Abstract
Type-II topoisomerases are responsible for untangling DNA during replication by removing supercoiled and interlinked DNA structures. Using a single-molecule micromanipulation setup, we follow the real-time decatenation of two mechanically braided DNA molecules by Drosophila melanogaster topoisomerase (Topo) II and Escherichia coli Topo IV. Although Topo II relaxes left-handed (L) and right-handed (R-) braids similarly at a rate of ≈2.9 s–1, Topo IV has a marked preference for L-braids, which it relaxes completely and processively at a rate of ≈2.4 s–1. However, Topo IV can unlink R-braids at about half that rate when they supercoil to form L-plectonemes. These results imply that the preferred substrate for unlinking by Topo IV has the symmetry of an L-crossing and shed new light on the decatenation of daughter strands during DNA replication, which are usually assumed to be linked in an R-braid.
Keywords: DNA replication
Type-II topoisomerases are ubiquitous ATP-dependent enzymes that catalyze the transport of one DNA segment [the transport (T) segment] through a transient double-stranded break in a second [the gate (G) segment] (1, 2). Their role in the cell is to control the supercoiling of DNA and to untangle the catenanes that arise during replication or recombination (3–6); their malfunction at mitosis or meiosis ultimately causes cell death. In vitro experiments have shown that these enzymes can decatenate DNA molecules well below the thermodynamic entanglement equilibrium (7). The mechanism by which this local action on DNA results in unknotting and decatenation (a global topological property) is not fully understood.
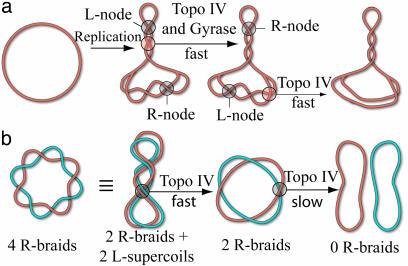
In contrast with eukaryotic topoisomerases that act similarly on positively and negatively supercoiled DNA, prokaryotes have two type-II topoisomerases: DNA gyrase and topoisomerase (Topo) IV, which act differently on (+) and (–) supercoiled DNA [respectively, left-handed (L-) and right-handed (R-) nodes; see Fig. 1a]. Gyrase removes (+) supercoils and actively generates (–) supercoils, whereas Topo IV preferentially relaxes (+) supercoils (8). The chiral discrimination of Topo IV, which prevents its interference with the winding activity of gyrase, was suggested (8) to result from a preferred interaction of Topo IV with the particular orientation of the G and T segments encountered in (+) supercoils: an L-crossing with acute angle (see Fig. 1b). However, the main role of Topo IV in bacteria is the unlinking of the sister chromosomes during replication (6, 9). Experiments in vitro have suggested that the decatenation of the sister strands could occur either by relaxing the (+) supercoils on the mother strand or by unlinking the braided daughter strands (10). Relaxation of the (+) supercoils arising in front of the replication fork (by gyrase and Topo IV) is thought to be the main pathway of unlinking in the early stages of replication, whereas decatenation of the entangled sister chromosomes becomes increasingly important as replication proceeds to the terminal stage (9, 10). But if these so-called precatenanes wrap around each other in a right-handed fashion [like (–) supercoils; see Fig. 1a], as is generally assumed (6, 10, 11), they should present an inefficient substrate for relaxation by Topo IV. Hence unlinking should become less and less efficient as replication proceeds to termination, a riddle we call the Topo IV decatenation paradox.
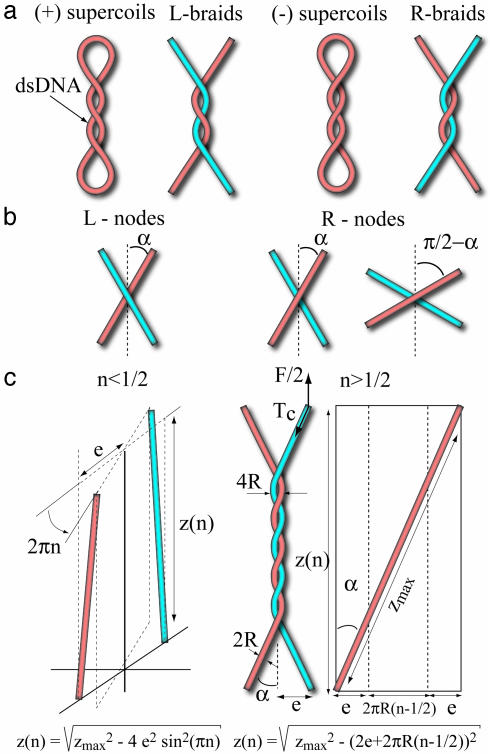
Fig. 1.
