Abstract
Cul1, a member of the cullin ubiquitin ligase family, forms a multiprotein complex known as SCF and plays an essential role in numerous cellular and biological activities. A Cul1 homologue, p185 (Cul7), has been isolated as an simian virus 40 large T antigen-binding protein. To understand the physiological role of p185, we generated mice lacking p185. p185–/– embryos are runted and die immediately after birth because of respiratory distress. Dermal and hypodermal hemorrhage is detected in mutant embryos at late gestational stage. p185–/– placentas show defects in the differentiation of the trophoblast lineage with an abnormal vascular structure. We demonstrate that p185 forms an SCF-like complex with Skp1, Rbx1, Fbw6 (Fbx29), and FAP68 (FAP48, glomulin). FAP68 has recently been identified as a gene responsible for familial glomuvenous malformation. These results suggest that p185 forms a multiprotein complex and plays an important role in vascular morphogenesis.
The rapid destruction of regulatory proteins by ubiquitin-mediated proteolysis plays an important role in various biological processes. Protein ubiquitination is carried out by the sequential action of three enzymes, namely E1, E2, and E3 (1). E3 ubiquitin ligases determine the specificity of the substrate protein, and it is likely that protein ubiquitination in vivo is controlled primarily by regulating E3 activity or E3–substrate interaction (2). The Skp1–Cul1–F-box (SCF) complex is a well characterized E3 ubiquitin ligase that plays an essential role in a wide variety of activities, including cell cycle control (3, 4), muscle atrophy (5, 6), and protein quality control (7). To date, the human cullin protein family consists of six members: CUL1, CUL2, CUL3, CUL4A, CUL4B, and CUL5 (8). Although the physiological function of each cullin protein is largely unknown, mouse knockout evidence has demonstrated their biological significance. Cul1-, Cul3-, and Cul4A-deficient mice show early embryonic lethality (9–12), indicating their crucial role in early mammalian development.
F box proteins are the variable subunits of the SCF complex, and a large number of human F box proteins have been reported (13, 14). F box proteins contain an F box motif that binds to Skp1 and a carboxyl-terminal domain that specifies association with substrate proteins (15, 16). SCF targets a number of different substrate proteins by associating with specific F box proteins (16, 17). Among the human cullin proteins, SKP1 protein has been reported to bind specifically to CUL1 (18). Degradation of some F box proteins, such as Cdc4, Grr1, and Met-30 in yeast (19–21) and SKP2 (22) in humans, are mediated by the SCF complex itself.
Simian virus 40 large T antigen transforms a variety of rodent cells in vitro and in vivo. p185 (KIAA0076, p193, Cul7) was isolated as an simian virus 40 large T antigen-associated protein in murine cells (23, 24). Although it has been reported that overexpression of p185 induces apoptosis in NIH 3T3 cells (24), the biological and molecular functions of p185 remain unclear. Here we demonstrate that p185 has extensive sequence similarity to Cul1 and forms a specific SCF-like complex with Skp1, Fbw6 (Fbx29), Rbx1, and FAP68 (FAP48, glomulin). Surprisingly, p185 associates with Skp1 in an Fbw6-dependent manner and the stability of Fbw6 protein is decreased in the absence of p185. In addition, p185-deficient mice are neonatal lethal and show vascular defects in both the embryo and the placenta. Consistent with these findings, a recent article has reported that loss-of-function mutations of FAP68 are responsible for familial glomuvenous malformation, a hereditary syndrome characterized by abnormalities in vascular morphogenesis (25).
Materials and Methods
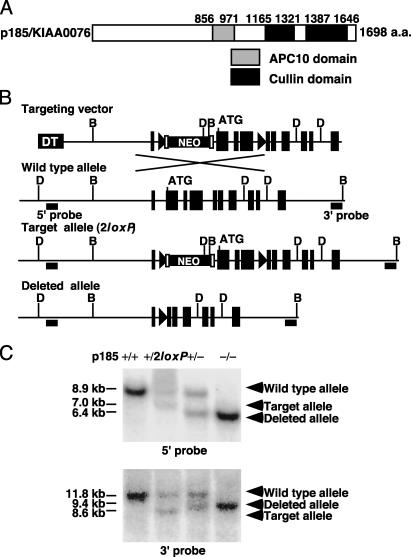
Generation of p185–/– Mice. A 9.7-kbp fragment of mouse genomic DNA, spanning from the promoter region to exon 10, was directly amplified by PCR from TC-1 embryonic stem cell genomic DNA. The exon sequences were verified by mouse genome sequence from the National Center for Biotechnology Information database. A 3.7-kb EcoRI–XhoI fragment and a 6-kb XhoI–NotI fragment were inserted into the targeting vector (26). A 3′ loxP site was inserted into intron 4 by PCR. A total of 384 colonies surviving G418 selection were screened. Three positive clones were isolated and confirmed by two different Southern blot analyses. To generate chimeras, two correctly targeted embryonic stem clones were injected into C57BL/6 blastocysts and subsequently transferred into pseudopregnant foster mothers. The chimeric mice were backcrossed with C57BL/6 mice. Heterozygous mice carrying the 2loxP allele were bred with EIIA-Cre/CD1 mice (27) and backcrossed with C57BL/6 mice to remove the Cre transgene. Primer sequences and PCR conditions for constructs and p185 genotyping will be provided on request.
