Abstract
The murine JNK-interacting protein 3 (JIP3) protein (also known as JSAP1) is expressed exclusively in neurons and has been identified as a scaffold protein for the c-Jun NH2-terminal kinase (JNK) signaling pathway and as an adapter protein for cargo transport by the microtubule motor protein kinesin. To investigate the physiological function of JIP3, we examined the effect of Jip3 gene disruption in mice. The Jip3–/– mice were unable to breathe and died shortly after birth. Microscopic analysis demonstrated that Jip3 gene disruption causes severe defects in the morphogenesis of the telencephalon. Jip3–/– mice lack the telencephalic commissure, a major connection between the left and right hemispheres of the brain. The central nervous system abnormalities of Jip3–/– mice may be accounted for in part by a reduction in signal transduction by RhoA and its effector ROCK.
The c-Jun NH2-terminal kinase-interacting protein 3 (JIP3) scaffold protein (also known as JSAP1) is expressed selectively in neurons (1–3). Orthologs of JIP3 have been identified in model organisms, and genetic analysis has identified important roles for JIP3 in neuronal function. Thus, the Drosophila JIP3 ortholog sunday driver is implicated in kinesin-dependent axonal vesicular transport (4). Similarly, the Caenorhabditis elegans ortholog UNC-16 regulates kinesin-dependent transport of vesicular cargo in neurons (5). The mechanism of JIP3 function in vesicular cargo trafficking seems to be mediated by an interaction of JIP3 with kinesin light chain (4). JIP3 therefore may function as an adaptor protein that mediates, in part, kinesin-dependent vesicular transport in neurons (6).
In addition to the function of JIP3 as a cargo adapter for the microtubule motor protein kinesin, biochemical studies have demonstrated that JIP3 also functions as a putative scaffold for the JNK signaling pathway (1, 2). JIP3 binds all JNK isoforms but seems to preferentially interact with the neuron-specific isoform JNK3. JIP3 also binds the mitogen-activated protein kinase (MAPK) kinase 7 (MKK7) and members of the mixed-lineage protein kinase (MLK) family of MKK kinases. JIP3 can assemble a functional signaling module and strongly potentiates JNK activation caused by MLK. Together, these data indicate that JIP3 is a candidate scaffold protein for the JNK signal transduction pathway in neurons (7).
JIP3 may be a multifunctional protein that serves both as a MAPK scaffold protein and as a cargo adapter for kinesin-mediated axonal transport (6, 7). However, these biological processes may be related. For example, JIP3 may contribute to the regulation of the subcellular distribution of JNK signaling modules within neurons. It is also possible that JNK signaling modules tethered to JIP3 act as a mechanism of localized regulation of kinesin-mediated axonal transport. This latter possibility is supported by genetic analysis of genes that encode orthologs of JIP3 and JNK pathway components in C. elegans (5). Nevertheless, the physiological role of JIP3 in mammals has not been established.
Insight into the possible function of JIP3 in mammals has been obtained from studies of DLK, a neuron-specific MLK that activates JNK (8). It has been demonstrated that DLK causes JNK activation during brain development and that this signaling pathway contributes to morphogenesis of the telencephalon (8). Because JIP3 can assemble a functional MLK signaling module, JIP3 may also be required for the normal development of the telencephalon. The role of DLK signaling in telencephalon morphogenesis has been proposed to be mediated in part by regulation of the neuronal microtubule network (8). However, the target of this pathway that is relevant to telencephalon morphogenesis has not been defined.
The purpose of this study was to examine the role of JIP3 in mammals. Our approach was to investigate the effect of Jip3 gene disruption in mice. We show that Jip3 is not required for embryonic viability. However, Jip3 is essential for life after birth. Jip3–/– mice are unable to breathe and exhibit severe defects in the development of the telencephalon, consistent with the proposed role of JIP3 as a regulator of MLK function. We show that this defect in central nervous system morphogenesis may be caused in part by reduced signal transduction by RhoA and its effector ROCK.
Methods
Mice. Jip3–/+ embryonic stem cells were prepared by homologous recombination and used to create mice by using standard methods. A murine strain 129 genomic clone was isolated from a bacterial artificial chromosome library by hybridization with a probe derived from the JIP3 cDNA. Sequence analysis confirmed that the clone encoded the Jip3 gene. A targeting vector was designed to replace exon 1 with a NeoR cassette (Fig. 1A). The targeting vector was linearized with NotI and electroporated into embryonic stem cells and subsequently selected by using G418 and gancyclovir. Positive clones were identified by Southern blot analysis, and two clones were used to create chimeric mice by blastocyst injection. Both clones transmitted the disrupted Jip3 gene through the germ line. The mice were backcrossed for 10 generations to the C57BL/6 strain (The Jackson Laboratory) and maintained by mating Jip3–/+ mice. Jnk1–/– (9) and Jnk2–/– (10) mice were as described and backcrossed to the C57BL/6 strain for 10 generations (11). The animals were housed in a facility accredited by the American Association for Laboratory Animal Care, and the animal studies were approved by the Institutional Animal Care and Use Committee of the University of Massachusetts.
