Abstract
While studying the import of the hepatitis B virus genome into the nucleus of permeabilized tissue culture cells, we found that viral capsids were imported in intact form through the nuclear pore into the nuclear basket. Import depended on phosphorylation of the capsid protein and was mediated by the cellular transport receptors importin α and β. Virus-derived capsids that contained the mature viral genome were able to release the viral DNA and capsid protein into the nucleoplasm. The uncoating reaction was independent of Ran, a GTP-binding enzyme responsible for dissociating other imported cargoes from the inner face of the nuclear pore. Immature capsids that did not contain the mature viral genome reached the basket but did not release capsid proteins nor immature genomes into the nucleoplasm. The different fate of mature and immature capsids after passing the nuclear pore indicates that the outcome of a nuclear import event may be regulated within the nuclear basket.
The nucleoplasm is separated from the cytoplasm by the nuclear envelope, a double membrane contiguous with the endoplasmic reticulum. Macromolecular traffic between the two compartments occurs through nuclear pore complexes (NPCs). For most macromolecules, transport through the NPC is tightly regulated. Import is typically mediated by soluble cytosolic transport receptors of the importin β (Impβ) (karyopherin β) superfamily. Importins facilitate the binding of the cargo–receptor complex to the NPC and its translocation through the pore into the nuclear basket. The complex stays in the basket until the Ras-related nuclear protein (Ran) in its GTP-associated form dissociates the cargo from the receptor (1), allowing diffusion of the cargo into the karyoplasm.
Many viruses that replicate within the nucleus of nondividing cells use importins to ensure import of their genomes into the nucleus (2). Because of the limited size of the NPC, capsids of large viruses such as herpes virus, adenovirus, and influenza virus release their genome before nuclear import. It has been suggested that small viruses such as parvoviruses may enter through the NPC in intact form (3). With diameters of 32 or 36 nm, the hepatitis B virus (HBV) capsids analyzed in this study are close to the size limit of the NPC but enter the nuclei in intact form (4).
HBV is an important pathogen that causes acute and chronic hepatitis and hepatocellular carcinoma. The early events of HBV infection are hard to analyze because no susceptible cell line exists. HBV particles contain a DNA genome. They replicate via a pregenomic RNA (PG) synthesized in the nucleus of the infected cell. After export from the nucleus to the cytosol, the PG is trapped within a newly formed isometric capsid (core particle) together with a cellular protein kinase (5–10). The shell of the capsid contains 180 or 240 identical capsid protein subunits of 21.5 kDa (11). Reverse transcription of the PG and subsequent synthesis of an incomplete second strand of DNA generate the mature genome inside the newly formed cytoplasmic capsids (12). Phosphorylation at C-terminal sites of the capsid proteins seems to be inextricably linked with genome maturation because mutations of the target sites for phosphorylation abolish generation of mature viral DNA (13).
The cytoplasmic capsids may enter two different pathways. They can deliver their encapsidated genome into the nucleus of the same cell, leading to amplification of the episomal nuclear viral DNA and persistent infection of the cell. Alternatively, they can be enveloped by the viral surface proteins in the endoplasmic reticulum, forming progeny viruses that are released into the extracellular space. At least the latter pathway requires that genome maturation has already occurred (14), indicating a difference between immature capsids (ImmatC) and mature capsids (MatC).
Using Escherichia coli-expressed capsids that contain RNA and their in vitro-phosphorylated equivalents, we have previously shown that capsids contain a classical nuclear localization signal (NLS) that allows binding to nuclei of permeabilized cells. Only in those capsids that contain phosphorylated capsid protein subunits was the NLS capable of interacting with the importin α (Impα)/Impβ pathway, which mediates binding to the NPC (15). Divergent from the in vivo situation (16, 17), these capsids failed to generate intranuclear capsids, indicating functional differences from authentic HBV capsids. Microinjection of in vitro-phosphorylated capsids into Xenopus oocytes showed that the capsids not only bound to the NPC but were imported into the nuclear basket (4).
In the present study, we analyzed the fate of authentic capsids and the encapsidated viral genome. Using permeabilized cells, we found that MatC were not only transported to and through the NPC but were also capable of genome uncoating and DNA release as in infection and the in vivo amplification process. ImmatC were transported through the NPC but remained arrested within the nuclear basket and did not show release of the immature genome. The different fate of the capsids could be correlated with structural changes of the capsids' surface. The data show that transport and genome release occurred in a regulated fashion and allow a detailed picture of the nuclear entry process. In addition, the arrest of the ImmatC within the basket indicates that nuclear import of physiological karyophilic cargoes may involve additional regulation steps beyond the initial cargo–import receptor interaction that have not been previously described.
Experimental Procedures
Transport Assays. The preparation of HBV capsids was done from a stably HBV-transfected cell line known to produce infectious virus by using routine methods described in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org. Polymerase-negative capsids were prepared from transiently transfected cells by using the same procedure. The capsids or the control proteins were subjected to digitonin-permeabilized cells (18), giving the cargoes access to the nucleus. The capsids were localized by indirect immunofluorescence, and the genomes were visualized by native fluorescence in situ hybridization using a FITC-labeled single-stranded RNA probe. The BSA control conjugates were already FITC-labeled. The details of these procedures are given in Supporting Text. The electron microscopy import assays in Xenopus laevis oocytes were done as described (4).