DNA crossings and geometrical model of DNA braiding. (a) Similarity of the crossings between a positively (negatively) supercoiled plasmid and L- (R-)braids. Note that overtwisting the (R-) double-stranded DNA (dsDNA) molecule generates (+) supercoils that are left-handed. (b) Diagram of the crossing of two DNA strands. 2α denotes the angle between strands; notice that an R-node with an obtuse angle π – 2α is identical to an L-node with an acute angle 2α < π/2, which is characteristic of the geometry of crossings in (+) supercoiled DNA. (c Left) Twisting two molecules by less than half a turn (0 < n < 1/2) so they do not cross. 2e denotes the distance between the molecules' anchoring points, zmax denotes their length, and z(n) denotes their vertical extension; a direct application of Pythagoras' theorem yields the equation for z(n). (Right) Case n > 1/2 when the two molecules are wrapped around each other. The axis of each molecule traces a helicoidal path on an imaginary cylinder of radius R. By describing that path in periodic cylindrical coordinates (schema on the far right), one notices that its azimuthal projection is 2πR(n – 1/2) + 2e. The equation for its vertical projection z(n) is then obtained from Pythagoras' theorem. The tension Tc in each molecule is given by Tc = F/(2 cos α).
Puzzled by these arguments, we decided to study the unbraiding of two DNA molecules by a single type-II topoisomerase (eukaryotic or prokaryotic) by using the single-molecule magnetic manipulation technique we used previously to study DNA uncoiling (8, 12). Our results confirm that, whereas the eukaryotic Topo II relaxes similarly L- and R-braids, the prokaryotic Topo IV preferentially relaxes L-braids [like (+) supercoils]. However, Topo IV can also unlink R-braids when they supercoil to form L-plectonemes. These observations suggest a solution to the aforementioned paradox.
Materials and Methods
DNA Constructs and Micromanipulation Setup. We multiply labeled a 16-kbp DNA (pXΔII, linking number Lk0 ≃ 1,500) at its extremities with biotin and digoxigenin (Dig) as described (13). The molecules were bound at one end to a streptavidin-coated magnetic bead [2.8 μm (Dynal, Great Neck, NY) or 1 μm (Biolabs, Northbrook, IL)] and at the other to an anti-Dig coated glass surface, previously incubated with BSA (Hoffmann–La Roche) to reduce nonspecific interactions. We translated small magnets placed above the sample to set the force pulling on the bead and rotated them to braid the two DNA molecules anchoring the bead to the surface. The extension of the vast majority (>99%) of beads tethered by single molecules was not affected by rotation of the magnets. Thus the anchoring DNA could not be supercoiled (see Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). We conclude that most of the molecules in our sample were nicked, probably because of shear stress. Bead tracking and force measurement were performed on an inverted microscope (14). The extension z was measured by tracking the 3D position of the tethered bead (13) with an error due to Brownian motion of ≈10 nm (with 1-s averaging). The bead's transverse fluctuations 〈δx2〉 allow for a determination of the stretching force using the equipartition theorem: F = kBTz/〈δx2〉, where kB is the Boltzmann constant and T, the temperature (at 25°C: kBT = 4 pN nm). F was measured with 10% accuracy. Notice that the magnetic trap implemented here is a natural force clamp: once the distance of the magnets to the sample (a few millimeters) is set, the force pulling on the bead is fixed. In plots showing DNA extension, the points correspond to the raw data and were obtained at 25 Hz. To eliminate microscope drifts, differential tracking with a second bead glued to the surface was performed.
Unbraiding Assays. Unbraiding experiments with Drosophila melanogaster topoisomerase II (Amersham Pharmacia; [Topo II] ≃ 12 ng/ml) were performed at 25°C in 10 mM Tris buffer (pH = 7.9) containing 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 15 μg/ml BSA, and 1 mM ATP. Unbraiding experiments with Topo IV [gift of N. Cozzarelli (University of California, Berkeley) and N. Crisona (University of California, Berkeley)] ([Topo IV] ≃ 20 ng/ml) were performed at 25°C in 25 mM Tris buffer (pH = 7.6) containing 100 mM potassium glutamate, 10 mM MgCl2, 0.5 mM DTT, 50 μg/ml BSA, and 1 mM ATP (8). Step measurements were performed in the same buffer with 5 μM ATP with 1-μm magnetic beads.