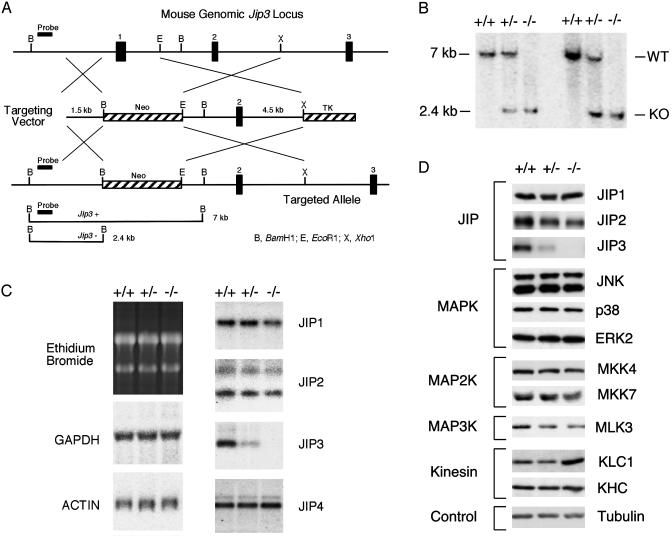
Fig. 1.
Targeted disruption of the murine Jip3 gene. (A) Schematic illustration of the strategy used for disruption of the Jip3 gene. A fragment of the wild-type gene that includes exons 1–3 is shown. The targeting vector was designed to replace exon 1 with a NeoR cassette. The diagram also indicates the probe used for distinguishing the wild-type and disrupted alleles by Southern blot analysis. TK, thymidine kinase. (B) Mice with homozygous and heterozygous disruptions of the Jip3 gene were identified by Southern blot analysis. WT, wild type; KO, knockout. (C) Total RNA isolated from the brains of E18.5 mice was examined by Northern blot analysis by using probes for GAPDH, actin, JIP1, JIP2, JIP3, and JIP4. Equal loading of RNA isolated from wild-type, Jip3–/+, and Jip3–/– embryos was confirmed by staining of ribosomal RNA with ethidium bromide. (D) E18.5 embryonic brains from wild-type, Jip3–/+, and Jip3–/– mice were examined by immunoblot analysis. Control experiments were performed by probing the blots with an antibody to tubulin. The increased expression of kinesin light chain in Jip3–/– mice shown in the figure was not reproducibly observed in independent experiments. KLC1, kinesin light chain 1; KHC, kinesin heavy chain.
Histology. Heads were partitioned by coronal sectioning into three regions, fixed in 10% neutral buffered formalin, processed, paraffin-embedded, and prepared as standard tissue sections with hematoxylin/eosin staining. Sections of lungs obtained from newborn mice were also prepared. Tissues were examined microscopically by a board-certified veterinary pathologist.
Immunocytochemistry. Heads from embryonic day (E)15.5 embryos were fixed in 10% neutral buffered formalin and processed in paraffin wax following standard procedures. Serial sections were cut through the heads, and selected sections were stained with antibodies raised against phospho-c-Jun (Cell Signaling Technology, Beverly, MA) and phospho-JNK (Cell Signaling Technology). The primary antibodies were visualized by using biotinylated MultiLink secondary antibodies (BioGenex Laboratories, San Ramon, CA), streptavidin peroxidase (BioGenex Laboratories), and diaminobenzidine (Vector Laboratories). Sections were counterstained with Mayer's hematoxylin.
Immunoblot Analysis. Tissue extracts were prepared by using lysis buffer (25 mM Hepes, pH 7.5/0.3 M NaCl/0.2 mM EDTA/0.1% Triton X-100/0.5 mM DTT/20 mM β-glycerophosphate/0.1 mM vanadate/2 μg/ml leupeptin/2 μg/ml aprotinin/1 mM PMSF). The extracts were clarified by using an Eppendorf microcentrifuge (14,000 rpm for 10 min at 4°C). The concentration of total soluble protein in the supernatant was quantitated by the Bradford method (Bio-Rad). Proteins were resolved by SDS/PAGE (10% gel) and electrophoretically transferred to an Immobilon-P membrane (Millipore). The membranes were incubated with 5% nonfat dry milk (4°C for 5 h) and then probed with primary antibodies. Immune complexes were detected by enhanced chemiluminescence (NEN).
