative analysis.
NEXUS file of malate esterase alignment. This file presents the alignment of the six malate esterase sequences used for comparative analysis.
NEXUS file of myosin 2 (light chain) alignment. This file presents the alignment of the six myosin 2 (light chain) sequences used for comparative analysis.
NEXUS file of opsin alignment. This file presents the alignment of the six opsin sequences used for comparative analysis.
NEXUS file of polyubiquitin alignment. This file presents the alignment of the six polyubiquitin sequences used for comparative analysis.
Acknowledgments
Acknowledgements
This work was supported by NSF grant (IOB0344789) to KLS and PDD and the Maryland Neuroethology Training Grant in support of PDD and SPM. JQ and FL are supported by a grant from the National Science Foundation (DBI-0552416) and support from the Dana-Farber Cancer Institute High Tech Fund. We are very grateful to S. Salzberg for assisting in this collaboration. S. Lesnik and three anonymous reviewers provided valuable comments on drafts of this manuscript.
Contributor Information
Patrick D Danley, Email: pdanley@umd.edu.
Sean P Mullen, Email: spm23@umd.edu.
Fenglong Liu, Email: fliu@jimmy.harvard.edu.
Vishvanath Nene, Email: nene@tigr.org.
John Quackenbush, Email: johnq@jimmy.harvard.edu.
Kerry L Shaw, Email: kerryshaw@umd.edu.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YHC, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Miklos GLG, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies A, de Pablos B, Delcher A, Deng ZM, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong FC, Gorrell JH, Gu ZP, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston DA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke ZX, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai ZW, Lasko P, Lei YD, Levitsky AA, Li JY, Li ZY, Liang Y, Lin XY, Liu XJ, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RDC, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AHH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang GG, Zhao Q, Zheng LS, Zheng XQH, Zhong FN, Zhong WY, Zhou XJ, Zhu SP, Zhu XH, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JMC, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai ZW, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chatuverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu ZP, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke ZX, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O'Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao HG, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun JT, Thomasova D, Ton LQ, Topalis P, Tu ZJ, Unger MF, Walenz B, Wang AH, Wang J, Wang M, Wang XL, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang HY, Zhao Q, Zhao SY, Zhu SPC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Consortium TCS. Genome sequence of the nematode C-elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Weinstock GM, Robinson GE, Gibbs RA, Weinstock GM, Weinstock GM, Robinson GE, Worley KC, Evans JD, Maleszka R, Robertson HM, Weaver DB, Beye M, Bork P, Elsik CG, Evans JD, Hartfelder K, Hunt GJ, Robertson HM, Robinson GE, Maleszka R, Weinstock GM, Worley KC, Zdobnov EM, Hartfelder K, Amdam GV, Bitondi MMG, Collins AM, Cristino AS, Evans JD, Lattorff HMG, Lobo CH, Moritz RFA, Nunes FMF, Page RE, Simoes ZLP, Wheeler D, Carninci P, Fukuda S, Hayashizaki Y, Kai C, Kawai J, Sakazume N, Sasaki D, Tagami M, Maleszka R, Amdam GV, Albert S, Baggerman G, Beggs KT, Bloch G, Cazzamali G, Cohen M, Drapeau MD, Eisenhardt D, Emore C, Ewing MA, Fahrbach SE, Foret S, Grimmelikhuijzen CJP, Hauser F, Hummon AB, Hunt GJ, Huybrechts J, Jones AK, Kadowaki T, Kaplan N, Kucharski R, Leboulle G, Linial M, Littleton JT, Mercer AR, Page RE, Robertson HM, Robinson