Braiding Model. The geometrical model considered here is the twisting of a swing by n turns (see Fig. 1c). Up to the first crossing, 0 < n < 0.5, we observe a sharp decrease in the vertical extension z of the molecules of radius R, whose extremities are separated by a distance 2e (e can be different for different complexes):
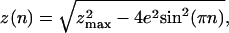 |
[1] |
where zmax is the extension at n = 0. As the molecules are further braided, n > 1/2, their extension z is better understood by describing the braids in periodic cylindrical coordinates as shown in Fig. 1c); this leads to
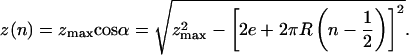 |
[2] |
Eq. 2 is valid so long as the distance between the axes of the
interlinked molecules is larger than their diameter, which implies cos
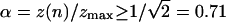 (or α ≤ αc = 45°). Notice that for a
given force, the number of turns, n uniquely determines the braid
angle α (when it is ≤45°). Further braiding increases the torque
until it is large enough to induce the braids to supercoil
(15). We observe this to
happen when z(n)/zmax ≡
zb/zmax ∼ 0.64.
(or α ≤ αc = 45°). Notice that for a
given force, the number of turns, n uniquely determines the braid
angle α (when it is ≤45°). Further braiding increases the torque
until it is large enough to induce the braids to supercoil
(15). We observe this to
happen when z(n)/zmax ≡
zb/zmax ∼ 0.64.
Results
Braiding Two DNA Molecules: Experiments and Model. We examined the decatenase activity of two different topoisomerases [Topo II from D. melanogaster (12) and Topo IV from Escherichia coli (8)] by monitoring in real time the increase in extension of the molecules as they were unlinked. The molecules' extension z at a given force F was measured as described in Materials and Methods. Because the elastic behavior of a single stretched DNA molecule is well known (16–18), we verified that the bead was tethered by two molecules by comparing the measured extension zmax in abscence of braiding with the expected extension for a bead tethered by two molecules (see Fig. 8a, which is published as supporting information on the PNAS web site).
The molecules were then braided around each other n times by rotating the magnets (Fig. 2), resulting in a decrease in z. The variation of z with n is nicely described by the geometric braiding of two ropes of radius R and maximal extension zmax anchored a distance 2e apart as in a swing (see Materials and Methods and Figs. 1 and 2). This model implies that, in contrast with previous single-molecule (12) (and bulk) supercoiling experiments, the braid angle α is related to the molecules' extension by cosα = z/zmax (Fig. 2). At any constant force F, the extension z shrinks with n, whereas α increases until the strands are in close contact at n = nc where α = αc = 45°. As we further twist the molecules and bring them into closer contact, the braiding torque increases until at n = nb, the braids buckle and form plectonemic supercoils. This transition is observed when z/zmax = zb/zmax ∼ 0.64 and is characterized by an increase in the fluctuations in extension (data not shown) and by a discontinuity in the slope of z/zmax vs. n (see Fig. 2). That buckling transition was also observed in numerical simulations of the braiding of two stretched DNA molecules (data not shown). On further twisting, the extension z shortened by a constant amount δz ∼ 42 nm per turn (see Fig. 2), as expected for plectonemic braids (15).
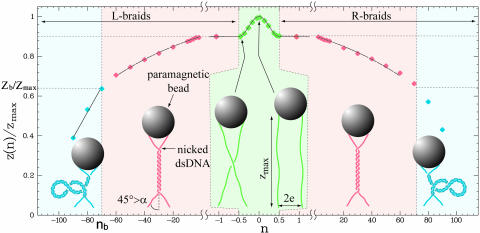
Fig. 2.
Braiding of two DNA molecules. Shown are two DNA molecules anchored to a magnetic bead. By rotating the bead by n turns, the molecules are braided around each other, and their extension z decreases from their maximal value zmax at n = 0. Plotted: the molecules' relative extension z/zmax vs. n at F = 1.15 pN (points). Notice the change in the horizontal scale when –1 < n < 1. There is a sharp decrease in extension for the first half turn (|n| < 1/2, green shaded area), i.e., until the two molecules cross, followed (for |n| > 1/2, red shaded area) by a more gradual decrease as they wrap around each other. The plain black lines are a best fit to the data using the geometrical model (described in Fig. 1c and Materials and Methods). For relative extensions z/zmax < zb/zmax ∼ 0.64 (blue shaded area), the braids in close contact supercoil leading to a decrease in extension δz ∼ 42 nm per turn (15).
At each force F, we determined the values of the intermolecular distance 2e (which varies from bead to bead) and the DNA's effective radius R that best fit the data for cosα to the model when α < 45°. That the best-fit values of 2e (for a given bead) varied little (0.8 ± 0.08 μm) over a factor 10 variation in F further validates our model. Furthermore, as predicted by Marko (15), at large forces (F > 0.2 pN) the braid radius R decreased as F–3/4 due to a reduction in entropic confinement (see Fig. 8b). Its value (R ∼ 5 nm) at F = 0.3 pN is compatible with values measured on free plasmids in similar salt conditions (19, 20). The excellent agreement between the braiding data and the simple geometric model just described allows for a detailed understanding of DNA unbraiding by type-II topoisomerases.