Tryptic Digestion of Capsids. To determine the surface exposure of the arginine-rich C termini on capsid surface, capsids were digested with trypsin. To prevent entry into capsids, trypsin was first immobilized onto 40-nm gold particles according to the manual of the vendor of the gold particles (British Biocell, Cardiff, U.K.). The digestion products were separated by SDS/PAGE and visualized by immunoblotting using an antibody against denatured capsid protein.
Results
Nuclear Transport of MatC and ImmatC. To analyze the interactions between HBV capsids and the nuclear membrane, we purified six different forms of capsids. (i) Recombinant capsids, devoid of viral polymerase (P-C); like the authentic capsids, these P-C were expressed in a hepatoma cell line. (ii) ImmatC isolated from the cytoplasm of a stably transfected hepatoma cell line (HepG2.2.15) known to produce infectious HBV (19). This preparation contained a mixture of capsids with a variety of encapsidated RNA and DNA intermediates. (iii) Capsids derived from HepG2.2.15 cells treated with foscarnet, an inhibitor of reverse transcription (ImmatC-Inh) (20). Because the DNA-containing capsids within the cells are secreted, the ImmatC-Inh should consist of capsids containing the viral PG and the viral polymerase. (iv) MatC derived by extraction from isolated, intact virus particles. These contained the viral DNA and had undergone all posttranslational modifications, including phosphorylation of the capsid proteins. Real-time PCR revealed that MatC contained one DNA genome per capsid, whereas the ImmatC had only 1 in 10 (data not shown). Hence, most of the ImmatC represented capsids in which reverse transcription had not taken place. (v) Recombinant capsids obtained by expressing the capsid protein in E. coli (EcC), and (vi) the same E. coli-derived recombinant capsids with some of the capsid proteins phosphorylated with trapped PKC (EcPC). These particles contain bacterial RNA, randomly trapped during synthesis.
The different capsid preparations were incubated with cells that had been digitonin-permeabilized to provide access to the nucleus (18). Incubation was performed in the presence of ATP and reticulocyte lysate. The bound capsids were then visualized by confocal laser scanning microscopy after indirect double immunofluorescence using anticapsid antibodies and antibodies against the NPCs (Fig. 1). The anticapsid antibodies used were specific for assembled forms of the capsid protein: they do not interact with nonassembled capsid protein denatured by heat, SDS treatment, or acid (15).
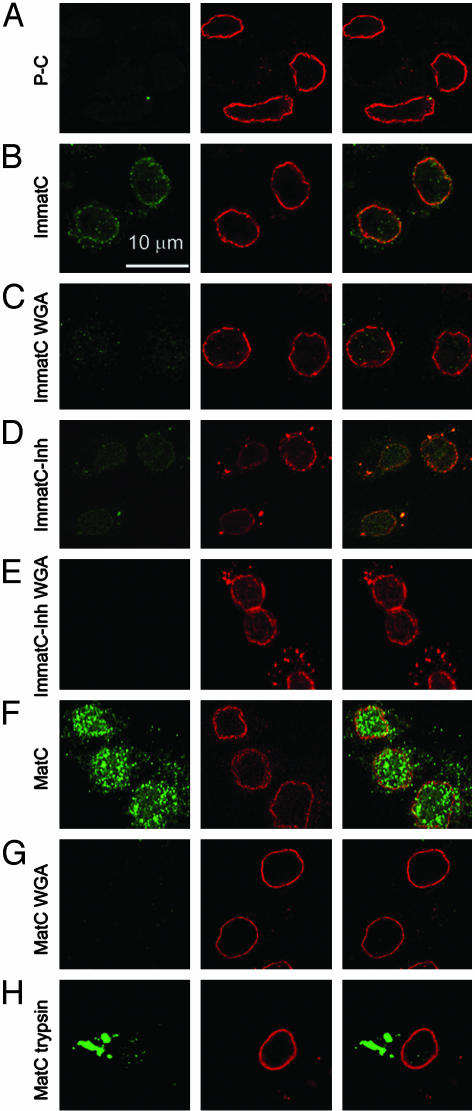
Fig. 1.
Localization of hepatitis B capsids in digitonin-permeabilized HuH-7 cells by confocal laser scanning microscopy after immune staining. Capsids (green, Left) and NPCs (red, Center) were stained by indirect immunofluorescence. (Right) Merged images are shown. In all samples, ATP and an ATP-generating system were present, and rabbit reticulocyte lysate was used as the source of cytosolic proteins. (A) P-C do not bind to cellular structures. (B) Nuclear binding of ImmatC. (C) ImmatC binding is inhibited when WGA is added. (D) ImmatC-Inh shows a weaker nuclear binding than ImmatC. (E) Like that of ImmatC, this binding is inhibited when WGA is added. (F) MatC generated intranuclear capsids. (G) The formation of intranuclear capsids is inhibited by WGA. (H) Tryptic digestion of MatC prevents nuclear binding and import. All pictures were taken at the same magnification.
We have previously shown that the E. coli-derived recombinant capsids bind to the NPC but only when some of the capsid proteins have a phosphorylated C-terminal sequence, which harbors an NLS (15). In these experiments, however, the capsids neither showed intranuclear localization nor evidence of uncoating. Thus, a substantial difference between E. coli-expressed and authentic core particles was expected.