GE, Richmond TA, Rodriguez-Zas SL, Rubin EB, Sattelle DB, Schlipalius D, Schoofs L, Shemesh Y, Sweedler JV, Velarde R, Verleyen P, Vierstraete E, Williamson MR, Beye M, Ament SA, Brown SJ, Corona M, Dearden PK, Dunn WA, Elekonich MM, Elsik CG, Foret S, Fujiyuki T, Gattermeier I, Gempe T, Hasselmann M, Kadowaki T, Kage E, Kamikouchi A, Kubo T, Kucharski R, Kunieda T, Lorenzen M, Maleszka R, Milshina NV, Morioka M, Ohashi K, Overbeek R, Page RE, Robertson HM, Robinson GE, Ross CA, Schioett M, Shippy T, Takeuchi H, Toth AL, Willis JH, Wilson MJ, Robertson HM, Zdobnov EM, Bork P, Elsik CG, Gordon KHJ, Letunic I, Hackett K, Peterson J, Felsenfeld A, Guyer M, Solignac M, Agarwala R, Cornuet JM, Elsik CG, Emore C, Hunt GJ, Monnerot M, Mougel F, Reese JT, Schlipalius D, Vautrin D, Weaver DB, Gillespie JJ, Cannone JJ, Gutell RR, Johnston JS, Elsik CG, Cazzamali G, Eisen MB, Grimmelikhuijzen CJP, Hauser F, Hummon AB, Iyer VN, Iyer V, Kosarev P, Mackey AJ, Maleszka R, Reese JT, Richmond TA, Robertson HM, Solovyev V, Souvorov A, Sweedler JV, Weinstock GM, Williamson MR, Zdobnov EM, Evans JD, Aronstein KA, Bilikova K, Chen YP, Clark AG, Decanini LI, Gelbart WM, Hetru C, Hultmark D, Imler JL, Jiang HB, Kanost M, Kimura K, Lazzaro BP, Lopez DL, Simuth J, Thompson GJ, Zou Z, De Jong P, Sodergren E, Csuros M, Milosavljevic A, Johnston JS, Osoegawa K, Richards S, Shu CL, Weinstock GM, Elsik CG, Duret L, Elhaik E, Graur D, Reese JT, Robertson HM, Robertson HM, Elsik CG, Maleszka R, Weaver DB, Amdam GV, Anzola JM, Campbell KS, Childs KL, Collinge D, Crosby MA, Dickens CM, Elsik CG, Gordon KHJ, Grametes LS, Grozinger CM, Jones PL, Jorda M, Ling X, Matthews BB, Miller J, Milshina NV, Mizzen C, Peinado MA, Reese JT, Reid JG, Robertson HM, Robinson GE, Russo SM, Schroeder AJ, St Pierre SE, Wang Y, Zhou PL, Robertson HM, Agarwala R, Elsik CG, Milshina NV, Reese JT, Weaver DB, Worley KC, Childs KL, Dickens CM, Elsik CG, Gelbart WM, Jiang HY, Kitts P, Milshina NV, Reese JT, Ruef B, Russo SM, Venkatraman A, Weinstock GM, Zhang L, Zhou PL, Johnston JS, Aquino-Perez G, Cornuet JM, Monnerot M, Solignac M, Vautrin D, Whitfield CW, Behura SK, Berlocher SH, Clark AG, Gibbs RA, Johnston JS, Sheppard WS, Smith DR, Suarez AV, Tsutsui ND, Weaver DB, Wei XH, Wheeler D, Weinstock GM, Worley KC, Havlak P, Li BS, Liu Y, Sodergren E, Zhang L, Beye M, Hasselmann M, Jolivet A, Lee S, Nazareth LV, Pu LL, Thorn R, Weinstock GM, Stolc V, Robinson GE, Maleszka R, Newman T, Samanta M, Tongprasit WA, Aronstein KA, Claudianos C, Berenbaum MR, Biswas S, de Graaf DC, Feyereisen R, Johnson RM, Oakeshott JG, Ranson H, Schuler MA, Muzny D, Gibbs RA, Weinstock GM, Chacko J, Davis C, Dinh H, Gill R, Hernandez J, Hines S, Hume J, Jackson L, Kovar C, Lewis L, Miner G, Morgan M, Nazareth LV, Nguyen N, Okwuonu G, Paul H, Richards S, Santibanez J, Savery G, Sodergren E, Svatek A, Villasana D, Wright R. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte D, Naskrecki P. Orthoptera Species Online http://viceroy.eeb.uconn.edu/Orthoptera
- Flook PK, Klee S, Rowell CHF. Combined molecular phylogenetic analysis of the Orthoptera (Arthropoda, insecta) and implications for their higher systematics. Systematic Biology. 1999;48:233–253. doi: 10.1080/106351599260274. [DOI] [PubMed] [Google Scholar]
- Jost MC, Shaw KL. Phylogeny of Ensifera (Hexapoda : Orthoptera) using three ribosomal loci, with implications for the evolution of acoustic communication. Molecular Phylogenetics and Evolution. 2006;38:510–530. doi: 10.1016/j.ympev.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Hertl PT, Brandenburg RL. Effect of soil moisture and time of year on mole cricket (Orthoptera : Gryllotalpidae) surface tunneling. Environmental Entomology. 2002;31:476–481. [Google Scholar]