Cellular Analysis. Mouse embryonic fibroblasts (MEFs) were established from embryos isolated at embryonic day 12.5 (E12.5) and cultured in DMEM containing 10% FBS. For the growth study, equal numbers of cells (1 × 105) from passage 2 MEFs were plated on 60-mm dishes. MEFs were counted at indicated time points in triplicate by using a hemocytometer.
Histological Analysis. Embryos and placentas from timed mating of p185+/– mice were fixed in Bouin's fixative (Sigma) and embedded in paraffin. Sections were stained with hematoxylin/eosin. For mRNA in situ hybridization, frozen sections were fixed with 4% paraformaldehyde for 12 h and hybridized with digoxigenin (Roche Molecular Biochemicals)-labeled Tpbp RNA riboprobe and detected by anti-digoxigenin (diluted 1:500 in PBS, Roche Molecular Biochemicals) followed by streptavidin biotinperoxidase (Santa Cruz Biotechnology). For morphometric analysis of the labyrinth, the outline of each individual vessel was identified and traced on National Institutes of Health image software. The percentage area of total maternal and fetal vessels was calculated.
Isolation of p185-Associated Protein and Binding Assay. U-2 OS cells stably expressing hemagglutinin (HA)-p185 (A2P2 cells) were generated by transfecting HA-p185-pcDNA3 construct, followed by G418 selection. Cells were labeled in methionine-free DMEM supplemented with 333 μCi (1 Ci = 37 GBq) of [35S]methionine per ml for2hafter1-hstarvation. Peptides were isolated from a silver-stained gel, digested with trypsin, and analyzed by microcapillary reverse-phase HPLC/nanoelectrospray tandem mass spectrometry. The p58 bands contained two peptide sequences, TWNYLEFEEEENK and ILVYSLEAGR, which match the sequence from human FAP68 and FBW6, respectively. To generate specific monoclonal antibody to p185 and polyclonal antibody to Fbw6, the amino-terminal 127- and 130-aa fragments, respectively, were cloned into the pGEX4T-3 vector (Amersham Pharmacia) and injected into RBF/DnJ mice, and hybridoma fusion was performed (28). FAP68 (29), SKP1 (Neomarkers, Fremont, CA), RBX1 (Neomarkers), and T7 (Novagen) antibodies were used for Western blotting and immunoprecipitation.
Clones. Human p185 and FBW6 constructs were PCR-amplified with full-length human p185/KIAA0076 and human FBW6 as a template. Mouse Fbw6 was PCR-amplified from EST clone 3158218 (American Type Culture Collection) and cloned with HA tag into pcDNA3 and pBabe/puro retroviral vector. The sequence of fragments generated by PCR was verified by sequencing.
Northern and Western Blot Analysis for MEFs. For Northern blot analysis, total RNA was extracted from MEFs with RNeasy Mini Kit (Qiagen, Valencia, CA). Each sample (14 μg) was electrophoresed in a 1% agarose/formaldehyde gel, blotted to Duralon-UV membrane (Stratagene), and probed with a 0.9-kb fragment of mouse p185 cDNA and a 0.6-kb fragment of mouse Fbw6 cDNA.
p185+/+ and p185–/– MEFs stably expressing HA-tagged mouse Fbw6 were generated by retroviral infection with HA-Fbw6-pBabe/puro construct. Protein stability of HA-Fbw6 signals was quantified and the half-life was calculated by using image gauge software (Fuji).
Results
Generation of p185–/– Mice. Database searches revealed that the carboxyl-terminal and middle regions of p185 show significant homology to Cul1 and a subunit of the anaphase-promoting complex/cyclosome, anaphase-promoting complex 10 (APC10)/DOC1 (30, 31) (Fig. 1A). To understand the biological role of p185, mice lacking p185 were generated by embryonic stem cell technology. The gene-targeting vector was designed to delete exons 2–4 including the translation start codon after Cre-mediated recombination (Fig. 1B). Residues encoded by exons 2 and 3 are highly conserved between human and mouse p185 (91.4% identity), suggesting a functional significance to the amino terminus. Heterozygous p185+/2loxP mice were crossed with an EIIa-Cre transgenic strain (27) to delete exons 2–4 and the Neo cassette.
Fig. 1.