Antibodies. JIP1, JIP2, and JIP3 antibodies have been described (1, 12). The antibodies to kinesin light chain 1 and MLK3 were provided by L. S. Goldstein (University of California, San Diego) and K. Gallo (Michigan State University, East Lansing), respectively. The antibodies to α-tubulin (Sigma), JNK (PharMingen), p38 MAPK (Santa Cruz Biotechnology), extracellular signal-regulated kinase 2 (Upstate Biotechnology, Lake Placid, NY), MKK4 (Santa Cruz Biotechnology), MKK7 (Zymed), kinesin heavy chain (Abcam, Cambridge, U.K.), GluR1 (Sigma), RhoA (Santa Cruz Biotechnology), ROCK (Santa Cruz Biotechnology), Munc18-1 (BD Transduction Laboratories, San Diego), Snap25 (Sigma), Synaptotagmin (Sigma), and Neuropilin (Sigma) were purchased from the indicated suppliers.
RNA Analysis. Total RNA isolated from E18.5 embryonic brains was examined by Northern blot analysis by using standard procedures. Microarray analysis was performed by using U74 chips in triplicate following manufacturer recommendations (Affymetrix, Santa Clara, CA).
Results
Disruption of the Murine Jip3 Gene. A targeting vector was designed to replace exon 1 of the Jip3 gene with a NeoR cassette by using homologous recombination in murine embryonic stem cells (Fig. 1 A). Two clones were identified by Southern blot analysis. These clones were injected into blastocysts to create chimeric mice. Germ-line transmission was obtained with both clones. The Jip3–/+ mice were found to be viable and fertile with no detected anatomical abnormalities. Time-course experiments indicated that the life span of the heterozygous animals was normal.
Jip3 Is an Essential Gene. Matings of Jip3–/+ mice yielded wild-type and Jip3–/+ littermates at weaning, but no Jip3–/– mice were obtained (Table 1). This observation suggested that homozygous inactivation of the Jip3 gene is a lethal event. We therefore examined the genotype of embryos. Southern blot analysis led to the identification of wild-type, heterozygous, and homozygous knockout embryos (Fig. 1B). Northern blot analysis of RNA isolated from embryonic brains demonstrated that Jip3 gene disruption caused loss of Jip3 mRNA but did not cause altered expression of mRNA encoding other JIP isoforms (Fig. 1C). Immunoblot analysis confirmed that JIP3 was not expressed in Jip3–/– embryos and demonstrated that no marked alteration in the expression of other JIP proteins was detected (Fig. 1D). Immunoblot analysis of embryonic brains also demonstrated no marked alterations in the expression of JNK pathway components (JNK, MKK4, MKK7, and MLK3), other MAPKs (p38 and extracellular signal-regulated kinase), or the expression of kinesin heavy chain (KHC) and kinesin light chain 1 (KLC1) (Fig. 1D).
Table 1. Genotype analysis of littermates obtained after crossing Jip3-/+ mice.
| No. of mice | +/+ | -/+ | -/- | |
|---|---|---|---|---|
| E13.5 | 32 | 8 | 17 | 7 |
| E15.5 | 22 | 6 | 12 | 4 |
| E17.5 | 448 | 119 | 214 | 115 |
| Weaned | 232 | 101 | 131 | 0 |
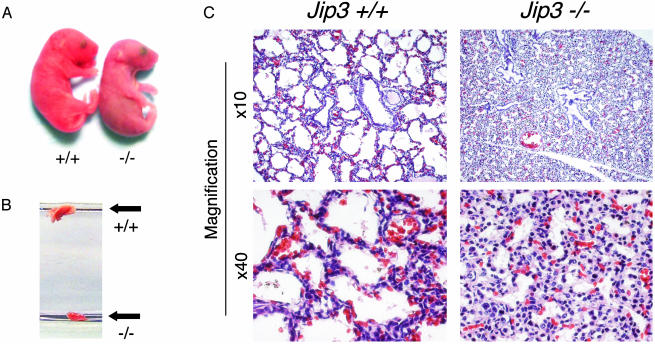
Analysis of E13.5, E15.5, and E17.5 embryos demonstrated the expected Mendelian ratios of wild-type, heterozygous, and homozygous knockout embryos (Table 1). Because no Jip3–/– mice were detected at weaning, these data indicated that Jip3–/– mice may die shortly after birth. Indeed, newborn Jip3–/– mice became rapidly anoxic and died (Fig. 2A). Visual inspection suggested that newborn Jip3–/– mice died because they did not breathe. To test this hypothesis, we removed the lungs from wild-type and Jip3–/– mice and placed the lungs in PBS (Fig. 2B). As expected, the wild-type lungs were inflated and floated. In marked contrast, the Jip3–/– lungs did not float. This observation suggested that the Jip3–/– lungs did not inflate after birth. Microscopic analysis confirmed this conclusion (Fig. 2C). Together, these data indicate that a failure to breathe after birth is likely to be a major cause of death of newborn Jip3–/– mice.
Fig. 2.