- Ji R, Xie BY, Li DM, Li Z, Zhang X. Use of MODIS data to monitor the oriental migratory locust plague. Agriculture Ecosystems & Environment. 2004;104:615–620. doi: 10.1016/j.agee.2004.01.041. [DOI] [Google Scholar]
- Lorch PD, Sword GA, Gwynne DT, Anderson GL. Radiotelemetry reveals differences in individual movement patterns between outbreak and non-outbreak Mormon cricket populations. Ecological Entomology. 2005;30:548–555. doi: 10.1111/j.0307-6946.2005.00725.x. [DOI] [Google Scholar]
- Zavala JA, Barrera JF, Morales H, Rojas-Wiesner ML. Design and evaluation of traps for Idiarthron subquadratum (Orthoptera : Tettigoniidae) with farmer participation in coffee plantations in Chiapas, Mexico. Journal of Economic Entomology. 2005;98:821–835. doi: 10.1603/0022-0493-98.3.821. [DOI] [PubMed] [Google Scholar]
- Barbara KA, Buss EA. Integration of insect parasitic nematodes (Rhabditida Steinernematidae) with insecticides for control of pest mole crickets (Orthoptera : Gryllotalpidae : Scapteriscus spp.) Journal of Economic Entomology. 2005;98:689–693. doi: 10.1603/0022-0493-98.3.689. [DOI] [PubMed] [Google Scholar]
- Stride B, Shah A, Sadeed SM. Recent history of Moroccan locust control and implementation of mechanical control methods in northern Afghanistan. International Journal of Pest Management. 2003;49:265–270. doi: 10.1080/0967087031000101098. [DOI] [Google Scholar]
- Tunstall DN, Pollack GS. Temporal and directional processing by an identified interneuron, ON1, compared in cricket species that sing with different tempos. Journal of Comparative Physiology a-Neuroethology Sensory Neural and Behavioral Physiology. 2005;191:363–372. doi: 10.1007/s00359-004-0591-7. [DOI] [PubMed] [Google Scholar]
- Farris HE, Mason AC, Hoy RR. Identified auditory neurons in the cricket Gryllus rubens: temporal processing in calling song sensitive units. Hearing Research. 2004;193:121–133. doi: 10.1016/j.heares.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Ronacher B, Franz A, Wohlgemuth S, Hennig RM. Variability of spike trains and the processing of temporal patterns of acoustic signals-problems, constraints, and solutions. Journal of Comparative Physiology a-Neuroethology Sensory Neural and Behavioral Physiology. 2004;190:257–277. doi: 10.1007/s00359-004-0494-7. [DOI] [PubMed] [Google Scholar]
- Uemura H, Tomioka K. Postembryonic changes in circadian photo-responsiveness rhythms of optic lobe interneurons in the cricket Gryllus bimaculatus. Journal of Biological Rhythms. 2006;21:279–289. doi: 10.1177/0748730406288716. [DOI] [PubMed] [Google Scholar]
- Castaneda LE, Nespolo RE, Roff DA. Dissecting the variance-covariance structure in insect physiology: The multivariate association between metabolism and morphology in the nymphs of the sand cricket (Gryllus firmus) Integrative and Comparative Biology. 2005;45:1116–1116. doi: 10.1016/j.jinsphys.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Stanley D. Prostaglandins and other eicosanoids in insects: Biological significance. Annual Review of Entomology. 2006;51:25–44. doi: 10.1146/annurev.ento.51.110104.151021. [DOI] [PubMed] [Google Scholar]
- Zera AJ, Borcher CA, Gaines SB. Juvenile-Hormone Degradation in Adult Wing Morphs of the Cricket, Gryllus-Rubens. Journal of Insect Physiology. 1993;39:845–856. doi: 10.1016/0022-1910(93)90117-A. [DOI] [Google Scholar]
- Adamo SA, Linn CE, Hoy RR. The Role of Neurohormonal Octopamine During Fight or Flight Behavior in the Field Cricket Gryllus-Bimaculatus. Journal of Experimental Biology. 1995;198:1691–1700. doi: 10.1242/jeb.198.8.1691. [DOI] [PubMed] [Google Scholar]
- Kanou M, Konishi A, Suenaga R. Behavioral analyses of wind-evoked escape of the cricket, Gryllodes sigillatus. Zoological Science. 2006;23:359–364. doi: 10.2108/zsj.23.359. [DOI] [PubMed] [Google Scholar]