Targeted disruption of p185 gene. (A) Domain structure of human p185 predicted by National Center for Biotechnology Information Conserved Domain Search. The residue for the beginning and end of each domain is shown at the top. (B) Genomic organization of the amino-terminal region of mouse p185, targeting vector and mutant allele after homologous and Cre recombination. Exon 2 contains the translation start site. The filled triangle and open rectangle indicate loxP and Frt sites, respectively. D, DraI; B, BspHI; DT, diphtheria toxin; NEO, neomycin acetyltransferase. (C) Southern blot analysis of p185+/+, p185+/2loxP, and p185+/– mice and p185–/–embryo. DraI- and BspHI-digested genomic DNA was hybridized with 5′ and 3′ probes, respectively.
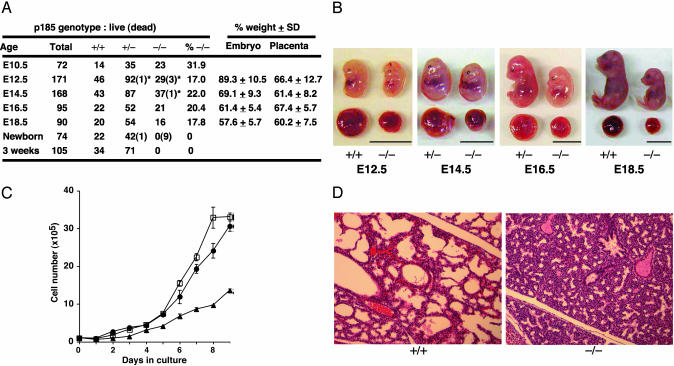
Neonatal Lethality and Growth Retardation of p185–/– Embryo. Heterozygous p185+/– mice were fertile and indistinguishable in appearance from wild-type littermates up to 6 months of age. When p185+/– mice were intercrossed, however, no p185–/– offspring were found on weaning. Analysis of p185–/– embryos at different gestational stages revealed neonatal lethality of mutant mice, although a small fraction of mutant embryos were probably lost during E10.5–E12.5 (Fig. 2A). At E12.5, p185–/– embryos were similar in size and weight to wild-type littermates, whereas p185–/– placentas were significantly reduced in size (Fig. 2 A and B). The growth retardation of p185–/– embryos became increasingly apparent relative to wild-type littermates in later gestational stages (Fig. 2 A and B).
Fig. 2.
Neonatal death and growth retardation of p185–/– embryo. (A) Genotype and phenotype of p185+/– intercrosses. Dead embryos are defined as necrotic or partially absorbed fetuses (indicated by *). %–/–, proportion of viable p185–/–embryos or pups at each stage; % weight, average of percentage weight of each p185–/– embryo against the average weight of p185+/– and p185+/+ littermates. The weights of p185+/– and p185+/+ embryos were indistinguishable (data not shown). (B) Gross appearance of p185+/+ and p185–/– embryos and placentas at indicated embryonic day. (Scale bar, 10 mm.) (C) Growth kinetics of MEFs. Passage-2 MEFs (1 × 105) were plated in 60-mm dishes and counted every 24 h. •, p185+/+; □, p185+/–; ▴, p185–/–. (D) The lung of E18.5 mutant embryo was unable to inflate, compared with a wild-type littermate. (×20.)
To understand the cause of death and the retarded growth of p185–/– embryos, cellular and histological analysis was performed. MEFs were generated from E12.5 embryos, and p185–/– MEFs had a significantly slower growth rate than wild-type and heterozygous littermate MEFs (Fig. 2C), suggesting that a cell-autonomous defect may contribute to the runted phenotype in addition to the placental defect. At E18.5, all mutant embryos recovered from the uteri of p185+/– intercrosses were alive. However, mutant embryos quickly turned cyanotic and lost their response to pain stimuli within 15 min, whereas wild-type and heterozygous littermates maintained strong and regular breathing. Histological examination of mutant E18.5 embryos and neonates revealed that p185–/– lungs failed to inflate, with the alveolar space markedly reduced (Fig. 2D). No abnormality was detected in other sites of the respiratory tract in the p185–/– embryos and neonates (data not shown).
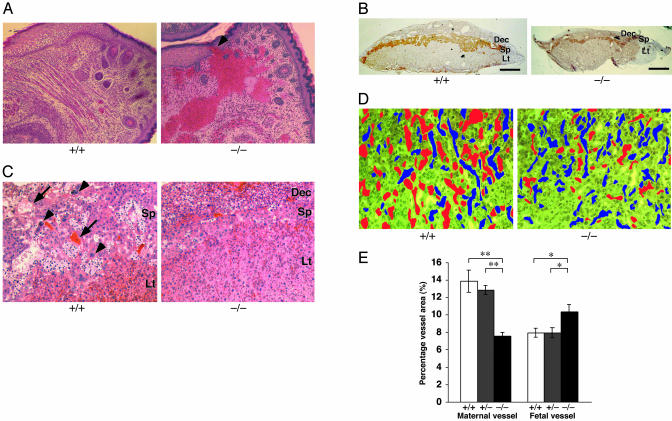
Vascular Abnormality in p185–/– Embryo and Placenta. Another histological abnormality found in mutant embryos was dermal and hypodermal hemorrhage in the lower lip (Fig. 3A). Histological analysis revealed that 4 of 21 (19.0%) p185–/– embryos at E16.5 and E18.5 had blood accumulation that was not surrounded by von Willebrand factor-positive endothelium (data not shown). In two cases, hemorrhage was located in the dermis (Fig. 3A, arrowhead) and hypodermis, suggesting that the probable primary site of hemorrhage was located in the dermis with blood later spreading into the hypodermis. In contrast, no hemorrhage was detected in any of the 18 wild-type littermates. Other vital organs in p185–/– embryos including the musculoskeletal, hematopoietic, and central and peripheral nervous systems displayed no obvious abnormality, although the size of organs was reduced proportionally.