Jip3 gene disruption causes defective lung development. (A) Newborn wild-type and Jip3–/– littermates obtained immediately after birth. The mutant mice appear to be morphologically normal but exhibit symptoms of severe anoxia and die shortly after birth. (B) The lungs of newborn wild-type and Jip3–/– littermates were dissected and placed in PBS. The wild-type and mutant lungs are indicated. (C) Microscopic comparison of lungs from newborn wild-type and Jip3–/– littermates. Wild-type alveolar spaces are expanded, indicating respiration; however, Jip3–/– alveoli are still collapsed, and the septae are hypercellular. The sections were stained with hematoxylin/eosin.
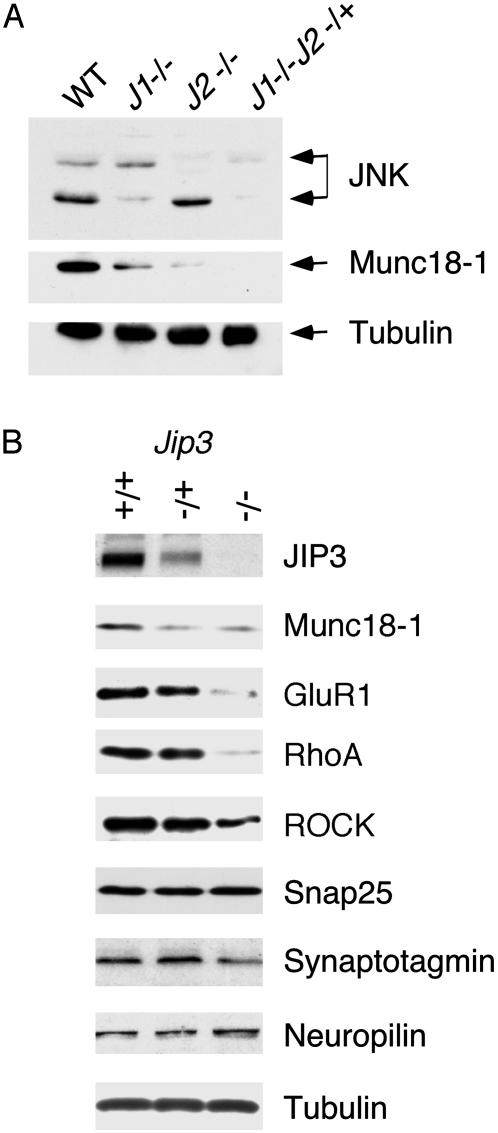
Munc18-1 Expression Is Reduced in JIP3-Deficient and JNK-Deficient Mice. JIP3 is selectively expressed in neurons. The failure of Jip3–/– mice to breathe therefore may reflect a defect in neuronal function, including defects in neuronal gene expression. One candidate gene is the Unc18/Sec1 ortholog Munc18-1, which is expressed at low levels in the brain of JNK-deficient mice (Fig. 3A). Indeed, immunoblot analysis indicated that Munc18-1 was also expressed at low levels in the brain of Jip3–/– embryos (Fig. 3B). Munc18-1 binds syntaxin 1 and is essential for synaptic vesicle fusion with the presynaptic membrane (13, 14). Munc18-1 knockout mice completely lack neurotransmitter secretion and synaptic activity (15). The reduced expression of Munc18-1 in Jip3–/– mice therefore may cause decreased synaptic activity and may contribute to the failure of these mice to breathe. However, the decreased expression of Munc18-1 by itself is not sufficient to account for the failure to breathe, because reduced Munc18-1 expression was also detected in Jip3–/+ mice (Fig. 3B).
Fig. 3.
Effect of Jip3 and Jnk gene disruption on protein expression in the brain. (A) Brain extracts were examined by immunoblot analysis by using antibodies to JNK, Munc18-1, and tubulin. The results of immunoblot analysis of wild-type (WT), Jnk1–/– (J1–/–), Jnk2–/– (J2–/–), and Jnk1–/–Jnk2–/+ (J1–/–J2–/+) mice are shown. The compound mutant Jnk1–/–Jnk2–/– was not examined because these mice die during early embryonic development. (B) E18.5 embryonic brains from wild-type, Jip3–/+, and Jip3–/– mice were examined by immunoblot analysis. Control experiments were performed by probing the blots with an antibody to tubulin.
Gene Expression in Jip3–/– Mice. We performed microarray analysis using mRNA isolated from wild-type and Jip3–/– brains from E18.5 embryos to examine changes in gene expression. This analysis demonstrated that the gene-expression profile in wild-type and knockout brains was substantially identical. However, altered expression of a limited group of genes was detected in the Jip3–/– brains (Table 2) including the Rho-GEF Net1 and the Rho-associated protein kinase ROCK. Furthermore, decreased expression of a number of glutamate receptor subunits was observed including the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunit Gria4, the N-methyl-d-aspartate receptor subunit Grin2A, and the kainate receptor subunit Grik5 (Table 2). To confirm these changes in gene expression identified by microarray analysis, we performed immunoblot analysis using lysates prepared from E18.5 embryonic brains. Decreased expression of RhoA, ROCK, and the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunit Gria1 (GluR1) was detected in Jip3–/– brains (Fig. 3B). In contrast, the expression of a number of other neuronal proteins was not changed markedly by Jip3 gene disruption including Snap25, Synaptotagmin, Neuropilin, and tubulin (Fig. 3B).