The Eukaryotic Topo Decatenates L- and R-Braids at the Same Rate as It
Relaxes Supercoiled DNA. The addition of eukaryotic Topo II in an
appropriate reaction buffer (see Material and Methods) led to an
increase in extension corresponding to decatenation of L- or R-braids
(Fig. 3a). To ensure
that a single enzyme was monitored, we set the concentration of topoisomerase
so that the typical waiting time for an unbraiding event
Tw was larger than the relaxation time
Tr. The probability that two enzymes acted
simultaneously was then negligibly small
(12). In these conditions, we
observed the enzyme to unlink L- and R-braids at similar rates (see
Fig. 3b) and with a
high processivity: all of the braids (usually ≈40, but as many as 80) were
unlinked in one burst of activity. Knowing the variation of z with
n (see Fig. 2), we
estimated the average decatenation rate (for both L- and R-braids):
vd = 2.7 ± 0.3 cycles/s at F =
0.3 pN. Notice that the tension Tc in each
molecule varied slightly with the braiding angle:
Tc = F/(2 cos α), where 0.7 ≤
cos α ≤ 0.9; the lower bound was reached at close contact (α =
45°) and the upper one at first contact (n = 1/2). As was the
case for supercoil removal
(12), the decatenation rate
was slowed down exponentially with the averaged molecular tension
〈Tc〉 ≃ F/1.6:
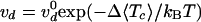 with Δ = 1.9 nm and
with Δ = 1.9 nm and 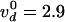 cycles/s; see
Fig. 3c.
cycles/s; see
Fig. 3c.
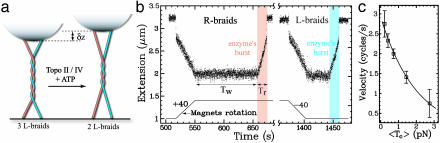
Fig. 3.
Decatenation by Topo II. (a) The sketch shows that when braids are
decatenated, the distance z of the bead to the surface increases by
δz per braid removed. The kinetics of decatenation can thus be
monitored in real-time by measuring z. (b) Recording of a
decatenation experiment using Topo II (raw extension signal z at 25
Hz). Rotation of the magnets by 40 turns (bottom trace) leads to a decrease in
the DNA's extension z; see Fig.
2. After waiting for a time Tw, the
processive decatenation of the two molecules by a single enzyme is observed
(shaded time interval) as an increase in extension within a relaxation time
Tr < Tw. The data
shown are time traces of the decatenation by Topo II of R-braids (red shaded
area) and L-braids (blue shaded area) starting at |n| =
40 (F = 1.15 pN). No significant difference was observed between
relaxation of R- and L-nodes. Notice the sharp increase in z at the
end of the relaxation run resulting from the decatenation of the last braid.
(c) Variation with the average tension
〈Tc〉 of the decatenation velocity
vd. Each point is an average over at least 10
runs at a given force. The data are fit by an exponential curve (plain line):
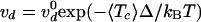 with
with 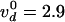 cycles/s, Δ = 1.9 nm
(12). The error bars here, as
in all plots, indicate the statistical error.
cycles/s, Δ = 1.9 nm
(12). The error bars here, as
in all plots, indicate the statistical error.
Topo IV Unlinks L-Braids Completely but R-Braids Only Partially.
Whereas eukaryotic Topo II is insensitive to the chirality of the DNA
crossing, bacterial Topo IV relaxes (–) supercoils 20 times slower than
(+) ones. Furthermore, a 20× higher enzymatic concentration is required
to observe those negative uncoiling events
(8). A chiral discrimination is
also observed in Topo IV unbraiding. As shown in
Fig. 4a, Topo IV
relaxed L-braids completely and with a high processivity but did not relax
R-braids below a final catenation number nS.
However, although uncoiling of (+) and (–) supercoils occurs at widely
different rates, Topo IV decatenated L- and R-braids similarly when n
> nS: 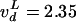 cycles per s and
cycles per s and 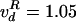 cycles per s,
respectively, for L- and R-nodes (at the same [TopoIV] ∼ 20 ng/ml); see
Fig. 4b. These rates,
which varied little with the relative extension
z/zmax, are closer to the rates estimated in
vivo [≈1 cycle/s (9)
than to those deduced from bulk in vitro data (0.1 cycles/s
(21,
22)]. The uncoiling rates of
Topo IV in single-molecule experiments were similarly reported to be larger
than the bulk measurements (8).