Fig. 3.
Histological abnormality of p185–/– embryos and placentas. (A) Sagittal section of a mutant embryo showed dermal (arrowhead) and hypodermal hemorrhage in lower lip region. (×20.) (B) In situ hybridization with spongiotrophoblast-specific marker, Tpbp. The spongiotrophoblast (Sp) and decidual layers (Dec) were reduced in E18.5 mutant placenta compared with the wild-type littermate. Lt, labyrinthine. (Scale bar, 1 mm.) (C) Hematoxylin/eosin-stained E18.5 placenta reveals secondary trophoblast giant cells (arrowhead) were present in wild-type placenta, whereas they were virtually absent in p185–/– placenta. Dilated maternal vessels (arrow) were detected in spongiotrophoblast layer of wild-type placenta, and only small vessels were detected in p185–/– placenta. (×2.) (D) E12.5 mutant labyrinthine contains fewer maternal (red) and more fetal (blue) vessels than wild type. Fetal and maternal vessels are distinguished by the size and nucleation of red blood cells. (×20.) (E) The total area of fetal and maternal vessels in labyrinthine was measured by NIH image software. Twelve different fields (×100) for each genotype were randomly selected for analysis, and the average of percentage area in each field was calculated. Significant differences between p185–/– and p185+/+ or p185+/– placenta were found in both fetal and maternal vessel areas. *, P < 0.05; **, P < 0.01.
The placentas for p185–/– embryos were consistently smaller than wild-type littermates from E12.5 to E18.5 (Fig. 2 A and B). In situ hybridization analysis of an E18.5 mutant placenta with a spongiotrophoblast-specific marker, Tpbp (32), revealed that the decidua and spongiotrophoblast layers were reduced in size (Fig. 3B). Wild-type placentas had numerous dilated vessels in the decidua, but the mutant decidua had fewer vessels, suggesting that maternal vascular development was severely affected (Fig. 3B). Hematoxylin/eosin-stained sections of an E18.5 mutant placenta showed very few secondary giant cells and no dilated maternal vessels in an abnormally thin spongiotrophoblast layer (Fig. 3C). Morphometric analysis of the placental blood vessels at E12.5 demonstrated that the labyrinthine layer of mutant placentas had a significantly decreased maternal and increased fetal blood vessel area (Fig. 3 D and E).
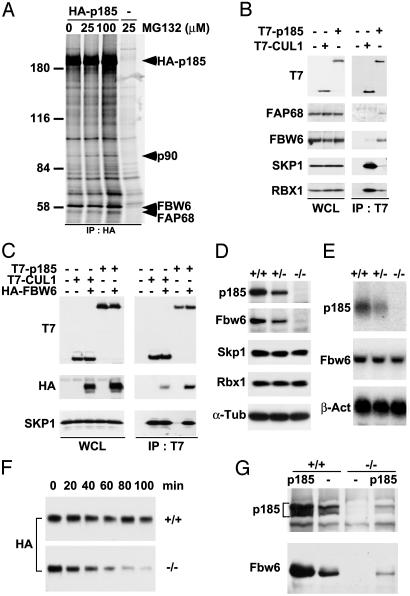
Isolation of p185-Associated Proteins. To understand the molecular function of p185, we sought to identify associated proteins. Cotransfection of T7-tagged p185 and HA-tagged RBX1, ROC2, or anaphase-promoting complex 11 followed by immunoprecipitation demonstrated specific binding between p185 and RBX1 (data not shown). To identify p185-binding proteins, U-2 OS cells stably expressing HA epitope-tagged human p185 (A2P2) were metabolically labeled with [35S]methionine and immunoprecipitated with anti-HA antibody (12CA5). During labeling, A2P2 cells were treated with increasing amounts of MG132 to inhibit proteasomal degradation of proteins. The 90- and 58-kDa protein bands were specifically detected from A2P2 cell lysates, but not from U-2 OS cell lysates (Fig. 4A). FBW6 (FBX29) and FAP68 peptide sequences were recovered from mass spectrometric analysis of the p58 bands. FBW6 is one of seven human F box proteins containing WD40 repeats (13, 33), whereas FAP68 has no recognizable functional domains.