The P-C were associated neither with the nucleus nor with cytoplasmic structures (Fig. 1A). This was expected, because like EcC, the capsid proteins in P-C could just encapsidate unspecific cellular RNA during synthesis. In contrast, the ImmatC showed a typical rim-like nuclear envelope staining (Fig. 1B). Some staining could be seen in the cytoplasm, but only a little staining was observed in the karyoplasm. The staining was greatly reduced if wheat germ agglutinin (WGA) was included in the incubation medium (Fig. 1C). Thus, the association with the nuclei most likely involved interaction with glycosylated proteins of the NPC (21). We concluded that the ImmatC mixture contained a fraction of capsids capable of association with the nuclear envelope. This was expected because the cells from which the ImmatC were purified must contain some MatC ready for envelopment into the surface proteins. The majority of the bound ImmatC, however, failed to move into the karyoplasm, being in accordance with the long-lasting maturation process of HBV. The staining pattern observed was similar to that previously observed for EcPC (15).
To analyze whether the observed rim-like stain was caused by DNA or viral RNA containing capsids, ImmatC-Inh were subjected to the transport assay. The binding was strongly reduced in comparison to ImmatC (Fig. 1D), indicating that genome maturation correlates with the interaction with the NPC. Addition of WGA abolished the binding (Fig. 1E).
MatC showed a much stronger immunofluorescence than ImmatC, and most of the staining was intranuclear (Fig. 1F). The loss of intranuclear capsid staining when WGA was present during the incubation (Fig. 1G) confirmed that transport of the capsids into the nucleus involved passage through the NPC. As in the case of the ImmatC, some binding to cytosolic structures was seen, and this was inhibited by WGA. The ≈5-fold higher signal obtained with the same amount of MatC than with ImmatC confirmed that only a fraction of the latter interacted with the nucleus.
Taken together, these results showed that the capacity of capsids to bind to NPCs and be internalized into the karyoplasm of permeabilized hepatoma cells depended on the maturation state. MatC were able to bind as well as to enter the karyoplasm efficiently. In contrast, only a small fraction of capsid intermediates in the ImmatC preparation bound, and even fewer entered the karyoplasm. The bound fraction apparently reflected capsids in which some genome maturation had occurred because binding of ImmatC-Inh was much weaker.
To determine whether these differences depended on liver cell-specific factors, we repeated the experiments with a cervical carcinoma cell line (HeLa). As shown in Fig. 6, which is published as supporting information on the PNAS web site, both MatC and ImmatC localized in a manner similar to that observed in the hepatoma cell line. Because the preparation of all capsid species involved the same purification steps (see Supporting Text), i.e., identical nuclease and detergent treatments, the diverse interactions were not caused by preparation artifacts but reflected intrinsic properties of the capsids, which were independent of host factors.
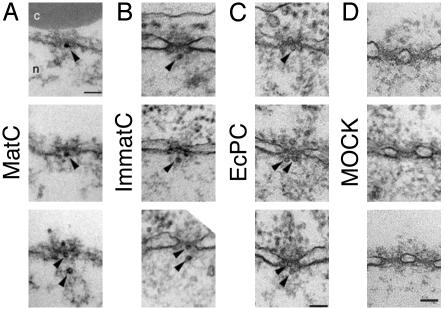
MatC and ImmatC Are Imported into the Nuclear Basket. To determine at which stage capsids were arrested during nuclear import, we microinjected ImmatC, EcPC, and MatC into X. laevis oocytes and analyzed the nuclear membranes in fixed sections by transmission electron microscopy (Fig. 2). HBV capsids were recognizable as 32- and 36-nm particles, most likely reflecting the T = 3 and T = 4 symmetric particles. The capsids were observed on the cytosolic side of the NPC, within the channel itself, and in the basket (Fig. 2 A–C). No capsid-like structures could be seen in sections from noninjected oocytes (Fig. 2D).
Fig. 2.
Intact HBV capsids are able to cross the nuclear pores. Views of nuclear envelope cross sections with adjacent cytoplasm (c) and nucleoplasm (n) from a Xenopus oocyte that has been microinjected with MatC (A), ImmatC (B), and EcPC (C). Arrowheads point to capsids associated with the nuclear face of the NPC. (D) Nuclear envelope cross section from a control (noninjected) Xenopus oocyte. (Scale bars = 100 nm.) Histograms showing the size distribution of the capsids associated with the cytoplasmic or the nuclear face of the nuclear pore are given in Fig. 7.
All three preparations contained capsids that were capable of translocation through the NPC into the nuclear basket. The diameters of the capsids found within the nuclear basket were similar to the diameters of the capsids at the cytosolic face of the NPC (Fig. 7, which is published as supporting information on the PNAS web site). This finding suggested that the rim staining seen for ImmatC and EcPC in permeabilized cells (Fig. 1B and ref. 15) was caused at least in part by accumulation of intact capsids in the basket and inside the NPC. The MatC were the only ones not trapped in the basket.
Electron microscopy has shown that the basket is a cage-like structure with a distal opening ≈44 nm in diameter (22). Having a diameter of 32 or 36 nm, HBV capsids should have no problem diffusing out of the basket unless retained by other factors. In fact, nucleoplasmin-coated gold particles of the same size travel from the basket into the nucleoplasm (4). The observation that the ImmatC and EcPC remained in the basket thus suggested that they interacted with basket-associated structures and/or basket components.