Table 2. Comparison of gene expression by microarray analysis of mRNA isolated from the brain of E18.5 wild-type and Jip3-/- mice.
| Description | Fold change, Jip3-/- vs. wild-type |
|---|---|
| BMP4 | -1.2 |
| Dynein intermediate chain 2 | 1.1 |
| Glial fibrillary acidic protein | -1.8 |
| Gria4 | -3 |
| Grik5 | -3 |
| Grin2A | -3.3 |
| Kinesin heavy chain member 5C | -1.4 |
| MAP5 | 1.1 |
| MAP2 | 1.8 |
| Munc-18 | -3 |
| Netrin-1 | 0 |
| Neurofilament heavy polypeptide | 1.4 |
| Neurofilament protein NF-L | -0.9 |
| Neuronal pentraxin 2 | -1.4 |
| Neuropeptide Y receptor type 5 | -1.4 |
| Notch 2 | -1.2 |
| Rho-GEF Net 1 | -2.9 |
| ROCK | -3.2 |
| Synaptotagmin I | -1.5 |
| Tau | 1.1 |
Together, these data indicate that Jip3 gene disruption causes selective changes in neuronal gene expression. The decreased expression of glutamate receptors and Munc18-1 suggests that synaptic activity in the Jip3–/– brain is likely to be impaired (13, 15). Furthermore, the decreased expression of RhoA, Net1, and ROCK suggests that Jip3 gene disruption may cause defects in neuronal growth-cone function and axon guidance (16, 17). It therefore is possible that decreased synaptic activity and/or altered axonal pathfinding may contribute to the inability of Jip3–/– mice to breathe (Fig. 2).
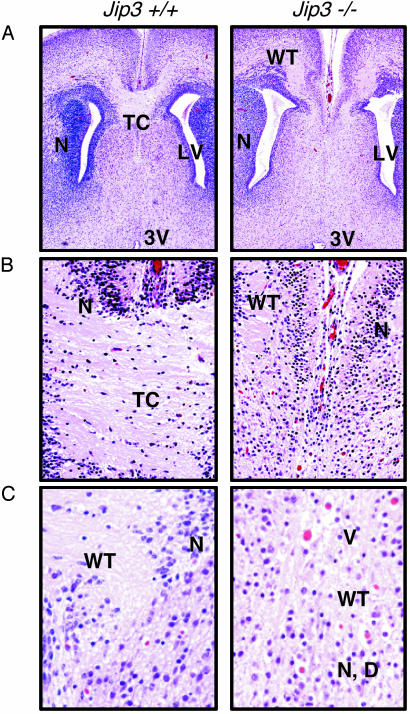
Jip3 Gene Disruption Prevents Normal Development of the Telencephalon. The Jip3–/– brains, although smaller than their wild-type counterparts, did not have any macroscopic abnormalities. Microscopic comparison of wild-type and Jip3–/– embryo brains suggested that many areas have a similar overall morphology and organization including the ependymal linings, neuronal rests, neuronal layers, and glial cell densities. However, serial sectioning to allow examination of similar structures at multiple depths revealed significant bilateral defects in the organization of the medial aspect of the telencephalon where the two hemispheres should be joined (Fig. 4). Wild-type animals had a well delineated, highly organized structure, composed of myelinated axons, that connects the medial aspect of the two brain hemispheres (telencephalic commissure). Many of the adjacent neurons are organized into layers and discrete clusters. In contrast, Jip3–/– brains lack this interhemispheric connection, and the adjacent white matter and neurons are relatively disorganized in comparison to the wild-type mice. Jip3–/– neurons are more randomly distributed and mixed with scattered debris. White-matter tracts in areas adjacent to the typical location of the telencephalic commissure were disorganized and contained many vacuoles (Fig. 4). These observations indicated that JIP3 has an essential role in the normal morphogenesis of the telencephalon.
Fig. 4.
Jip3 gene disruption causes defects in the formation of the telencephalon. (A) The wild-type E18.5 brain has compact neuronal layers (N) surrounding the lateral ventricles (LV) and a well demarcated thick band of axonal fibers [telencephalic commissure (TC)] connecting the hemispheres at the level of the lateral ventricles. (Magnification, ×5.) The Jip3–/– brain has a more diffuse, less compact zone of neurons (N) surrounding each dilated lateral ventricle, and bilateral white tract (WT) areas in each hemisphere have no connection to the opposite side. 3V, third ventricle. (B) An organized zone of axons connecting the hemispheres of wild-type brains. (Magnification, ×20.) In contrast, no white matter intrahemispheric connection in the Jip3–/– brain was observed. (C) Compact neuronal layers (N) and a well demarcated thick band of axonal fibers [white-matter tracts (WT)] connecting the hemispheres in the wild-type brain. (Magnification, ×40.) The Jip3–/– brain has more randomly distributed neurons (N) with scattered debris (D) and disorganized white tracts with frequent vacuoles (V). All slides were stained with hematoxylin/eosin.