Fig. 4.
p185 associates with FBW6, FAP68, SKP1, and RBX1. (A) Identification of [35S]methionine-labeled proteins that specifically coimmunoprecipitate with HA-p185. U-2 OS cells stably expressing HA-p185 were treated with the indicated amount of MG132 and immunoprecipitated with anti-HA antibody (12CA5). Retained proteins were resolved by SDS/PAGE and visualized by autoradiography. Subsequently, 90-kDa and 58-kDa bands were cut out from the silver-stained gel and subjected to mass spectrometric analysis. The 58-kDa band contained two different proteins, FBW6 and FAP68. No meaningful peptide sequence was recovered from the 90-kDa band. (B) T7-p185 associates with endogenous FBW6 and FAP68 in addition to SKP1 and RBX1. Lysates from HeLa cells transfected with indicated constructs were immunoprecipitated with anti-T7 antibody and analyzed by immunoblotting with anti-FBW6, FAP68, SKP1, and RBX1 antibodies. (C) p185 binds to SKP1 in an FBW6-dependent manner. T7-p185 or T7-Cul1 were transfected into HeLa cells with HA-Fbw6. Immunoblotting was performed with indicated antibodies for cell extract and immunoprecipitates. (D) Expression of Fbw6 protein in p185–/– MEFs. Immunoblotting was performed on whole cell lysates (100 μg per lane) prepared from p185+/+, p185+/–, and p185–/– MEFs at passage 2. The membranes were probed with anti-p185 and Fbw6 antibodies. (E) mRNA expression of Fbw6 in p185–/– MEFs. Total mRNA (14 μg per lane) from p185+/+, p185+/–, and p185–/– MEFs at passage 2 were analyzed by using radiolabeled p185 and Fbw6 probes. (F) p185+/+ and p185–/– MEFs stably expressing HA-tagged mouse Fbw6 were treated with 50 μg/ml cycloheximide and harvested every 20 min. The membrane was immunoblotted with anti-HA. Half-life of Fbw6 in p185–/– and p185+/+ MEFs was 51 and 111 min, respectively. (G) Lysates from p185+/+ and p185–/– MEFs that were transduced with a retroviral vector only (–) or containing wild-type mouse p185 were immunoblotted with p185 or Fbw6 antibodies.
To confirm the association, full-length T7-p185 and T7-CUL1 constructs were transfected into HeLa cells and immunoprecipitated with anti-T7 antibody. The transferred membrane was immunoblotted with antibodies to SKP1, RBX1, FBW6, and FAP68 as well as T7. p185 associated with all four proteins; however, p185 coprecipitated the endogenous SKP1 very weakly compared with CUL1 (Fig. 4B). CUL1 bound strongly to SKP1 and RBX1, as expected, and weakly to FBW6 (Fig. 4B).
To understand the weak affinity of p185 to SKP1, we examined if the association between p185 and SKP1 depended on FBW6 protein expression. T7-p185 and T7-CUL1 were transfected with HA-FBW6 and analyzed by immunoprecipitation and Western blotting (Fig. 4C). The association between p185 and SKP1 was significantly enhanced by the coexpression of FBW6, indicating that p185 binds to SKP1 in an FBW6-dependent manner. Similar results demonstrating the dependence on SKP1 binding to FBW6 were reported recently (34). Overexpression of FBW6 had no effect on the amount of SKP1 that was coprecipitated with CUL1.
Specific Binding of Fbw6 to p185. We observed that Fbw6 protein expression was significantly decreased in p185-null MEFs, whereas mRNA expression levels appeared unchanged relative to wild-type and heterozygous MEFs (Fig. 4 D and E). The expression of Skp1 and Rbx1 in p185-null MEFs was similar to that in wild-type cells (Fig. 4D). The stability of Fbw6 protein was determined by cycloheximide pulse chase. The half-lives of retrovirally introduced HA-Fbw6 in p185–/– and p185+/+ MEFs were 51 and 111 min, respectively, suggesting that instability of Fbw6 contributes to its low protein expression in p185-null MEFs (Fig. 4F). Addition of proteasome inhibitors to the cells before lysis did not increase the half-life of Fbw6 in either the wild-type or null MEFs (data not shown). To test whether Fbw6 could respond to increased expression of p185, a retroviral vector expressing mouse p185 was introduced into wild-type and p185-null MEFs. As shown in Fig. 4G, levels of Fbw6 increased when p185 expression was restored to the p185–/– MEFs. In addition, increased expression of Fbw6 was observed when levels of p185 were increased in wild-type MEFs.