- Brown WD, Smith AT, Moskalik B, Gabriel J. Aggressive contests in house crickets: size, motivation and the information content of aggressive songs. Animal Behaviour. 2006;72:225–233. doi: 10.1016/j.anbehav.2006.01.012. [DOI] [Google Scholar]
- deCarvalho TN, Shaw KL. Nuptial feeding of spermless spermatophores in the Hawaiian swordtail cricket, Laupala pacifica (Gryllidae : Triginodiinae) Naturwissenschaften. 2005;92:483–487. doi: 10.1007/s00114-005-0023-8. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Mito T, Sarashina I, Zhang HJ, Shinmyo Y, Ohuchi H, Noji S. Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mechanisms of Development. 2004;121:119–130. doi: 10.1016/j.mod.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Gu X, Zera AJ. Developmental Profiles and Characteristics of Hemolymph Juvenile-Hormone Esterase, General Esterase and Juvenile-Hormone Binding in the Cricket, Gryllus-Assimilis. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology. 1994;107:553–560. doi: 10.1016/0305-0491(94)90184-8. [DOI] [Google Scholar]
- Danley PD, Shaw KL. Differential developmental programs in two closely related Hawaiian crickets. Annals of the Entomological Society of America. 2005;98:219–226. doi: 10.1603/0013-8746(2005)098[0219:DDPITC]2.0.CO;2. [DOI] [Google Scholar]
- Bentley D, Hoy RR. Post-embryonic development of adult motor patterns in crickets: a neural analysis. Science. 1970;170 doi: 10.1126/science.170.3965.1409. [DOI] [PubMed] [Google Scholar]
- Bussiere LF, Hunt J, Jennions MD, Brooks R. Sexual conflict and cryptic female choice in the black field cricket, Teleogryllus commodus. Evolution. 2006;60:792–800. doi: 10.1554/05-378.1. [DOI] [PubMed] [Google Scholar]
- Fedorka KM, Mousseau TA. Female mating bias results in conflicting sex-specific offspring fitness. Nature. 2004;429:65–67. doi: 10.1038/nature02492. [DOI] [PubMed] [Google Scholar]
- Gwynne DT. Sexual differences in response to larval food stress in two nuptial feeding orthopterans - implications for sexual selection. Oikos. 2004;105:619–625. doi: 10.1111/j.0030-1299.2004.12857.x. [DOI] [Google Scholar]
- Howard DJ, Marshall JL, Hampton DD, Britch SC, Draney ML, Chu JM, Cantrell RG. The genetics of reproductive isolation: A retrospective and prospective look with comments on ground crickets. American Naturalist. 2002;159:S8–S21. doi: 10.1086/338369. [DOI] [PubMed] [Google Scholar]
- Shaw KL, Danley PD. Behavioral genomics and the study of speciation at a porous species boundary. Zoology. 2003;106:261–273. doi: 10.1078/0944-2006-00129. [DOI] [PubMed] [Google Scholar]
- Shaw KL, Herlihy DP. Acoustic preference functions and song variability in the Hawaiian cricket Laupala cerasina. Proceedings of the Royal Society of London Series B-Biological Sciences. 2000;267:577–584. doi: 10.1098/rspb.2000.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KL, Khine AH. Courtship behavior in the Hawaiian cricket Laupala cerasina: Males provide spermless spermatophores as nuptial gifts. Ethology. 2004;110:81–95. doi: 10.1046/j.1439-0310.2003.00946.x. [DOI] [Google Scholar]
- Zuk M, Rotenberry JT, Simmons LW. Geographical variation in calling song of the field cricket Teleogryllus oceanicus: the importance of spatial scale. Journal of Evolutionary Biology. 2001;14:731–741. doi: 10.1046/j.1420-9101.2001.00329.x. [DOI] [Google Scholar]
- Willett CS, Ford MJ, Harrison RG. Inferences about the origin of a field cricket hybrid zone from a mitochondrial DNA phylogeny. Heredity. 1997;79:484–494. doi: 10.1038/sj.hdy.6882430. [DOI] [PubMed] [Google Scholar]
- Shaw KL. Sequential radiations and patterns of speciation in the Hawaiian cricket genus Laupala inferred from DNA sequences. Evolution. 1996;50:237–255. doi: 10.2307/2410796. [DOI] [PubMed] [Google Scholar]