Discussion
JIP3 has been reported to be a multifunctional protein that may act both as a scaffold protein for a JNK signaling module and as an adapter protein for kinesin-mediated axonal transport (1, 2, 4, 5). Genetic analysis of model organisms indicates that orthologs of JIP3 in Drosophila (sunday driver) and C. elegans (unc-16) are required for normal axonal transport (4, 5). The mechanism of transport seems to be mediated by an interaction of JIP3 with the light chain of the microtubule motor protein kinesin 1 (6). Mutant worms and flies lacking JIP3 mislocalize and accumulate vesicular cargo within axon and dendritic processes. For example, unc-16 mutant worms mislocalize both synaptic vesicles and glutamate receptors, suggesting that UNC-16 is required for transport of synaptic vesicles or synaptic vesicle precursors (5). This transport function may account for the observation that sunday driver and unc-16 are essential genes in flies and worms, respectively. Studies of JIP3-deficient mice demonstrate that Jip3 is also an essential gene (Table 1). However, the critical role of JIP3 that is required for mouse viability may not be the same as that in flies and worms, because the transport functions of murine JIP3 may be partially redundant with those of other cargo adapter proteins (6).
Jip3 Is an Essential Gene. Disruption of the Jip3 gene in mice causes death shortly after birth. Failure to breathe represents one cause of newborn death (Fig. 2). The mechanism that accounts for the breathing failure is unclear. Because JIP3 is selectively expressed in neurons, it is likely that a neuronal defect is a major cause of the failure to breathe. Two different types of neuronal defects were identified in Jip3–/– mice that may be relevant to this phenotype: altered expression of proteins that are required for synaptic transmission (Fig. 3) and defective axonal pathfinding and target cell innervation (Fig. 4). Further studies are required to determine whether these classes of neuronal defects contribute to the breathing failure observed in Jip3–/– mice.
The finding that Jip3 is an essential gene contrasts with previous studies of Jip1. Like JIP3, JIP1 was identified as a scaffold protein for a JNK signaling module that binds the microtubule motor protein kinesin (12, 18, 19). However, JIP1 and JIP3 are structurally distinct proteins (7). Jip1 is not an essential gene, and Jip1–/– mice are viable and fertile with no detected developmental defects (12), although Jip1 gene disruption may cause a preimplantation lethal phenotype in some mouse-strain genetic backgrounds (20). The viable Jip1–/– mice exhibit defects in neuronal apoptosis caused by excitotoxins and ischemia (12). These observations indicate that JIP1 and JIP3 serve nonredundant functions in mice. Further studies of JIP2- and JIP4-deficient mice will be required to define the spectrum of activities of the JIP group of MAPK scaffold proteins that interact with the microtubule motor protein kinesin (7).
Comparative immunocytochemical studies of wild-type and Jip3–/– mice performed by staining serial sections of embryonic brains with antibodies that bind phospho-JNK or phospho-Jun did not reveal major defects in JNK activation in the Jip3–/– embryos (unpublished observation). Similarly, obvious abnormal accumulations of axonal vesicles were not detected by this analysis. Several mechanisms could account for these observations including the possibility that JIP3 function is developmental stage-specific, that it is restricted to a specific subregion of the brain, or that immunocytochemical methods are insufficiently sensitive to detect the Jip3–/– phenotype. It is possible that measurement of a robust defect in JNK activation in Jip3–/– mice may require neuronal stimulation. For example, defects in JNK activation in Jip1–/– brains were only observed in studies of excitotoxin stimulation or exposure to anoxia (12). The lethal phenotype of Jip3 gene disruption did not enable similar studies of Jip3–/– mice to be performed.