This is not surprising, because bulk assays measure a rate averaged over a
population of enzymes, only a small fraction of which may be active at any
time.
cycles per s,
respectively, for L- and R-nodes (at the same [TopoIV] ∼ 20 ng/ml); see
Fig. 4b. These rates,
which varied little with the relative extension
z/zmax, are closer to the rates estimated in
vivo [≈1 cycle/s (9)
than to those deduced from bulk in vitro data (0.1 cycles/s
(21,
22)]. The uncoiling rates of
Topo IV in single-molecule experiments were similarly reported to be larger
than the bulk measurements (8).
This is not surprising, because bulk assays measure a rate averaged over a
population of enzymes, only a small fraction of which may be active at any
time.
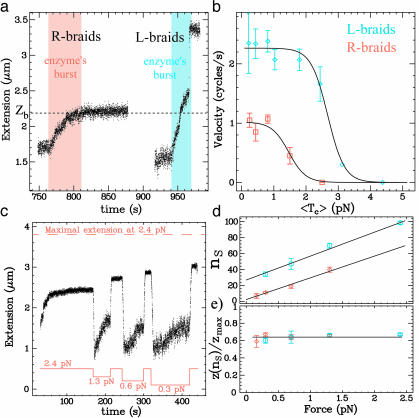
Fig. 4.
Decatenation by Topo IV. (a) Time traces of relaxation by Topo IV
of R-braids (red shaded area; initial n = 45) and L-braids (blue
shaded area; initial n = 55) at F = 0.7 pN. Whereas L-braids
are completely relaxed, R-braids are relaxed only partially: the reaction
stops at a final n ≡ nS = 19 at
which z(nS)/zmax =
0.66. The value of the extension zb where braids
begin to supercoil (see Fig. 2)
is shown for comparison. (b) Variation with the averaged tension
〈Tc〉 of the decatenation rate
vd (averaged over at least 10 runs at each force)
for R-braids (red, □) and L-braids (blue, ⋄) and fit (plain line) to
a model where the rate-limiting step of the reaction is force-independent
(30):
vd = v0/(1 + k
exp(〈Tc〉Δ/kBT));
with 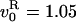 cycle/s, Δ = 15.4
nm, kR= 3.10–3(R-braids) and
cycle/s, Δ = 15.4
nm, kR= 3.10–3(R-braids) and
 cycle/s, Δ = 15.4 nm,
kL= 3.10–5(L-braids).
(c) Time trace obtained when varying the force during decatenation of
R-braids by Topo IV. After the decatenation stopped at F = 2.4 pN,
the force was reduced to F = 1.3 pN. Decatenation resumed, then
stopped again. The force was increased back to F = 2.4 pN to allow
for comparison of the molecule's initial and final extension. The procedure
was repeated at F = 0.6 pN and F = 0.3 pN. (d) The
remaining catenation number at arrest nS vs.
F for two different sets of experiments (different tethered beads,
i.e., different anchoring distances 2e).
nS is proportional to F and increases as
e decreases. Red (⋄), e = 1.1 μm; blue (○),
e = 0.8 μm. (e) The relative extension at arrest
z(nS)/zmax varies
little with F or e. Its averaged value is:
z(nS)/zmax = 0.65
± 0.02, which is about the same as the value where the braids
supercoil.
cycle/s, Δ = 15.4 nm,
kL= 3.10–5(L-braids).
(c) Time trace obtained when varying the force during decatenation of
R-braids by Topo IV. After the decatenation stopped at F = 2.4 pN,
the force was reduced to F = 1.3 pN. Decatenation resumed, then
stopped again. The force was increased back to F = 2.4 pN to allow
for comparison of the molecule's initial and final extension. The procedure
was repeated at F = 0.6 pN and F = 0.3 pN. (d) The
remaining catenation number at arrest nS vs.
F for two different sets of experiments (different tethered beads,
i.e., different anchoring distances 2e).
nS is proportional to F and increases as
e decreases. Red (⋄), e = 1.1 μm; blue (○),
e = 0.8 μm. (e) The relative extension at arrest
z(nS)/zmax varies
little with F or e. Its averaged value is:
z(nS)/zmax = 0.65
± 0.02, which is about the same as the value where the braids
supercoil.
In an effort to understand the relaxation of R-braids by Topo IV, we
measured the final catenation number nS at
successively smaller forces F and for two different anchoring
distances 2e; see Fig.
4c. We noticed that nS is
proportional to F and increases as e decreases
(Fig. 4d). However, in
all cases the molecules' relative extension at arrest
z(nS)/zmax is the
same whatever the values of F or e:
z(nS)/zmax = 0.65
± 0.02 (Fig.