Structural Differences Between Capsids. To explain the different transport behavior, we analyzed the composition and conformational state of the various capsids in more detail. Although there are remarkable differences with regard to the encapsidated nucleic acid, it is unlikely that the RNA and DNA content as such would influence nuclear transport. More likely structural changes on the capsid surface determine different interactions with cellular proteins.
The capsid protein consists of a rigidly structured N-terminal domain formed by five α-helices (11, 23, 24) and a flexible C-terminal peptide comprising ≈36 residues necessary for RNA packaging. In addition to four arginine clusters and five potential phosphorylation sites, the C terminus contains a classical NLS, the exposure of which may be regulated by phosphorylation.
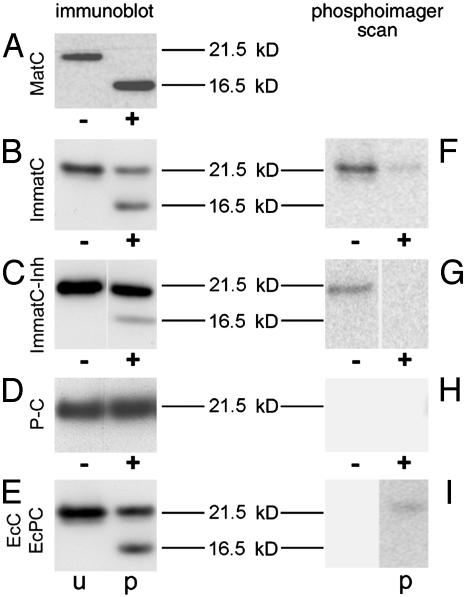
To test whether this peptide sequence was exposed in the different preparations, we treated capsids with trypsin immobilized onto 40-nm colloidal gold particles so that it could reach only the surface of the capsids. A time-course experiment revealed that digestions were complete and comparable between the different samples (Fig. 8, which is published as supporting information on the PNAS web site). When MatC were digested, immunoblots after SDS/PAGE showed that all capsid proteins were processed to a shorter 16.5-kDa form consistent with the removal of ≈30 residues (Fig. 3A). In contrast, only ≈50% of the capsid protein in ImmatC was similarly digested (Fig. 3B). ImmatC-Inh showed the same digestion pattern but were digested to an even lower extent (≈15%, Fig. 3C).
Fig. 3.
Maturation-dependent structural changes of capsids. +, Capsids digested with immobilized trypsin after phosphorylation with 32P. The capsid proteins were visualized by immunoblotting (A–E), using an antibody against denatured capsid protein, or by phosphorimaging (F–I) after SDS/PAGE. Nondigested controls (–) were visualized in the same way. (A) In MatC, all capsid proteins were cleaved to 16.5 kDa. (B) The digestion of ImmatC resulted predominantly in capsid proteins of the size of undegraded protein (21.5 kDa), but the same degradation products of 16.5 kDa. (C) When ImmatC-Inh were digested, the same pattern was observable, but the amount of degradation product was reduced. (D and E) The capsid protein of the P-C was not degraded (D) as the unphosphorylated EcC (E, lane u). In contrast, tryptic digestion of EcPC generated the same degradation products as ImmatC (E, lane p).
To determine the cleavage sites, ImmatC and ImmatC-Inh were incubated with [γ-32P]ATP before the digestion. The 32P is transferred by the endogenous protein kinase to the C terminus of the capsid proteins. Phosphorimaging of the immune blot revealed that all radioactive phosphates were removed from the digested capsid protein fragments (Fig. 3 F and G), confirming the cleavage at the C terminus. However, whereas the remaining full-length protein of ImmatC-Inh was devoid of radioactive phosphates, some phosphates remained at full-length ImmatC for unknown reasons. Consistent with the same binding and transport properties, digestion of EcPC (Fig. 3 E and I) showed the same reaction pattern as the ImmatC and ImmatC-Inh. When comparing the amount of transferred phosphates, ImmatC-Inh were phosphorylated to only 30% of ImmatC. This observation indicates that genome maturation is related to phosphorylation.
The P-C were not affected by incubation with the trypsin beads nor were the unphosphorylated EcC cleaved (Fig. 3 D and E), suggesting that their C termini were not exposed. The addition of [γ-32P]ATP to P-C did not lead to a phosphorylation of the capsid proteins (Fig. 3H), which supports the correlation between phosphorylation and structural change of the capsid. The difference between P-C and ImmatC-Inh, however, indicates that either the encapsidation of specific HBV RNA sequences or, more likely, the encapsidation of the polymerase induces some phosphorylation.
The size of cleaved capsid proteins in SDS/PAGE was compared with a capsid mutant C-terminally deleted at amino acid 145 (Fig. 8). This mutant showed a migration slightly faster (15.5 kDa) than the trypsin cleavage products (16.5 kDa). This finding indicates that the cleavage site was located within the first Arg cluster at the beginning of the flexible region (amino acids 149–154), upstream of the capsid NLS (amino acids 158–168; ref. 15).
To confirm that the C terminus is required for the interaction with the NPC, we prepared trypsin-treated MatC and isolated them by ultracentrifugation. When added to permeabilized cells these capsids did not show any binding or nuclear import (Fig. 1H).