Jip3 Is Required for Morphogenesis of the Telencephalon. Previous studies have established that the brain-specific MLK isoform DLK functions as an MKK kinase that causes JNK activation in the developing mouse telencephalon and that altered DLK/JNK signaling disrupts telencephalon morphogenesis (8). The JIP3 scaffold protein can regulate JNK signaling modules that are initiated by MLK isoforms (7). Regulation of the MLK signaling pathway by the JIP3 scaffold protein therefore may contribute to the development of the telencephalon. To test this hypothesis, we performed microscopic analysis of brains obtained from wild-type and Jip3–/– embryos (Fig. 4). This analysis demonstrated disrupted formation of the telencephalon in Jip3–/– mice. Two major defects were observed. First, the Jip3–/– telencephalon contained poorly organized neurons with scattered debris and disorganized white tracts. Second, the telencephalic commissure (a major interhemispheric connection in the brain) was absent in Jip3–/– mice. The disorganized telencephalon in Jip3–/– mice indicates that JIP3 is required for telencephalon morphogenesis. Furthermore, the absence of the telencephalic commissure suggests that these Jip3–/– neurons exhibit severe axonal pathfinding defects. This axon guidance defect may reflect a transport role for JIP3 in the normal function of growth cones during development. JIP3 may also influence growth-cone migration by regulating a DLK signaling module. Whether altered JNK function is sufficient to account for the axonal pathfinding defect is unclear because compound mutant mice that lack JNK expression die early in development before morphogenesis of the telencephalon (21). Future studies of conditional JNK knockout mice will be required to provide an answer to this question.
Jip3 Is Required for Normal Neuronal Gene Expression. Analysis of gene expression by microarray analysis did not demonstrate widespread differences between mRNA isolated from wild-type and Jip3–/– mice. Nevertheless, significant differences in gene expression were detected (Table 2). These changes in gene expression may reflect defects in DLK signaling caused by JIP3 deficiency. Three functionally distinct groups of genes were found to be down-regulated in the brains of Jip3–/– mice.
First, decreased expression of several classes of glutamate receptor subunits was observed (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid, N-methyl-d-aspartate, and kainate). The decreased expression of glutamate receptors may cause inefficient synaptic function in Jip3–/– mice. Second, decreased expression of the Unc-18/Sec1 ortholog Munc18-1 was observed in Jip3–/– brains. Munc18-1 binds syntaxin 1 and is essential for synaptic vesicle fusion with the presynaptic membrane (13, 14). Munc18-1 knockout mice completely lack neurotransmitter secretion and synaptic activity (15). Interestingly, Munc18-1 deficiency does not impair the morphological development of the brain. The reduced expression of Munc18-1 in Jip3–/– mice therefore may contribute to decreased synaptic activity. The failure of Jip3–/– mice to breathe may reflect changes in neuronal function caused by decreased expression of glutamate receptors and Munc18-1.
The third group of genes found to be down-regulated correspond to genes that have been implicated in growth-cone function and the control of axon guidance during development. Specifically, Jip3–/– brains expressed lower levels of RhoA, the Rho-GEF Net1, and the Rho-associated protein kinase ROCK. The decreased expression of RhoA and Net1 may cause reduced levels of GTP-loaded Rho proteins in Jip3–/– brains during embryonic development. This reduced activation of Rho, together with reduced expression of the Rho effector ROCK, would be expected to cause defects in axon pathfinding and migration (16, 17, 22). Indeed, the observation that the telencephalic commissure is absent in Jip3–/– mice may reflect abnormal Rho signaling because it is known that altered Rho signaling causes defects in the morphogenesis of commissural neurons (23, 24). Furthermore, cell surface Eph receptors that are required for axonal pathfinding by commissural neurons (25, 26) are known to function by regulating RhoA by a mechanism that involves the Rho-GEF ephexin (27). Together, these data indicate that the observed defects in RhoA and its effector ROCK may account in part for the effects of Jip3 gene disruption on the development of the telencephalic commissure.
Acknowledgments
We thank Drs. L. S. Goldstein and K. Gallo for providing antibodies; Judith Riley and Linda Lesco for technical assistance; and K. Gemme for administrative assistance. This study was supported in part by a grant from the National Cancer Institute. R.A.F. and R.J.D. are investigators of the Howard Hughes Medical Institute.
Abbreviations: JNK, c-Jun NH2-terminal kinase; JIP3, JNK-interacting protein 3; MAPK, mitogen-activated protein kinase; MKK, MAPK kinase; MLK, mixed-lineage protein kinase; En, embryonic day n.