4e). This value is very close to the critical extension
zb/zmax ≈ 0.64 below which
the braided molecules supercoil (Fig.
2). Furthermore, linear extrapolation of
nS to F = 0 and e = 0 (as
expected for catenated plasmids in bulk experiments) yields
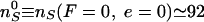 ,
or a catenation number
,
or a catenation number
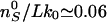 , close to
Marko's estimate for the onset of supercoiled braids in catenated DNAs
(15). These considerations
imply that R-braids are relaxed by Topo IV when they supercoil to form
L-plectonemes.
, close to
Marko's estimate for the onset of supercoiled braids in catenated DNAs
(15). These considerations
imply that R-braids are relaxed by Topo IV when they supercoil to form
L-plectonemes.
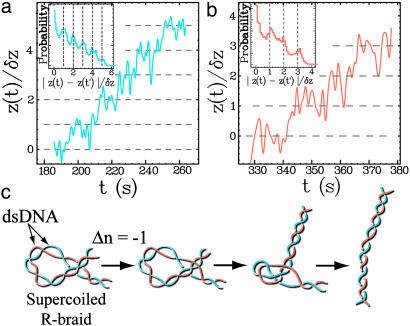
By reducing the concentration of ATP ([ATP] = 5 μM), we slowed down the enzymatic activity and were able to observe the step-by-step unlinking of supercoiled braids (i.e., when z < zb)by a single Topo IV. The slow relaxation dynamics of these plectonemic braids cause an increase in the low-frequency fluctuations in extension, which hinders the direct observation of well defined single relaxation events; see Fig. 5 a and b. However, the histograms of the difference in extension at two times |z(t) – z(t′)| (Fig. 5 a and b Insets) reveals a series of peaks at values corresponding to an integer number of braids. This indicates that Topo IV relaxes both R- and L-braids one at a time.
Fig. 5.
Step-by-step decatenation by Topo IV. (a and b) Filtered (1-Hz) time traces of relaxation of L- (a) and R-braids (b) by Topo IV obtained at low ATP concentration (5 μM) in the regime of supercoiled braids (z/zmax < zb/zmax ∼ 0.64) at F = 0.9 pN. The real-time extension z(t) is normalized by the measured decrease in extension per turn: δz ≈ 42 nm (Fig. 2). The slow relaxation of braided supercoils results in a large increase in the low-frequency fluctuations of extension that hinder the observation of well defined decatenation steps at integer values of z(t)/δz (dashed lines). However, the histogram of the normalized distance between two points (|z(t) – z(t′)|/δz) displays peaks at integer values (Insets), implying that decatenation occurs by steps of Δn =–1. (c) Proposed model for the relaxation of R-braids supercoils by Topo IV: two enzymatic reactions are needed to transfer one braid through one DNA and thereby decrease the catenation number by one (Δn = –1). Thermal fluctuations then drive the relaxation of the loop to an extended topoisomer. The rate of supercoiled R-braid unlinking is thus expected to be half the rate of L-braid unlinking (see Fig. 4b).
Discussion
Our results confirm that the eukaryotic topoisomerase is insensitive to the chirality of the DNA braids. The rates of unbraiding and uncoiling measured for the fly Topo II and their variation with force are similar, implying that the different substrates (supercoiled DNA and braided nicked molecules) interact similarly with Topo II. The exponential decrease of the relaxation rates with tension indicates that work is done against the molecular tension during the enzymatic rate limiting step, suggesting that the religation step may be rate limiting (12).
Topo IV was previously reported to relax preferentially (+) rather than (–) supercoiled DNA. This observation could be explained either by a preferential binding and cooperative assembly of Topo IV on positively coiled DNA or by the intrinsic orientation of the T and G segments on the enzyme that is disfavored by (–) supercoiling. Our results on the unbraiding of nicked DNA molecules support the later model: they show that Topo IV has a preference for a particular orientation of the T and G segments (8), corresponding to an L-node with acute angle. Numerical simulations have shown that the preferred substrate of Topo IV, (+) supercoiled DNA also forms L-nodes with an acute angle 2α ≈ 60° (23). Because the supercoiling of R-braids generates L-crossings, their unbraiding by Topo IV may proceed by DNA transport through these crossings. Thus the removal of one R-node implies two enzymatic cycles (the successive transport of two DNA segments through a third one; see Fig. 5c. Supercoiled R-braids should therefore be removed at half the rate of L-braids, as indeed is observed (see Fig. 4b).
Interestingly, the force dependence of the Topo IV unlinking rate differs from that of Topo II. At low forces, the Topo IV rate is constant, suggesting that (in contrast to Topo II) no work is performed against the force during its rate-limiting step. Thus not only does Topo IV differ from Topo II in its chiral discrimination capability, but these enzymes also have different rate-limiting steps.
