Abstract
Phosphatidylinositol-5-phosphate (PI-5-P) is a newly identified phosphoinositide with characteristics of a signaling lipid but no known cellular function. PI-5-P levels are controlled by the type II PI-5-P 4-kinases (PIP4K IIs), a family of kinases that converts PI-5-P into phosphatidylinositol-4,5-bisphosphate (PI-4,5-P2). The PI-5-P pathway is an alternative route for PI-4,5-P2 synthesis as the bulk of this lipid is generated by the canonical pathway in which phosphatidylinositol-4-phosphate (PI-4-P) is the intermediate. Here we examined the effect of activation of the PI-5-P pathway on phosphoinositide 3-kinase (PI3K) signaling by expressing PIP4K IIβ in cells that lack this enzyme. Although PIP4K II generates PI-4,5-P2, a substrate for PI3K, expression of this enzyme reduced rather than increased phosphatidylinositol-3,4,5-trisphosphate (PI-3,4,5-P3) levels in cells stimulated with insulin or cells expressing activated PI3K. This reduction in PI-3,4,5-P3 levels resulted in decreased activation of the downstream protein kinase, Akt/PKB. Consistent with these results, expression of IpgD, a bacterial phosphatase that converts PI-4,5-P2 to PI-5-P, resulted in Akt activation, and this effect was partially reversed by PIP4K IIβ. PIP4K IIβ expression did not impair insulin-dependent association of PI3K with insulin receptor substrate 1 (IRS1) but abbreviated Akt activation, indicating that PIP4K II regulates PI-3,4,5-P3 degradation rather than synthesis. These data support a model in which the PI-5-P pathway controls insulin signaling that leads to Akt activation by regulating a PI-3,4,5-P3 phosphatase.
Phosphoinositides are important mediators of cellular responses to growth factors including proliferation, survival, migration, and glucose uptake. Phosphatidylinositol-3,4,5-trisphosphate (PI-3,4,5-P3) synthesis by phosphoinositide 3-kinase (PI3K) is an essential step in the transduction of the insulin signal that leads to Akt phosphorylation and GLUT4 transport (1). Phosphatidylinositol-4,5-bisphosphate (PI-4,5-P2) is a critical regulator of several cellular functions and the main precursor of PI-3,4,5-P3 (2).
PI-4,5-P2 can be synthesized through phosphorylation of phosphatidylinositol-4-phosphate (PI-4-P) by type I phosphatidylinositol phosphate kinase (PIPK) (or PIP5K I) or phosphorylation of phosphatidylinositol-5-phosphate (PI-5-P) by type II PIPK (or PIP4K II) (3). Both type I and II PIPKs are present in the genomes of a wide range of multicellular organisms including Caenorhabditis elegans, Drosophila, and mammals, but only type I is present in the genome of unicellular eukaryotes such as Saccharomyces cerevisiae. It is generally accepted that the majority of the PI-4,5-P2 in mammalian cells comes from PI-4-P phosphorylation by the PIP5K I (4). Expression of PIP5K Is can cause dramatic changes in cytoskeleton, whereas PIP4K IIs do not affect cytoskeletal rearrangement (5). Thus, it is likely that PIP4K IIs evolved to perform a specialized function in multicellular organisms and that the PI-4-P and PI-5-P pathways for PI-4,5-P2 synthesis have nonredundant physiological functions in cells. However, the cellular functions for PI-5-P and the PIP4K II remain unknown.
PI-5-P levels in mammalian cells are low (at least 50-fold lower than the levels of PI-4-P) and comparable with the levels of other signaling phosphoinositides such as phosphatidylinositol-3-phosphate (6). Growth factor treatment can increase PI-5-P levels (ref. 7 and L.E.R., unpublished data), suggesting a role for PI-5-P in signaling. A dramatic increase in PI-5-P levels was shown recently to follow Shigella flexneri infection due to the activity of the virulence factor IpgD. IpgD is a 4-phosphatase that specifically converts PI-4,5-P2 to PI-5-P. IpgD expression in mammalian cells results in significant loss of PI-4,5-P2 and dramatic changes in actin cytoskeleton, cell morphology, and adhesion. Infection of HeLa cells with S. flexneri also causes a substantial increase in the levels of PI-3,4,5-P3 (8).