References
- 1.Kelkar, N., Gupta, S., Dickens, M. & Davis, R. J. (2000) Mol. Cell. Biol. 20, 1030–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito, M., Yoshioka, K., Akechi, M., Yamashita, S., Takamatsu, N., Sugiyama, K., Hibi, M., Nakabeppu, Y., Shiba, T. & Yamamoto, K. I. (1999) Mol. Cell. Biol. 19, 7539–7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akechi, M., Ito, M., Uemura, K., Takamatsu, N., Yamashita, S., Uchiyama, K., Yoshioka, K. & Shiba, T. (2001) Neurosci. Res. 39, 391–400. [DOI] [PubMed] [Google Scholar]
- 4.Bowman, A. B., Kamal, A., Ritchings, B. W., Philp, A. V., McGrail, M., Gindhart, J. G. & Goldstein, L. S. (2000) Cell 103, 583–594. [DOI] [PubMed] [Google Scholar]
- 5.Byrd, D. T., Kawasaki, M., Walcoff, M., Hisamoto, N., Matsumoto, K. & Jin, Y. (2001) Neuron 32, 787–800. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein, L. S. (2001) Proc. Natl. Acad. Sci. USA 98, 6999–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrison, D. & Davis, R. J. (2003) Annu. Rev. Cell Dev. Biol., in press. [DOI] [PubMed]
- 8.Hirai, S., Kawaguchi, A., Hirasawa, R., Baba, M., Ohnishi, T. & Ohno, S. (2002) Development (Cambridge, U.K.) 129, 4483–4495. [DOI] [PubMed] [Google Scholar]
- 9.Dong, C., Yang, D. D., Wysk, M., Whitmarsh, A. J., Davis, R. J. & Flavell, R. A. (1998) Science 282, 2092–2095. [DOI] [PubMed] [Google Scholar]
- 10.Yang, D. D., Conze, D., Whitmarsh, A. J., Barrett, T., Davis, R. J., Rincon, M. & Flavell, R. A. (1998) Immunity 9, 575–585. [DOI] [PubMed] [Google Scholar]
- 11.Weston, C. R., Wong, A., Hall, J. P., Goad, M. E. P., Flavell, R. A. & Davis, R. J. (2003) Genes Dev. 17, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitmarsh, A. J., Kuan, C. Y., Kennedy, N. J., Kelkar, N., Haydar, T. F., Mordes, J. P., Appel, M., Rossini, A. A., Jones, S. N., Flavell, R. A., et al. (2001) Genes Dev. 15, 2421–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizo, J. & Sudhof, T. C. (2002) Nat. Rev. Neurosci. 3, 641–653. [DOI] [PubMed] [Google Scholar]
- 14.Toonen, R. F. & Verhage, M. (2003) Trends Cell Biol. 13, 177–186. [DOI] [PubMed] [Google Scholar]
- 15.Verhage, M., Maia, A. S., Plomp, J. J., Brussaard, A. B., Heeroma, J. H., Vermeer, H., Toonen, R. F., Hammer, R. E., van den Berg, T. K., Missler, M., et al. (2000) Science 287, 864–869. [DOI] [PubMed] [Google Scholar]
- 16.Amano, M., Fukata, Y. & Kaibuchi, K. (2000) Exp. Cell Res. 261, 44–51. [DOI] [PubMed] [Google Scholar]
- 17.Bito, H., Furuyashiki, T., Ishihara, H., Shibasaki, Y., Ohashi, K., Mizuno, K., Maekawa, M., Ishizaki, T. & Narumiya, S. (2000) Neuron 26, 431–441. [DOI] [PubMed] [Google Scholar]
- 18.Whitmarsh, A. J., Cavanagh, J., Tournier, C., Yasuda, J. & Davis, R. J. (1998) Science 281, 1671–1674. [DOI] [PubMed] [Google Scholar]
- 19.Verhey, K. J., Meyer, D., Deehan, R., Blenis, J., Schnapp, B. J., Rapoport, T. A. & Margolis, B. (2001) J. Cell Biol. 152, 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson, N. A., Haefliger, J. A., Senn, A., Tawadros, T., Magara, F., Ledermann, B., Nicod, P. & Waeber, G. (2001) J. Biol. Chem. 276, 27745–27748. [DOI] [PubMed] [Google Scholar]
- 21.Davis, R. J. (2000) Cell 103, 239–252. [DOI] [PubMed] [Google Scholar]
- 22.Wittmann, T. & Waterman-Storer, C. M. (2001) J. Cell Sci. 114, 3795–3803. [DOI] [PubMed] [Google Scholar]
- 23.Brouns, M. R., Matheson, S. F., Hu, K. Q., Delalle, I., Caviness, V. S., Silver, J., Bronson, R. T. & Settleman, J. (2000) Development (Cambridge, U.K.) 127, 4891–4903. [DOI] [PubMed] [Google Scholar]
- 24.Brouns, M. R., Matheson, S. F. & Settleman, J. (2001) Nat. Cell Biol. 3, 361–367. [DOI] [PubMed] [Google Scholar]
- 25.Orioli, D., Henkemeyer, M., Lemke, G., Klein, R. & Pawson, T. (1996) EMBO J. 15, 6035–6049. [PMC free article] [PubMed] [Google Scholar]
- 26.Henkemeyer, M., Orioli, D., Henderson, J. T., Saxton, T. M., Roder, J., Pawson, T. & Klein, R. (1996) Cell 86, 35–46. [DOI] [PubMed] [Google Scholar]
- 27.Shamah, S. M., Lin, M. Z., Goldberg, J. L., Estrach, S., Sahin, M., Hu, L., Bazalakova, M., Neve, R. L., Corfas, G., Debant, A. & Greenberg, M. E. (2001) Cell 105, 233–244. [DOI] [PubMed] [Google Scholar]