- Ross CL, Harrison RG. A fine-scale spatial analysis of the mosaic hybrid zone between Gryllus firmus and Gryllus pennsylvanicus. Evolution. 2002;56:2296–2312. doi: 10.1554/0014-3820(2002)056[2296:AFSSAO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Marshall DC, Cooley JR. Reproductive character displacement and speciation tn periodical cicadas, with description of a new species, 13-year Magicicada neotredecim. Evolution. 2000;54:1313–1325. doi: 10.1554/0014-3820(2000)054[1313:RCDASI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Holtmeier CL, Zera AJ. Differential Mating Success of Male Wing Morphs of the Cricket, Gryllus Rubens. American Midland Naturalist. 1993;129:223–233. doi: 10.2307/2426502. [DOI] [Google Scholar]
- Harrison RG, Bogdanowicz SM. Mitochondrial-DNA Phylogeny of North-American Field Crickets - Perspectives on the Evolution of Life-Cycles, Songs, and Habitat Associations. Journal of Evolutionary Biology. 1995;8:209–232. doi: 10.1046/j.1420-9101.1995.8020209.x. [DOI] [Google Scholar]
- Britch SC, Cain ML, Howard DJ. Spatio-temporal dynamics of the Allonemobius fasciatus-A. socius mosaic hybrid zone: a 14-year perspective. Molecular Ecology. 2001;10:627–638. doi: 10.1046/j.1365-294x.2001.01215.x. [DOI] [PubMed] [Google Scholar]
- Braswell WE, Andres JA, Maroja LS, Harrison RG, Howard DJ, Swanson WJ. Identification and comparative analysis of accessory gland proteins in Orthoptera. 49. 2006. pp. 1069–1080. [DOI] [PubMed]
- Andres JA, Maroja LS, Bogdanowicz SM, Swanson WJ, Harrison RG. Molecular evolution of seminal proteins in field crickets. Molecular Biology and Evolution. 2006;23:1574–1584. doi: 10.1093/molbev/msl020. [DOI] [PubMed] [Google Scholar]
- Kang L, Chen XY, Zhou Y, Liu BW, Zheng W, Li RQ, Wang J, Yu J. The analysis of large-scale gene expression correlated to the phase changes of the migratory locust. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17611–17615. doi: 10.1073/pnas.0407753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uvarov B. Grasshoppers and locusts, a handbook of general acridology. Vol. 1. London , Cambridge University Press; 1966. p. 481. [Google Scholar]
- Pener MP. Locust Phase Polymorphism and Its Endocrine Relations. Advances in Insect Physiology. 1991;23:1–79. [Google Scholar]
- Simpson SJ, McCaffery AR, Hagele BF. A behavioural analysis of phase change in the desert locust. Biological Reviews of the Cambridge Philosophical Society. 1999;74:461–480. doi: 10.1017/S000632319900540X. [DOI] [Google Scholar]
- Huber F. Uber Die Funktion Der Pilzkorper (Corpora-Pedunculata) Beim Gesang Der Keulenheuschrecke Gomphocerus Rufus L (Acrididae) Naturwissenschaften. 1955;42:566–567. doi: 10.1007/BF00623792. [DOI] [Google Scholar]
- Hedwig B. Control of cricket stridulation by a command neuron: Efficacy depends on the behavioral state. Journal of Neurophysiology. 2000;83:712–722. doi: 10.1152/jn.2000.83.2.712. [DOI] [PubMed] [Google Scholar]
- Hedwig B. Pulses, patterns and paths: neurobiology of acoustic behaviour in crickets. Journal of Comparative Physiology a-Neuroethology Sensory Neural and Behavioral Physiology. 2006;192:677–689. doi: 10.1007/s00359-006-0115-8. [DOI] [PubMed] [Google Scholar]
- Hennig RM. Neuronal Control of the Forewings in 2 Different Behaviors - Stridulation and Flight in the Cricket, Teleogryllus-Commodus. Journal of Comparative Physiology a-Sensory Neural and Behavioral Physiology. 1990;167:617–627. [Google Scholar]
- Otto D. Central Nervous Control of Sound Production in Crickets. Zeitschrift Fur Vergleichende Physiologie. 1971;74:227–271. doi: 10.1007/BF00297729. [DOI] [Google Scholar]