To assess the possibility that the PI-5-P pathway supplies PI-4,5-P2 as substrate for PI3K in a concerted pathway for generation of PI-3,4,5-P3, we expressed the PIP4K IIβ gene in Chinese hamster ovary cells that express the human insulin receptor (CHO-IR cells), which have no detectable levels of endogenous PIP4K IIβ. Surprisingly, CHO-IR cells expressing PIP4K IIβ had lower levels of insulin-induced PI-3,4,5-P3 compared with control cells and, as a consequence, lower levels of insulin-induced Akt phosphorylation. Conversely, cells expressing IpgD had higher levels of basal and insulin-stimulated Akt phosphorylation. PIP4K IIβ expression partially reversed the effect of IpgD on Akt phosphorylation. PIP4K IIβ expression did not reduce IR or IR substrate phosphorylation, PI3K recruitment, or PI-3,4-P2 levels, indicating that the effect of PIP4K IIβ is downstream of PI3K activation. PIP4K IIβ expression caused total depletion of PI-3,4,5-P3 in cells expressing the constitutively active PI3K, whereas it increased PI-3,4-P2 levels, suggesting that the PI-5-P pathway regulates a PI-3,4,5-P3-specific 5-phosphatase. Consistent with this hypothesis, the PI3K signal that leads to Akt phosphorylation was terminated prematurely in cells expressing PIP4K IIβ. Together, these results suggest that the PI-5-P pathway for PI-4,5-P2 synthesis controls insulin signaling by promoting PI-3,4,5-P3 dephosphorylation.
Methods
Cell Culture. CHO-IR cells were maintained in RPMI medium 1640 supplemented with 10% FBS. COS cells were maintained in DMEM supplemented with 10% FBS.
DNA Constructs. The hemagglutinin (HA)-Akt construct was a gift from J. Blenis (Harvard Medical School). The pcDNA3-PIP4K IIβ construct was a gift from Moses Chao (Cornell University Medical College, New York). The pcDNA3-His-Ship2 was a gift from Shonna Moodie (Metabolex, Hayward, CA). The GFP-p85-Flag was a gift from J. Luo (Harvard Medical School). The p110-caax construct was a gift from Julian Downward (London Research Institute, London). The GFP-IpgD constructs were gifts from P. Sansonetti (Institut Pasteur, Paris), K. Nieburh (Institut Pasteur), H. Tronchère (Institut National de la Santé et de la Recherche Médicale, Toulouse, France), and C. Pendaries (Institut National de la Santé et de la Recherche Médicale).
Transfections. Transfections were performed by using Lipofectamine plus reagent (Invitrogen). Cells were plated on 60-mm dishes and transfected the next day with various constructs according to each experiment. The total amount of DNA in each plate was kept constant by the addition of empty vector as control. Lipids or protein lysates were collected 48 h after the beginning of the transfection.
Akt Phosphorylation Assay. Cells were transfected with HA-Akt and with different constructs as indicated in each experiment. The amount of HA-Akt DNA transfected was 1/10 or 1/20 of the total DNA. Cells were serum-starved for 24 h or not and stimulated or not according to each experiment. Protein lysates were prepared in buffer containing 50 mM Tris·HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 10% glycerol, and protease and phosphatase inhibitors. The cells were scraped, and the lysates were centrifuged at 18,000 × g to remove the insoluble fraction. Samples of the lysates were collected for Western blotting, and the remaining lysate was incubated with anti-HA antibody (HA-11, Babco, Richmond, CA) for 2 h and with protein-G Sepharose beads (Pharmacia) for 1 h. The beads were centrifuged and washed by using phosphate-buffer saline containing 1% Nonidet P-40. The immunocomplexes and total lysates were resuspended in SDS-loading buffer and resolved by SDS/PAGE. The proteins were transferred to nitrocellulose membrane. Activated Akt was detected by using phospho-specific antibody against T308 or S473 (Cell Signaling Technology, Beverly, MA). Total Akt was detected by using anti-HA antibody (Babco). PIP4K II was detected by using a polyclonal antibody against PIP4K IIβ (a gift from Moses Chao), exogenous Ship2 was detected by using X-press antibody (Invitrogen), and endogenous Ship2 was detected by using the polyclonal anti-Ship2 antibody (Santa Cruz Biotechnology).