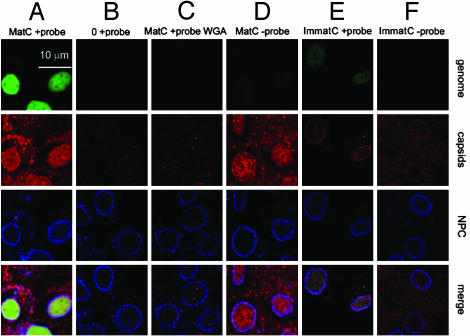
MatC Release Their DNA into the Nucleus. The viral DNA has to be released from the capsids into the nucleus to allow transcription and replication. To analyze the uncoating process, we determined whether digitonin-permeabilized cells support capsid uncoating. We used fluorescence in situ hybridization under native conditions with the single-stranded region of the viral genome as the target for the probe. To ensure that only uncoated viral genomes were detected, the probe used was made so large (≈1 kb, >300 nm in length) that it was expected to be excluded from the interior of intact capsids. The holes in the capsid shell are 2 nm in diameter (11). To evaluate the specificity of hybridization, we also generated a probe of the opposite orientation.
MatC were allowed to enter the nucleus of permeabilized cells before we hybridized with the positive-stranded probe. We observed a strong, intranuclear signal indicating that the viral DNA was indeed exposed inside the nucleus (Fig. 4A, first panel). Immunofluorescence with the capsid antibodies (Fig. 4A, second panel) showed the presence of intranuclear capsid staining most likely resulting from intranuclear capsid shells or fragments. When the images were merged it was evident that the capsids and the viral DNA did not colocalize (Fig. 4A, fourth panel), indicating that the DNA was no longer associated with capsids. Control experiments showed that no hybridization occurred if no capsids were added to the cells (Fig. 4B). Also, there were no detectable signals from genomes and capsid particles when the nuclear import of the capsids was inhibited by WGA (Fig. 4C). The control for the strand specificity with the counterstrand probe did not show hybridization (Fig. 4D, first panel), but it revealed intranuclear capsids (Fig. 4D, second panel).
Fig. 4.
Release of HBV genomes from the capsids in the presence of cytosolic proteins. Released HBV genomes were detected by native fluorescence in situ hybridization (FITC-labeled probe; green); capsids (in red) and NPCs (in blue) were detected by immunostaining. The merges are shown in the bottom row. (A) MatC were subjected to transport conditions, and hybridization was performed by using a probe against the single-stranded region of the mature HBV genome. (B) No capsids were added. (C) Inhibition of the nuclear transport of MatC by WGA using the same probe as in A.(D) Nuclear transport was as in A, but the hybridization was performed with the counterstrand probe. (E and F) ImmatC were added to the import reaction. (E) A weak hybridization signal was observed by using the probe against the single-stranded region of the mature HBV genome. (F) No signal was observed when the hybridization was performed with the counterstrand probe, indicating that capsids containing the RNA precursor do not release the genome.
With ImmatC, only a weak fluorescence in situ hybridization signal could be seen with the probe of the positive orientation (Fig. 4E, first panel), most likely resulting from a small fraction of MatC present in the preparation. When the hybridization was performed with the counterstrand probe no signal of released viral genome could be observed (Fig. 4F), indicating that the RNA precursors were not released.
We concluded that genome uncoating of incoming MatC was taking place in permeabilized cells. It resulted in the separation of genomes and capsids inside the nucleus; a picture similar to that has been reported in the liver of infected humans. Surprisingly, the antibody, which depends on particle structure, recognized the intranuclear capsid protein. This observation indicates that either the capsids do not fully disintegrate to isolated proteins or, after reaching a threshold concentration, the capsid subunits spontaneously reassemble to particles (25).
However, ImmatC could enter the nuclear basket but were not capable of efficient genome release or entry to the karyoplasm.
Capsid Uncoating Is Ran-Independent. We had previously shown that binding of EcPC to the NPC is mediated by soluble transport receptors, Impα and Impβ. To determine whether the generation of intranuclear capsids and the release of DNA from MatC also required these receptors, we replaced the reticulocyte lysate with a medium to which isolated Impα, Impβ, and Ran could be added in controlled amounts and in different combinations. As a control import cargo (Fig. 5 Bottom), we used FITC-labeled BSA conjugated with a classical NLS peptide from simian virus 40 T antigen (26). This conjugate is known to be imported by the same importin-mediated pathway as the capsids (27).
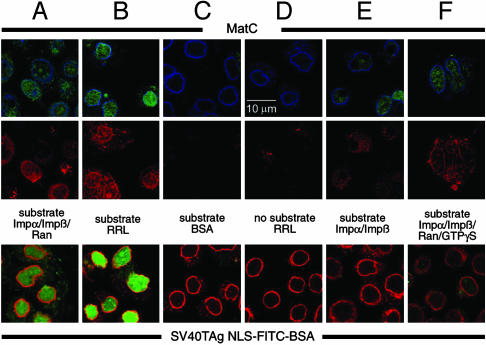
Fig. 5.