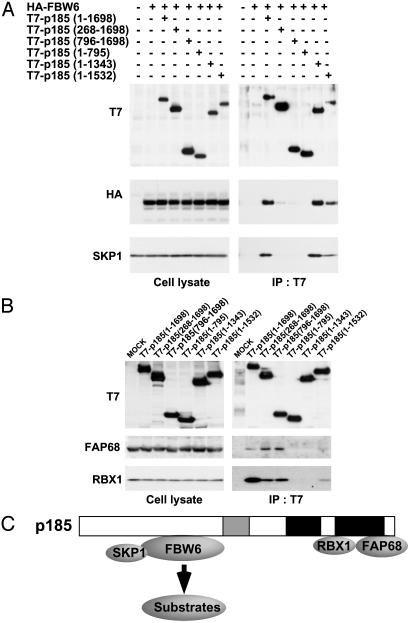
Overall Structure of p185-Based Complex. Structural analysis of SCF complex revealed that Cul1 binds to Skp1-F boxSkp2 through the amino-terminal portion of its first cullin repeat and to Rbx1 through the carboxyl-terminal globular a/b domain (35). We investigated whether the p185-based complex had an overall structural similarity to the SCF complex by determining the binding regions of p185 to SKP1, FBW6, FAP68, and RBX1. Several p185 truncation mutants were constructed and transfected into HeLa cells to examine whether they retained binding to the SCF components. Cotransfection experiments demonstrated that three p185 mutants (268–1698, 796–1698, and 1–795) were significantly reduced in FBW6- and SKP1-binding activity, suggesting that the N terminus was required for binding (Fig. 5A). As shown in Fig. 5B, two mutants (1–795 and 1–1343) failed to bind FAP68 and RBX1. A mutant (1–1532) containing the principle RBX1-binding region as predicted by homology weakly associates with both FAP68 and RBX1. Similar to CUL1, a carboxyl-terminal region of p185 was required for binding to RBX1.
Fig. 5.
Overall structure of p185-based complex. (A) FBW6 binds to amino-terminal region of p185. HeLa cells were cotransfected with HA-FBW6 and six different p185 constructs. Lysates of HeLa cells were immunoprecipitated with anti-T7 and analyzed by anti-HA and SKP1 antibodies. (B) FAP68 binds to the carboxyl-terminal region of p185. Lysates from HeLa cells transfected with p185 deletion constructs were immunoprecipitated with anti-T7 and analyzed by immunoblotting with anti-FAP68 and RBX1 antibodies. (C) A model of p185-based complex.
Discussion
Phenotype of p185–/– Mice. Early embryonic death has been observed after disruption of several cullins in mice, including Cul1, Cul3, and Cul4A (9–12). Lethality uniformly occurred at E4.5–E7.5, before the organogenesis stage. In contrast, p185–/– mice show neonatal lethality, indicating that p185 plays a role in mammalian development that is apparently distinct from that of Cul1, Cul3, and Cul4A. The growth of the p185–/– embryo is particularly impaired at late gestational stage. Because the reduced placental size precedes the retarded growth of p185–/– embryo (Fig. 2 A and B) and an imbalance of fetal to maternal blood flow is detected in the p185–/– placental labyrinthine layer (Fig. 3 D and E), placental insufficiency may be the principal cause of the growth retardation of the p185–/– embryo.
In p185–/– placentas, we found several abnormalities, including a thin spongiotrophoblast layer with a reduced number of secondary trophoblast giant cells and underdeveloped deciduas (Fig. 3 B and C). It has been assumed that the spongiotrophoblast is largely derived from the cells of the ectoplacental cone and secondary trophoblast giant cells from precursor cells in the spongiotrophoblast layer (36). Angiogenic factors produced by giant cells are critical for maternal vascular development (37). Therefore, a differentiation defect of trophoblasts in the ectoplacental cone may account for the histopathological abnormalities detected in p185–/– extraembryonic tissues. The precise molecular mechanism underlying the differentiation defect of the p185–/– trophoblasts remains to be determined.
In addition to the placental defects, p185–/– embryos exhibited dermal and hypodermal hemorrhage in the lower-lip region. Although no specific vascular lesions were found on hematoxylin/eosin-stained sections, it is possible that the endothelial or supporting cells (pericytes and smooth muscle cells) in the lower-lip region were malfunctioning. We found no specific evidence for glomus cells, typical for glomuvenous malformations, in any of the mutant embryos (data not shown).
p185-Based Complex. In this study, we present biochemical evidence that p185 forms an SCF-like complex with overall structural similarity to SCF. In SCF complexes, the variable F box protein subunit binds to the substrate protein (16) and Cul1/Rbx1 forms a catalytic core complex that serves to recruit an E2 ubiquitin-conjugating enzyme (38, 39). The analogy to the SCF complex suggests that a p185/Cul7–Skp1–Fbw6 complex targets a specific substrate for ubiquitination in a similar fashion. To date, we have been unable to confirm any candidate substrates, including FAP68, for this complex.