The results presented here shed new light on the possible mechanisms of sister chromosome decatenation during replication in bacteria. If the replication complex is anchored to the bacterial membrane (22), it will establish a topological barrier preventing the back-diffusion of the (+) supercoils generated downstream from the replication fork and preempting the formation upstream of R-catenanes. The separated sister strands will thus remain unlinked, with the role of Topo IV limited exclusively to removal of the (+) supercoils, which it can do very efficiently (8, 10). In Salmonella, a defect in the separation of the daughter chromosomes (a partition defect) is associated with a mutation in a membrane protein, ParF (24). This protein is thought to anchor the replication complex (22), in agreement with the previous scenario. This model is not contradicted by the observation in partially replicated plasmids of both supercoiling of the mother strand and catenation of the daughter DNAs (25), because any topological barrier would be removed during extraction of the plasmid, allowing equilibration of the torsional stress across the replication fork.
If there is no topological barrier at the replication fork [as might be the case in E. coli where mutations of the gene homologous to parF (plsC) does not result in a partition defect (22)], the joint action of Topo IV and gyrase could be sufficient to unwind the mother strand, allowing its melting (26) in the terminal stage of replication and generating L-catenanes that could efficiently be unlinked by Topo IV (see Fig. 6a). Indeed, the Topo IV rates of relaxation of (+) supercoils and L-nodes measured in the present single molecule experiments (≈2.5 cycles per s at 25°C) are compatible with the removal of the extra links generated during replication (9, 11). The observation in vivo and in vitro of L-precatenanes when both Topo IV and gyrase are present (25) and the partition defect reported in gyrase mutants (27, 28) provides supporting evidence for that scenario. It is also consistent with the role of Topo IV as the primary decatenating enzyme (of L-braids) and of gyrase as the enzyme required for elongation synthesis (11, 21). The observation of R-braids when Topo IV is inhibited is easily explained by the lower rate of (+) supercoil removal by gyrase alone (9). In that situation, removal of the R-precatenanes can be done efficiently by overexpression of Topo III (29).
Fig. 6.
Configurations of a circular replicating DNA. (a) Model for in vivo strand separation: the progression of the replication complex leads to the formation of (+) supercoils (L-nodes) in front of the replication fork and R-precatenanes behind. Topo IV removes (+) supercoils and gyrase generates (–) supercoils so that, under the action of both enzymes, the chirality of the precatenanes is inverted (25). Topo IV may then unlink the molecules by removing the L-catenanes. (b) Model for in vitro decatenation by Topo IV: R-catenated plasmids may form L-supercoils that are removed by Topo IV with a high rate [as observed in our experiments and some bulk assays (21)]. However, the removal of the last few links is slow (11, 22), because Topo IV will relax only a rare fluctuation of an R-node to an obtuse angle that is similar to its preferred substrate: an L-node with acute angle (see Fig. 1b).
Although we believe the previous scenario to be the main decatenation pathway in vivo, it has nevertheless been shown that Topo IV can unlink R-precatenanes in vitro (11, 22). However, the rates of unlinking measured in these bulk experiments (kcat = 0.1 cycles per s) are 30 times smaller than both the rates reported in vivo and those measured here for the complete decatenation of L-braids (and the partial unlinking of R-braids). These slow decatenation events may result from the fluctuations of the R-intertwined molecules that will juxtapose the T and G segments in the correct orientation (an R-node with obtuse angle; see Fig. 1b) for Topo IV to act on.
Alternatively, as the present data show, the supercoiling of R-braids can provide an efficient (L-) substrate for decatenation by Topo IV. These L-plectonemic braids are expected if the number of interlinks n is large enough: n > 0.06Lk0 (15); see Fig. 6b. Moreover, if the individual plasmids are supercoiled, their interlinks could be localized and concentrated enough to locally induce plectonemic braids. These considerations may explain why the rates of unlinking of supercoiled and multiply linked plasmids are higher than the unlinking of nicked and singly linked catenanes (21).
Supplementary Material
Acknowledgments
We acknowledge helpful discussions with N. Cozzarelli, A. Vologodskii, M. Stone, and Z. Bryant and thank them for sharing their preprint, in which they describe a work similar to that presented here. We also thank N. Cozzarelli and N. Crisona for a gift of Topo IV, G. Lia and J.-F. Allemand for DNA constructs, S. Neukirch for a helpful suggestion, and O. Saleh for careful reading of the manuscript. This work was supported by grants from Association pour la Recherche sur le Cancer, Centre National de la Recherche Scientifique, Ecole Normale Supérieure, and Universités Paris VI and VII.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Topo, topoisomerase; T, transport; G, gate; L-, left-handed; R-, right-handed.