The release of the HBV genome is Ran-independent. (Top) Localization of the released viral genomes (in green) from MatC. (Middle) Capsids in the same cells (in red). (Bottom) Corresponding nuclear import of a simian virus 40 T antigen (SV40TAg) NLS-FITC-labeled BSA conjugate (in green). (Top) The costain against the NPCs is depicted in blue. (Bottom) This control stain is shown in red. (A) Impα, Impβ, and Ran replaced the cytosolic proteins. Intranuclear genomes and capsids were generated, and the control conjugate was imported. (B) Positive control, in which rabbit reticulocyte lysate (RRL) was added. As in A, released intranuclear genomes and capsids could be observed. (C and D) Control reactions in which no transport factors were present (C) or in which no substrates were added (D). (E) Cytosolic proteins were replaced by Impα and Impβ, resulting in the generation of intranuclear genomes and capsids, but not in a nuclear import of the control conjugate. (F) Impα, Impβ, Ran, and GTP[γS] were added. Intranuclear genomes and capsids were generated to the same extent as in E, but the nuclear import of the control conjugate was strongly reduced.
With medium containing all three components (Impα, Impβ, and Ran), MatC entered the nucleoplasm, and viral DNA was released (Fig. 5A Top). Quantification showed that efficiency of uncoating was 40% of that observed with reticulocyte lysate (Fig. 5B Top and Table 1). The uptake of NLS-conjugated BSA was also reduced to 40% of that seen with the reticulocyte lysate (Fig. 5 A and B Bottom and Table 1). When Impα, Impβ, and Ran all were omitted, or no capsids were added, no uptake or uncoating could be observed (Fig. 5 C and D and Table 1).
Table 1. Quantification of the Ran-independent release of HBV genomes from mature virus-derived capsids.
| Sample | % Genomes | % Protein |
|---|---|---|
| No substrate (D) | 0 | 0 |
| Substrate + BSA + ATP + GTP (C) | 0 | 0 |
| Substrate + reticulocyte lysate (B) | 100 | 100 |
| Substrate + BSA + ATP + GTP + Impα + Impβ (E) | 28 | 0 |
| Substrate + BSA + ATP + GTP + Impα + Impβ + Ran GDP (A) | 40 | 40 |
| Substrate + BSA + ATP + GTP + Impα + Impβ + Ran GDP + GTP[γS] (F) | 28 | 6 |
In 50 cells per sample, the intranuclear hybridization signal was analyzed. The background, revealed from the sample in which no substrate was added, was subtracted and taken as 0%. The positive control, in which rabbit reticulocyte lysate was added, was taken as the 100% value. The letters in parentheses refer to the panels in Fig. 5. % Genomes, percentage of released intranuclear HBV genomes. % Protein, percentage of intranuclear control protein.
When Impα and Impβ were added to the medium without Ran, uptake of NLS-conjugated BSA was completely inhibited (Fig. 5 E Bottom and Table 1) as expected because, without Ran, the import of NLS-containing cargo usually comes to a halt (1, 28, 29). In contrast to the NLS-conjugated BSA, the MatC were internalized in the absence of Ran, and a significant amount of the DNA was uncoated (Fig. 5E Top). The efficiency of uncoating amounted to 70% of the amount observed in the presence of all three import factors (28% versus 40% of the positive control; see Table 1).
To confirm that capsid import could occur without functional Ran, we performed an assay in which the nonhydrolyzable GTP analogue GTP[γS] was added, in the presence of Impα, Impβ, and Ran, to the medium. This reagent inhibits the recycling of Ran (30, 31). The extent of uncoating was again 70% of the control value with all three import factors (Fig. 5F Top and Table 1), whereas the import of NLS-conjugated BSA was only 15% (6% of the positive control) (Fig. 5F Bottom and Table 1).
To confirm the Ran independence, the experiments were repeated by using another stoichiometry of capsids and transport factors. Increasing the transport factors 3-fold, the amount of released viral genomes was increased in the samples containing Impα/Impβ, Impα/Impβ + Ran GDP, and Impα/Impβ + Ran GDP + GTP[γS] by 60%, whereas the transport of the conjugate + Impα/Impβ remained at 0%. The data are given in the Supporting Text.
Discussion
In contrast to other DNA viruses with larger capsids such as herpes viruses, adenoviruses, and HIV, uncoating of the HBV genome takes place inside the nucleus, although the diameter of the capsid with 32 or 36 nm is barely smaller than the upper limit for transport through the pore (4).
The capsid nuclear import uses the classical pathway of a surface-exposed NLS and interactions with the nuclear transport receptor Impβ via the adapter protein Impα (15). Exposure of the NLS on the capsid is, however, a tightly regulated process coupled to the maturation of the genome. Newly synthesized capsid proteins, protein kinase, and the polymerase associate with each other and with the viral PG to form ImmatC. In this state the C terminus is known to be localized in the lumen of the capsid (32), and it binds to packaged RNA (33, 34). The specific encapsidation of the PG is, however, mediated by polymerase (35).
The protein kinase necessary for phosphorylation of the capsid protein is trapped inside the forming capsid. Phosphorylation results in the exposure of the C-terminal sequences on capsid surface as determined by the tryptic degradation of the capsid protein in EcPC. The linkage of phosphorylation and genome maturation (13) thus results in a maturation-correlated exposure. In MatC, all of the C-terminal sequences were accessible, whereas in the ImmatC pool only a small fraction was exposed. In ImmatC-Inh, in which genome maturation was inhibited, even fewer C termini were exposed. In P-C and EcC, in which no phosphorylation could be observed, the C-terminal sequences were protected from the immobilized trypsin. The exposure of C-terminal sequences is thus coupled to the presence of the viral polymerase and to genome maturation.