In Vivo Labeling of Lipids. Transfected cells were either labeled with inorganic 32P for 4 h in phosphate-free medium or [3H]inositol for 24 h in inositol-free medium. After labeling, cells were stimulated or not (according to each experiment) and lysed in 1 M HCl. The lipids were extracted in chloroform/methanol (1:1, vol/vol) and deacylated as described (9). Deacylated lipids were separated by anionic-exchange HPLC, detected by an online radiomatic detector, and quantified by using the flo-one analysis program (Packard). Each peak was identified by using in vitro-synthesized internal standard lipids. For the [32P]orthophosphate labeling, the counts in each peak were normalized against the sum of the counts present in the PI-4-P and PI-4,5-P2 peaks. For the [3H]inositol labeling, the counts present in each peak were normalized against the counts present in the phosphatidylinositol peak.
PI3K Recruitment Assay. Cells were transfected as described above and serum-starved for 24 h in RPMI medium 1640 containing 0.5% BSA. Cells were stimulated with 10 nM insulin for 10 min and lysed in buffer containing 50 mM Tris·HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 10% glycerol, and protease and phosphatase inhibitors. Lysates were cleared by centrifugation at 18,000 × g for 10 min, and supernatants were incubated with anti-phosphotyrosine (pTyr) antibody for2hand with protein A-Sepharose beads for 1 h. The beads were centrifuged and washed three times with phosphate-buffer saline containing 1% Nonidet P-40, two times with high-salt buffer containing 50 mM Tris·HCl, pH 7.5, and 0.5 M LiCl, and two times with low-salt buffer containing 50 mM Tris·HCl, 10 mM NaCl, and 1 mM EDTA, pH 8.0. The immunoprecipitation complexes were either resolved by SDS/PAGE and analyzed by Western blot with anti-pTyr antibody (4G10) or assayed for PI3K activity by incubation with PI-4,5-P2/phosphatidylserine in buffer containing 30 mM Hepes at pH 7.0, 10 mM MgCl2, and 2 μCi (1 Ci = 37 GBq) of [γ-32P]ATP. The lipids were separated by TLC and visualized by autoradiography.
Results
PIP4K II Down-Regulates Akt Phosphorylation. To determine whether stimulation of PI-4,5-P2 synthesis regulates the insulin-induced PI-3,4,5-P3 production that leads to Akt activation, we expressed type I and II PIPKs in CHO-IR cells. These cells are highly transfectable, respond to insulin, and have undetectable levels of endogenous PIP4K IIβ.
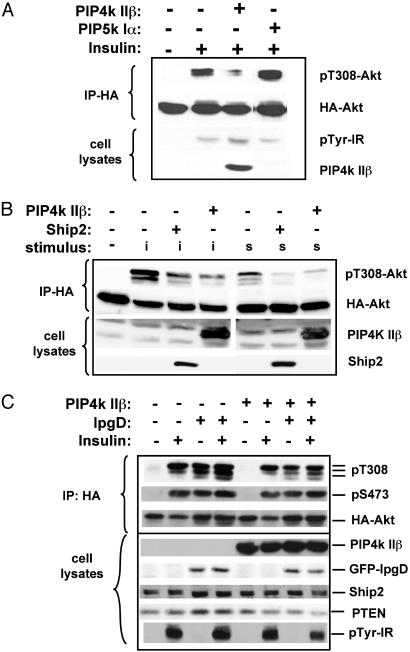
Fig. 1A shows that activation of the PI-4-P pathway for PI-4,5-P2 synthesis by overexpression of the PIP5K I resulted in an ≈2-fold increase in insulin-induced Akt phosphorylation, which can be explained by an increase in the amount of PI-4,5-P2 available as a substrate for PI3K. However, activation of the PI-5-P pathway for PI-4,5-P2 synthesis by PIP4K IIβ expression resulted in significantly diminished phosphorylation of Akt on threonine 308 (T308) after insulin stimulation (Fig. 1). The effect of PIP4K IIβ expression on insulin- or serum-induced Akt phosphorylation was comparable with the effect of overexpressing the PI-3,4,5-P3 phosphatase Ship2 in these cells (Fig. 1B). PIP4K IIβ expression did not decrease the tyrosine phosphorylation of the IR (Fig. 1 A).