Skp2, one of the most investigated F box proteins, is ubiquitinated by the SCFSkp2 complex in an autocatalytic manner (22). The decrease of Cul1 expression by antisense oligonucleotides led to increased protein level of Skp2 in quiescent cells (22). Unlike Skp2, Fbw6 expression in p185+/+ and p185–/– MEFs was not sensitive to proteasome inhibitor under standard culture conditions (data not shown) and Fbw6 protein was destabilized in the absence of p185 (Fig. 4F). The underlying mechanism that controls the stability of Fbw6 protein is apparently different from the mechanism for Skp2.
FAP48, a splice variant of FAP68, was originally isolated as an FKBP59-binding protein (40). However, recent articles have reported that FAP68 is the major gene product with the truncated FAP48 mRNA and protein expression not detected in several cell lines and tissues (25, 29). Based on the protein size, we concluded that the 58-kDa p185-associated band corresponds to FAP68 (Fig. 4A). Recently, 14 different germ-line mutations of FAP68 were identified in patients with inherited glomuvenous malformations (25). Therefore, gene inactivation of two components of the p185-based complex (p185 and FAP68) contributes to vascular defects in mice (Fig. 3 A, D, and E) and humans (25), respectively, supporting the idea that both components may be involved in a common signaling pathway.
The phenotype of p185 knockout mice and the human syndrome of glomuvenous malformation caused by mutation of one p185-associated protein, FAP68, suggest that p185 is involved in vascular morphogenesis. Further investigation of p185 may lead to a molecular understanding of vascular formation and the etiology for glomuvenous malformations. Although the precise mechanism of how vascular abnormality is caused by p185 and FAP68 gene knockout remains unknown, it is likely that substrate proteins ubiquitinated by p185-based complex play a crucial role in endothelial proliferation and/or differentiation.
Acknowledgments
We thank R. DePinho, J. Horner, N. Bardeesy, and A. Sharpe for knockout mouse production, and C. Schweitzer and J. Gan for anti-p185 monoclonal antibody. We also thank T. Crepaldi for providing anti-FAP68 antibody, M. Pagano and W. Harper for F box constructs, the Kazusa DNA Research Institute for KIAA clones, J. Rossant and D. Linzer for in situ hybridization probe, R. Bronson for histological analysis, and W. Lane for mass spectrometric analysis. J.A.D. is a scholar of the Leukemia Lymphoma Society. This work was supported in part by National Cancer Institute Grant RO1 CA093804.
Abbreviations: SCF, Skp1–Cul1–F-box; MEFs, mouse embryonic fibroblasts; E4.5, E7.5, E10.5, E12.5, E16.5, and E18.5, embryonic day 4.5, 7.5, 10.5, 12.5, 16.5, and 18.5, respectively.
References
- 1.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- 2.Deshaies, R. J. (1999) Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- 3.Zhang, H., Kobayashi, R., Galaktionov, K. & Beach, D. (1995) Cell 82, 915–925. [DOI] [PubMed] [Google Scholar]
- 4.Koepp, D. M., Harper, J. W. & Elledge, S. J. (1999) Cell 97, 431–434. [DOI] [PubMed] [Google Scholar]
- 5.Bodine, S. C., Latres, E., Baumhueter, S., Lai, V. K., Nunez, L., Clarke, B. A., Poueymirou, W. T., Panaro, F. J., Na, E., Dharmarajan, K., et al. (2001) Science 294, 1704–1708. [DOI] [PubMed] [Google Scholar]
- 6.Gomes, M. D., Lecker, S. H., Jagoe, R. T., Navon, A. & Goldberg, A. L. (2001) Proc. Natl. Acad. Sci. USA 98, 14440–14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida, Y., Chiba, T., Tokunaga, F., Kawasaki, H., Iwai, K., Suzuki, T., Ito, Y., Matsuoka, K., Yoshida, M., Tanaka, K., et al. (2002) Nature 418, 438–442. [DOI] [PubMed] [Google Scholar]
- 8.Kipreos, E. T., Lander, L. E., Wing, J. P., He, W. W. & Hedgecock, E. M. (1996) Cell 85, 829–839. [DOI] [PubMed] [Google Scholar]
- 9.Dealy, M. J., Nguyen, K. V., Lo, J., Gstaiger, M., Krek, W., Elson, D., Arbeit, J., Kipreos, E. T. & Johnson, R. S. (1999) Nat. Genet. 23, 245–248. [DOI] [PubMed] [Google Scholar]
- 10.Wang, Y., Penfold, S., Tang, X., Hattori, N., Riley, P., Harper, J. W., Cross, J. C. & Tyers, M. (1999) Curr. Biol. 9, 1191–1194. [DOI] [PubMed] [Google Scholar]