- Hoy RR, Paul RC. Genetic-Control of Song Specificity in Crickets. Science. 1973;180:82–83. doi: 10.1126/science.180.4081.82. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Hoy RR. Genetic-Control of Neuronal Network Generating Cricket (Teleogryllus-Gryllus) Song Patterns. Animal Behaviour. 1972;20:478–492. doi: 10.1016/S0003-3472(72)80012-5. [DOI] [PubMed] [Google Scholar]
- Shaw KL. Polygenic inheritance of a behavioral phenotype: Interspecific genetics of song in the Hawaiian cricket genus Laupala. Evolution. 1996;50:256–266. doi: 10.2307/2410797. [DOI] [PubMed] [Google Scholar]
- Mendelson TC, Shaw KL. Sexual behaviour: Rapid speciation in an arthropod. Nature. 2005;433:375–376. doi: 10.1038/433375a. [DOI] [PubMed] [Google Scholar]
- Shaw KL. Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: What mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16122–16127. doi: 10.1073/pnas.242585899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KL. Further acoustic diversity in Hawaiian forests: two new species of Hawaiian cricket (Orchoptera : Gryllidae : Trigonidiinae : Laupala) Zoological Journal of the Linnean Society. 2000;129:73–91. doi: 10.1006/zjls.1998.0201. [DOI] [Google Scholar]
- Mendelson TC, Siegel AM, Shaw KL. Testing geographical pathways of speciation in a recent island radiation. Molecular Ecology. 2004;13:3787–3796. doi: 10.1111/j.1365-294X.2004.02375.x. [DOI] [PubMed] [Google Scholar]
- Parsons YM, Shaw KL. Species boundaries and genetic diversity among Hawaiian crickets of the genus Laupala identified using amplified fragment length polymorphism. Molecular Ecology. 2001;10:1765–1772. doi: 10.1046/j.1365-294X.2001.01318.x. [DOI] [PubMed] [Google Scholar]
- Shaw KL. A nested analysis of song groups and species boundaries in the Hawaiian cricket genus Laupala. Molecular Phylogenetics and Evolution. 1999;11:332–341. doi: 10.1006/mpev.1998.0558. [DOI] [PubMed] [Google Scholar]
- Shaw KL. Interspecific genetics of mate recognition: Inheritance of female acoustic preference in Hawaiian crickets. Evolution. 2000;54:1303–1312. doi: 10.1554/0014-3820(2000)054[1303:IGOMRI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Shaw KL, Parsons YM. Divergence of mate recognition behavior and its consequences for genetic architectures of speciation. American Naturalist. 2002;159:S61–S75. doi: 10.1086/338373. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Band MR, Bonaldo MF, Kumar CG, Liu L, Pardinas JR, Robertson HM, Soares MB, Robinson GE. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Research. 2002;12:555–566. doi: 10.1101/gr.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia QY, Zhou ZY, Lu C, Cheng DJ, Dai FY, Li B, Zhao P, Zha XF, Cheng TC, Chai CL, Pan GQ, Xu JS, Liu C, Lin Y, Qian JF, Hou Y, Wu ZL, Li GR, Pan MH, Li CF, Shen YH, Lan XQ, Yuan LW, Li T, Xu HF, Yang GW, Wan YJ, Zhu Y, Yu MD, Shen WD, Wu DY, Xiang ZH, Yu J, Wang J, Li RQ, Shi JP, Li H, Li GY, Su JN, Wang XL, Li GQ, Zhang ZJ, Wu QF, Li J, Zhang QP, Wei N, Xu JZ, Sun HB, Dong L, Liu DY, Zhao SL, Zhao XL, Meng QS, Lan FD, Huang XG, Li YZ, Fang L, Li CF, Li DW, Sun YQ, Zhang ZP, Yang Z, Huang YQ, Xi Y, Qi QH, He DD, Huang HY, Zhang XW, Wang ZQ, Li WJ, Cao YZ, Yu YP, Yu H, Li JH, Ye JH, Chen H, Zhou Y, Liu B, Wang J, Ye J, Ji H, Li ST, Ni PX, Zhang JG, Zhang Y, Zheng HK, Mao BY, Wang W, Ye C, Li SG, Wang J, Wong GKS, Yang HM. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- Mita K, Kasahara M, Sasaki S, Nagayasu Y, Yamada T, Kanamori H, Namiki N, Kitagawa M, Yamashita H, Yasukochi Y, Kadono-Okuda K, Yamamoto K, Ajimura M, Ravikumar G, Shimomura M, Nagamura Y, Shin-I T, Abe H, Shimada T, Morishita S, Sasaki T. The genome sequence of silkworm, Bombyx mori. DNA Research. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- Miao XX, Xu SJ, Li MH, Li MW, Huang JH, Dai FY, Marino SW, Mills DR, Zeng PY, Mita K, Jia SH, Zhang Y, Liu WB, Xiang H, Guo QH, Xu AY, Kong XY, Lin HX, Shi YZ, Lu G, Zhang XL, Huang W, Yasukochi Y, Sugasaki T, Shimada T, Nagaraju J, Xiang ZH, Wang SY, Goldsmith MR, Lu C, Zhao GP, Huang YP. Simple sequence repeat-based consensus linkage map of Bombyx mori. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16303–16308. doi: 10.1073/pnas.0507794102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Riddiford LM. The origins of insect metamorphosis. NATURE. 1999;401:447–452. doi: 10.1038/46737. [DOI] [PubMed] [Google Scholar]