The correlation of genome maturation and phosphorylation may be explained by the inhibition of phosphorylation caused by RNA bound to the C-terminal sites that are phosphorylated (6). The residual phosphorylation observed for ImmatC-Inh is in accordance with the report that specific encapsidation of the PG with polymerase requires at least some phosphorylated capsid subunits (36), which may reflect a selection of capsid subunits interacting with the kinase.
Once the NLS is exposed it addresses the MatC via the classical importin pathway to the NPC. In agreement with this conclusion, only capsids that have trypsin-cleavable C termini associated with the nucleus, i.e., the MatC, EcPC, and some of the capsids in the ImmatC and ImmatC-Inh. Moreover, capsids in which the C termini were removed by trypsin did not show nuclear binding or import.
MatC were the only capsid form that was able to uncoat their DNA in the nucleus and to move their capsid proteins from the nuclear basket into the karyoplasm. Surprisingly, intranuclear uncoating and transport were independent of Ran. Ran-independent nuclear import has already been described for the HIV integrase protein, but this import requires neither Impα nor Impβ (37). Normally, large cargoes, which are imported via Impα and Impβ, require Ran GTP for dissociating incoming cargo from the import receptors of the Impβ superfamily (1, 28) thus allowing the cargo to move from the nuclear basket deeper into the nucleus.
ImmatC were surprisingly arrested within the basket, as shown by electron microscopy. Such an arrest has not been described for any other substrate to our knowledge. In fact, in the current model of nuclear import only the initial interaction of cargo and import receptor in the cytoplasm determines whether a karyophilic protein is imported into the karyoplasm or not.
The arrest indicates that the surface of ImmatC interacts with proteins of the basket. The only difference from the MatC we found is that they expose fewer C termini as determined by the tryptic digestions. It is thus likely that the C terminus does not cause the arrest. The remaining capsid domains, which form capsid surface, are rigid and identical in both capsid species. It is likely that these domains comprise the peptide sequence, which interacts with the basket. Only the MatC, however, generate intranuclear capsid protein and genomes. A plausible explanation is that the conversion of the single-stranded DNA to the partially double-stranded DNA genome may prime the capsid for subsequent disintegration in the basket of the NPC. It is known that single-stranded nucleic acids have a much higher affinity for the capsid protein than double-stranded DNA (38) and that the nucleic acid–capsid protein interaction stabilizes the capsid (25, 39). According to this model, the capsid is weakened after double-stranded DNA synthesis and may disassemble in the basket. Subsequently, the DNA, which is not bound by Impα/Impβ, would be free to move into the nucleoplasm. In addition, only a few importin molecules are associated with the capsid (6). Thus, not only the DNA, but also capsid protein, which is not associated with Impα/Impβ, would diffuse into the karyoplasm, establishing that the translocation is Ran-independent.
The described pathway of nuclear import and uncoating implies that two regulatory steps are required to establish persistent infection of the cell. The first step comprises the maturation-dependent exposure of the NLS in the C-terminal domain of the capsid protein. This exposure guarantees that only replication-competent capsids are transported into the nucleus. The second regulatory step involves the release of mature viral genomes from the capsids. This mechanism is essential because it has been shown that in the absence of capsid protein no genome maturation occurs (34). Thus, the selective disintegration of capsids with a mature genome prevents the disruption of the viral life cycle.
Supplementary Material
Acknowledgments
We thank S. Behrens, W. Gerlich, and B. Boschek for critically reading the manuscript; D. Görlich for providing Impβ, Impα, and Ran expression plasmids; A. Berting for expressing the polymerase-negative capsids; H. Will for providing the antibody against the denatured capsid protein; and P. Pumpens and G. Borisova for providing the purified E. coli-derived capsids. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 535 B5 (to M.K.) and grants from the Canadian Institute of Health Research and the Natural Sciences and Engineering Research Council of Canada (to N.P.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NPC, nuclear pore complex; Impβ, importin β; Impα, importin α; HBV, hepatitis B virus; PG, pregenomic RNA; NLS, nuclear localization signal; P-C, capsids devoid of viral polymerase; ImmatC, immature capsids; ImmatC-Inh, capsids derived from HepG2.2.15 cells treated with the reverse-transcription inhibitor foscarnet; MatC, mature capsids; EcC, capsid protein in E. coli; EcPC, E. coli-derived recombinant capsids with some of the capsid proteins phosphorylated with trapped PKC; WGA, wheat germ agglutinin.