- 11.Singer, J. D., Gurian-West, M., Clurman, B. & Roberts, J. M. (1999) Genes Dev. 13, 2375–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, B., Ruiz, J. C. & Chun, K. T. (2002) Mol. Cell. Biol. 22, 4997–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winston, J. T., Koepp, D. M., Zhu, C., Elledge, S. J. & Harper, J. W. (1999) Curr. Biol. 9, 1180–1182. [DOI] [PubMed] [Google Scholar]
- 14.Cenciarelli C, Chiaur D. S., Guardavaccaro D., Parks W., Vidal M. & Pagano M. (1999) Curr. Biol. 9, 1177–1179. [DOI] [PubMed] [Google Scholar]
- 15.Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W. & Elledge, S. J. (1996) Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- 16.Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J. & Harper, J. W. (1997) Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- 17.Patton, E. E., Willems, A. R. & Tyers, M. (1998) Trends Genet. 14, 236–243. [DOI] [PubMed] [Google Scholar]
- 18.Michel, J. J. & Xiong, Y. (1998) Cell Growth Differ. 9, 435–449. [PubMed] [Google Scholar]
- 19.Zhou, P. & Howley, P. M. (1998) Mol. Cell 2, 571–580. [DOI] [PubMed] [Google Scholar]
- 20.Galan, J. M. & Peter, M. (1999) Proc. Natl. Acad. Sci. USA 96, 9124–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouillon A., Barbey R., Patton E. E., Tyers M. & Thomas D. (2000) EMBO J. 19, 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirbelauer, C., Sutterluty, H., Blondel, M., Gstaiger, M., Peter, M., Reymond, F. & Krek, W. (2000) EMBO J. 19, 5362–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohrman, D. C. & Imperiale, M. J. (1992) J. Virol. 66, 1752–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai, S. C., Pasumarthi, K. B., Pajak, L., Franklin, M., Patton, B., Wang, H., Henzel, W. J., Stults, J. T. & Field, L. J. (2000) J. Biol. Chem. 275, 3239–3246. [DOI] [PubMed] [Google Scholar]
- 25.Brouillard, P., Boon, L. M., Mulliken, J. B., Enjolras, O., Ghassibe, M., Warman, M. L., Tan, O. T., Olsen, B. R. & Vikkula, M. (2002) Am. J. Hum. Genet. 70, 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardeesy, N., Sinha, M., Hezel, A. F., Signoretti, S., Hathaway, N. A., Sharpless, N. E., Loda, M., Carrasco, D. R. & DePinho, R. A. (2002) Nature 419, 162–167. [DOI] [PubMed] [Google Scholar]
- 27.Lakso, M., Pichel, J. G., Gorman, J. R., Sauer, B., Okamoto, Y., Lee, E., Alt, F. W. & Westphal, H. (1996) Proc. Natl. Acad. Sci. USA 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalal, S. N., Schweitzer, C. M., Gan, J. & DeCaprio, J. A. (1999) Mol. Cell. Biol. 19, 4465–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grisendi, S., Chambraud, B., Gout, I., Comoglio, P. M. & Crepaldi, T. (2001) J. Biol. Chem. 276, 46632–46638. [DOI] [PubMed] [Google Scholar]
- 30.Hwang, L. H. & Murray, A. W. (1997) Mol. Biol. Cell 8, 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossberger, R., Gieffers, C., Zachariae, W., Podtelejnikov, A. V., Schleiffer, A., Nasmyth, K., Mann, M. & Peters, J. M. (1999) J. Biol. Chem. 274, 14500–14507. [DOI] [PubMed] [Google Scholar]
- 32.Lescisin, K. R., Varmuza, S. & Rossant, J. (1988) Genes Dev. 2, 1639–1646. [DOI] [PubMed] [Google Scholar]
- 33.Koepp, D. M., Schaefer, L. K., Ye, X., Keyomarsi, K., Chu, C., Harper, J. W. & Elledge, S. J. (2001) Science 294, 173–177. [DOI] [PubMed] [Google Scholar]
- 34.Dias, D. C., Dolios, G., Wang, R. & Pan, Z. Q. (2002) Proc. Natl. Acad. Sci. USA 99, 16601–16606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., et al. (2002) Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]
- 36.Cross, J. C. (2000) Cell Dev. Biol. 11, 105–113. [DOI] [PubMed] [Google Scholar]
- 37.Cross, J. C., Hemberger, M., Lu, Y., Nozaki, T., Whiteley, K., Masutani, M. & Adamson, S. L. (2002) Mol. Cell. Endocrinol. 187, 207–212. [DOI] [PubMed] [Google Scholar]
- 38.Skowyra, D., Koepp, D. M., Kamura, T., Conrad, M. N., Conaway, R. C., Conaway, J. W., Elledge, S. J. & Harper, J. W. (1999) Science 284, 662–665. [DOI] [PubMed] [Google Scholar]
- 39.Seol, J. H., Feldman, R. M., Zachariae, W., Shevchenko, A., Correll, C. C., Lyapina, S., Chi, Y., Galova, M., Claypool, J., Sandmeyer, S., et al. (1999) Genes Dev. 13, 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambraud, B., Radanyi, C., Camonis, J. H., Shazand, K., Rajkowski, K. & Baulieu, E. E. (1996) J. Biol. Chem. 271, 32923–32929. [DOI] [PubMed] [Google Scholar]