- Peel AD, Telford MJ, Akam M. The evolution of hexapod engrailed-family genes: evidence for conservation and concerted evolution. Proceedings of the Royal Society B-Biological Sciences. 2006;273:1733–1742. doi: 10.1098/rspb.2006.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M. Genomes, phylogeny, and evolutionary systems biology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6630–6635. doi: 10.1073/pnas.0501984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson JA, Stern DL. The pea aphid, Acyrthosiphon pisum: an emerging genomic model system for ecological, developmental and evolutionary studies. Bioessays. 2006;28:747–755. doi: 10.1002/bies.20436. [DOI] [PubMed] [Google Scholar]
- Pittendrigh BR, Clark JM, Johnston JS, Lee SH, Romero-Severson J, Dasch GA. Sequencing of a new target genome: the Pediculus humanus humanus (Phthiraptera : Pediculidae) genome project. Journal of Medical Entomology. 2006;43:1103–1111. doi: 10.1603/0022-2585(2006)43[1103:SOANTG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Falciani F, Hausdorf B, Schroder R, Akam M, Tautz D, Denell R, Brown S. Class 3 Hox genes in insects and the origin of zen. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:8479–8484. doi: 10.1073/pnas.93.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DFCI Cricket Gene Index http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=cricket
- Kent WJ. BLAT - The BLAST-like alignment tool. Genome Research. 2002;12:656–664. doi: 10.1101/gr.229202. 10.1101/gr.229202. Article published online before March 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DFCI Human Gene Index http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=human
- DFCI Mouse Gene Index http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=mouse
- DFCI Rat Gene Index http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=rat
- Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. 1999:138–148. [PubMed] [Google Scholar]
- The Gene Ontology http://www.geneontology.org
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Index Project http://compbio.dfci.harvard.edu/tgi/
- LocustDB http://locustdb.genomics.org.cn/
- Renn SCP, Aubin-Horth N, Hofmann HA. Biologically meaningful expression profiling across species using heterologous hybridization to a cDNA microarray. BMC Genomics. 2004;5 doi: 10.1186/1471-2164-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov V, Dowdle J, Smirnoff N, Ford-Lloyd B, Newbury HJ, Macnair MR. Comparison of gene expression in segregating families identifies genes and genomic regions involved in a novel adaptation, zinc hyperaccumulation. Molecular Ecology. 2006;15:3045–3059. doi: 10.1111/j.1365-294X.2006.02981.x. [DOI] [PubMed] [Google Scholar]
- Dahm R, Geisler R. Learning from small fry: The zebrafish as a genetic model organism for aquaculture fish species. Marine Biotechnology. 2006;8:329–345. doi: 10.1007/s10126-006-5139-0. [DOI] [PubMed] [Google Scholar]