References
- 1.Wang, J. (1998) Q. Rev. Biophys. 31, 107–144. [DOI] [PubMed] [Google Scholar]
- 2.Wang, J. (2002) Nat. Rev. Mol. Cell Biol. 3, 430–440. [DOI] [PubMed] [Google Scholar]
- 3.Liu, L., Liu, C. & Alberts, B. (1980) Cell 19, 697–707. [DOI] [PubMed] [Google Scholar]
- 4.Roca, J. & Wang, J. (1992) Cell 71, 833–840. [DOI] [PubMed] [Google Scholar]
- 5.Roca, J. & Wang, J. (1996) Genes Cells 1, 17–27. [DOI] [PubMed] [Google Scholar]
- 6.Adams, D., Shekhtman, E., Zechiedrich, E., Schmid, M. & Cozzarelli, N. (1992) Cell 71, 277–288. [DOI] [PubMed] [Google Scholar]
- 7.Rybenkov, V., Ullsperger, C., Vologodskii, A. & Cozzarelli, N. (1997) Science 277, 690–693. [DOI] [PubMed] [Google Scholar]
- 8.Crisona, N., Strick, T., Bensimon, D., Croquette, V. & Cozzarelli, N. (2000) Genes Dev. 14, 2881–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zechiedrich, E. & Cozzarelli, N. (1995) Genes Dev. 9, 2859–2869. [DOI] [PubMed] [Google Scholar]
- 10.Postow, L., Crisona, N., Peter, B., Hardy, C. & Cozzarelli, N. (2001) Proc. Natl. Acad. Sci. USA 98, 8219–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiasa, H. & Marians, K. (1996) J. Biol. Chem. 271, 21529–21535. [DOI] [PubMed] [Google Scholar]
- 12.Strick, T., Croquette, V. & Bensimon, D. (2000) Nature 404, 901–904. [DOI] [PubMed] [Google Scholar]
- 13.Strick, T., Allemand, J.-F., Bensimon, D. & Croquette, V. (1998) Biophys. J. 74, 2016–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strick, T., Allemand, J., Bensimon, D., Bensimon, A. & Croquette, V. (1996) Science 271, 1835–1837. [DOI] [PubMed] [Google Scholar]
- 15.Marko, J. (1997) Phys. Rev. E 55, 1758–1772. [Google Scholar]
- 16.Smith, S., Finzi, L. & Bustamante, C. (1992) Science 258, 1122–1126. [DOI] [PubMed] [Google Scholar]
- 17.Bustamante, C., Marko, J., Siggia, E. & Smith, S. (1994) Science 265, 1599–1600. [DOI] [PubMed] [Google Scholar]
- 18.Marko, J. & Siggia, E. (1995) Macromolecules 28, 8759–8770. [Google Scholar]
- 19.Boles, T., White, J. & Cozzarelli, M. (1990) J. Mol. Biol. 213, 931–951. [DOI] [PubMed] [Google Scholar]
- 20.Rybenkov, V., Cozzarelli, N. & Vologodskii, A. (1993) Proc. Natl. Acad. Sci. USA 90, 5307–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ullsperger, C. & Cozzarelli, N. (1996) J. Biol. Chem. 271, 31549–31555. [DOI] [PubMed] [Google Scholar]
- 22.Levine, C., Hiasa, H. & Marians, K. (1998) Biochim. Biophys. Acta 1400, 29–43. [DOI] [PubMed] [Google Scholar]
- 23.Vologodskii, A. & Cozzarelli, N. (1996) Biophys. J. 70, 2548–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid, M. (1990) J. Bacteriol. 172, 5416–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peter, B., Ullsperger, C., Hiasa, H., Marians, K. & Cozzarelli, N. (1998) Cell 94, 819–827. [DOI] [PubMed] [Google Scholar]
- 26.Strick, T., Croquette, V. & Bensimon, D. (1998) Proc. Natl. Acad. Sci. USA 95, 10579–10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orr, E., Fairweather, N., Holland, I. & Pritchard, R. (1979) Mol. Gen. Genet. 177, 103–112. [DOI] [PubMed] [Google Scholar]
- 28.Steck, T. & Drlica, K. (1984) Cell 36, 1081–1088. [DOI] [PubMed] [Google Scholar]
- 29.Nurse, P., Levine, C., Hassing, H. & Marians, K. (2003) J. Biol. Chem. 278, 8653–8660. [DOI] [PubMed] [Google Scholar]
- 30.Wang, M., Schnitzer, M., Yin, H., Landick, R., Gelles, J. & Block, S. (1998) Science 282, 902–907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.