References
- 1.Görlich, D., Pante, N., Kutay, U., Aebi, U. & Bischoff, F. R. (1996) EMBO J. 15, 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 2.Whittaker, G. R., Kann, M. & Helenius, A. (2000) Annu. Rev. Cell Dev. Biol. 16, 627–657. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, J. S., Wilcher, R. & Samulski, R. J. (2000) J. Virol. 74, 2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pante, N. & Kann, M. (2002) Mol. Biol. Cell 13, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kann, M., Thomssen, R., Kochel, H. G. & Gerlich, W. H. (1993) Arch. Virol. 8, Suppl., 53–62. [DOI] [PubMed] [Google Scholar]
- 6.Kann, M. & Gerlich, W. H. (1994) J. Virol. 68, 7993–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kau, J. H. & Ting, L. P. (1998) J. Virol. 72, 3796–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duclos Vallee, J. C., Capel, F., Mabit, H. & Petit, M. A. (1998) J. Gen. Virol. 79, 1665–1670. [DOI] [PubMed] [Google Scholar]
- 9.Barrasa, M. I., Guo, J. T., Saputelli, J., Mason, W. S. & Seeger, C. (2001) J. Virol. 75, 2024–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daub, H., Blencke, S., Habenberger, P., Kurtenbach, A., Dennenmoser, J., Wissing, J., Ullrich, A. & Cotten, M. (2002) J. Virol. 76, 8124–8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowther, R. A., Kiselev, N. A., Bottcher, B., Berriman, J. A., Borisova, G. P., Ose, V. & Pumpens, P. (1994) Cell 77, 943–950. [DOI] [PubMed] [Google Scholar]
- 12.Seeger, C. & Mason, W. S. (2000) Microbiol. Mol. Biol. Rev. 64, 51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hui, E. K., Chen, K. L. & Lo, S. J. (1999) J. Gen. Virol. 80, 2661–2671. [DOI] [PubMed] [Google Scholar]
- 14.Gerelsaikhan, T., Tavis, J. E. & Bruss, V. (1996) J. Virol. 70, 4269–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kann, M., Sodeik, B., Vlachou, A., Gerlich, W. H. & Helenius, A. (1999) J. Cell Biol. 145, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuta, S., Nagata, A., Kiyosawa, K., Takahashi, T. & Akahane, Y. (1975) Gastroenterol. Jpn. 10, 208–214. [DOI] [PubMed] [Google Scholar]
- 17.Gudat, F., Bianchi, L., Sonnabend, W., Thiel, G., Aenishaenslin, W. & Stalder, G. A. (1975) Lab. Invest. 32, 1–9. [PubMed] [Google Scholar]
- 18.Adam, S. A., Marr, R. S. & Gerace, L. (1990) J. Cell Biol. 111, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sells, M. A., Zelent, A. Z., Shvartsman, M. & Acs, G. (1988) J. Virol. 62, 2836–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fourel, I., Saputelli, J., Schaffer, P. & Mason, W. S. (1994) J. Virol. 68, 1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finlay, D. R., Newmeyer, D. D., Price, T. M. & Forbes, D. J. (1987) J. Cell Biol. 104, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akey, C. W. & Radermacher, M. (1993) J. Cell Biol. 122, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conway, J. F., Cheng, N., Zlotnick, A., Wingfield, P. T., Stahl, S. J. & Steven, A. C. (1997) Nature 386, 91–94. [DOI] [PubMed] [Google Scholar]
- 24.Kenney, J. M., von Bonsdorff, C. H., Nassal, M. & Fuller, S. D. (1995) Structure (London) 3, 1009–1019. [DOI] [PubMed] [Google Scholar]
- 25.Seifer, M., Zhou, S. & Standring, D. N. (1993) J. Virol. 67, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Görlich, D., Prehn, S., Laskey, R. A. & Hartmann, E. (1994) Cell 79, 767–778. [DOI] [PubMed] [Google Scholar]
- 27.Makkerh, J. P., Dingwall, C. & Laskey, R. A. (1996) Curr. Biol. 6, 1025–1027. [DOI] [PubMed] [Google Scholar]
- 28.Ribbeck, K., Kutay, U., Paraskeva, E. & Görlich, D. (1999) Curr. Biol. 9, 47–50. [DOI] [PubMed] [Google Scholar]
- 29.Görlich, D. & Kutay, U. (1999) Annu. Rev. Cell Dev. Biol. 15, 607–660. [DOI] [PubMed] [Google Scholar]
- 30.Melchior, F., Paschal, B., Evans, J. & Gerace, L. (1993) J. Cell Biol. 123, 1649–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moroianu, J. & Blobel, G. (1995) Proc. Natl. Acad. Sci. USA 92, 4318–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zlotnick, A., Cheng, N., Stahl, S. J., Conway, J. F., Steven, A. C. & Wingfield, P. T. (1997) Proc. Natl. Acad. Sci. USA 94, 9556–9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatton, T., Zhou, S. & Standring, D. N. (1992) J. Virol. 66, 5232–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nassal, M. (1992) J. Virol. 66, 4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartenschlager, R. & Schaller, H. (1992) EMBO J. 11, 3413–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gazina, E. V., Fielding, J. E., Lin, B. & Anderson, D. A. (2000) J. Virol. 74, 4721–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Depienne, C., Mousnier, A., Leh, H., Le Rouzic, E., Dormont, D., Benichou, S. & Dargemont, C. (2001) J. Biol. Chem. 276, 18102–18107. [DOI] [PubMed] [Google Scholar]
- 38.Melegari, M., Bruss, V. & Gerlich, W. H. (1991) The Arginine-Rich Carboxyl-Terminal Domain Is Necessary for RNA Packaging by Hepatitis B Core Protein (Williams & Williams, Baltimore), pp. 164–168.
- 39.Birnbaum, F. & Nassal, M. (1990) J. Virol. 64, 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.