- Jung S, Main D, Staton M, Cho I, Zhebentyayeva T, Arus P, Abbott A. Synteny conservation between the Prunus genome and both the present and ancestral Arabidopsis genomes. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakefield PM. Evo-devo and constraints on selection. Trends in Ecology and Evolution. 2006;21:362–368. doi: 10.1016/j.tree.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Yandell M, Mungall CJ, Smith C, Prochnik S, Kaminker J, Hartzell G, Lewis S, Rubin GM. Large-scale trends in the evolution of gene structures within 11 animal genomes. Plos Computational Biology. 2006;2:113–125. doi: 10.1371/journal.pcbi.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopazo H, Dopazo J. Genome-scale evidence of the nematode-arthropod clade. Genome Biology. 2005;6 doi: 10.1186/gb-2005-6-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zahler AM. Conservation, regulation, synteny, and introns in a large-scale C-briggsae-C-elegans genomic alignment. Genome Research. 2000;10:1115–1125. doi: 10.1101/gr.10.8.1115. [DOI] [PubMed] [Google Scholar]
- Curole JP, Kocher TD. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends in Ecology and Evolution. 1999;14:394–398. doi: 10.1016/S0169-5347(99)01660-2. [DOI] [PubMed] [Google Scholar]
- Evans JD, Gundersen-Rindal D. Beenomes to Bombyx: future directions in applied insect genomics. Genome Biology. 2003;4 doi: 10.1186/gb-2003-4-3-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beldade P, Rudd S, Gruber JD, Long AD. A wing expressed sequence tag resource for Bicyclus anynana butterflies, an evo-devo model. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Animal Genome Size Database http://www.genomesize.com
- Gregory TR. Synergy between sequence and size in large-scale genomics. Nature Reviews Genetics. 2005;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A laboratory manual. Cold Spring Harbor, New York , CSHL Press; 1996. [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Research. 2001;29:159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. Journal of Computational Biology. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Madan A. CAP3: A DNA sequence assembly program. Genome Research. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dbEST: database of "Expressed Sequence Tags" http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BLAT best hits results. This is a text file that lists the top BLAT matches for each of the 5,225 unique sequences with significant sequence similarity to known proteins.
Characters used in the gene alignments for comparative analysis. This file identifies the characters used in the comparative analysis of the 10 unigenes presented in Table 5.
NEXUS file of Actin alignment. This file presents the alignment of the six actin sequences used for comparative analysis.
NEXUS file of alpha-tubulin alignment. This file presents the alignment of the six alpha-tubulin sequences used for comparative analysis.
NEXUS file of alpha-tubulin alignment. This file presents the alignment of the six alpha-tubulin sequences used for comparative analysis.
NEXUS file of dynein (light chain) alignment. This file presents the alignment of the six dynein (light chain) sequences used for comparative analysis.
NEXUS file of histone 2a alignment. This file presents the alignment of the six histone 2a sequences used for comparative analysis.
NEXUS file of HSP40 alignment. This file presents the alignment of the six HSP40 sequences used for comparative analysis.
NEXUS file of malate esterase alignment. This file presents the alignment of the six malate esterase sequences used for comparative analysis.
NEXUS file of myosin 2 (light chain) alignment. This file presents the alignment of the six myosin 2 (light chain) sequences used for comparative analysis.
NEXUS file of opsin alignment. This file presents the alignment of the six opsin sequences used for comparative analysis.
NEXUS file of polyubiquitin alignment. This file presents the alignment of the six polyubiquitin sequences used for comparative analysis.