Fig. 1.
The PI-5-P pathway for PI-4,5-P2 synthesis regulates the insulin-induced Akt phosphorylation. CHO-IR cells were transfected with HA-Akt and with the genes indicated above each panel. Cells were serum-starved for 24 h and stimulated (as indicated) with insulin (i, 10 nM) or serum (s, 20%) for 10 min. Phosphorylation of the transfected HA-Akt was assayed by using phospho-specific antibodies against pT308 or pS473 of Akt. Also shown are the Western blots of the total cell lysates with antibodies against the transfected proteins or against pTyr. Results are representative of three or more experiments. IP, immunoprecipitation.
Expression of the PI-4,5-P2-specific 4-phosphatase IpgD in CHO-IR cells led to a dramatic increase in basal and insulin-stimulated Akt phosphorylation (Fig. 1C), indicating that the reported increase in PI-3,4,5-P3 after S. flexneri infection (8) is due to IpgD. IpgD-induced Akt phosphorylation was partially reversed by PIP4K IIβ expression (Fig. 1C). Neither PIP4K IIβ nor IpgD affected expression of endogenous Ship2 and PTEN (Fig. 1C).
Together, these results indicate that the PI-5-P pathway for PI-4,5-P2 synthesis plays a role in PI3K signaling that leads to Akt phosphorylation.
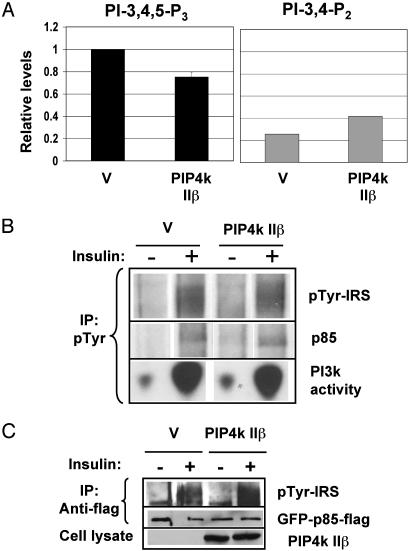
PIP4K IIβ Effect Is Downstream of PI3K Activation. To determine which events downstream of insulin stimulation and upstream of Akt phosphorylation are affected by PIP4K IIβ, we investigated the effect of PIP4K IIβ expression on insulin-induced PI-3,4,5-P3 levels. CHO-IR cells were transfected with PIP4K IIβ or empty vector, serum-starved for 24 h, and labeled with [32P]orthophosphate. Fig. 2A shows that PIP4K IIβ expression in CHO-IR cells decreased insulin-induced PI-3,4,5-P3 levels by ≈25%. Because PI-3,4,5-P3 is essential for the induction of Akt phosphorylation at T308, this decrease in PI-3,4,5-P3 levels is consistent with the decrease in insulin-induced Akt phosphorylation shown in Fig. 1. The effect of PIP4K IIβ expression on PI-3,4,5-P3 levels was not as dramatic as its effect on Akt phosphorylation. This can be explained by the fact that the PI-3,4,5-P3 detected in our assay corresponds to the PI-3,4,5-P3 induced in the total population of cells, but only 60–80% of the cells expressed PIP4K IIβ, as estimated by the percentage of cells that express transfected GFP (data not shown). Measurements of HA-Akt phosphorylation, on the other hand, are restricted to the transfected cells. Interestingly, Fig. 2 A Right shows that, unlike PI-3,4,5-P3, PI-3,4-P2 levels did not decrease with expression of PIP4K IIβ (discussed below).
Fig. 2.
PIP4K IIβ decreases PI-3,4,5-P3 levels downstream of PI3K activation. CHO-IR cells were transfected with PIP4K IIβ or empty vector (V) as indicated and serum-starved for 24 h. (A) Cells were labeled for 4 h with [32P]orthophosphate and stimulated with insulin (10 nM) for 5 min. The deacylated lipids were analyzed by HPLC, quantified, and normalized as described. The graph represents the relative levels of PI-3,4,5-P3 (black) or PI-3,4-P2 (gray) when compared with the PI-3,4,5-P3 lvels in cells transfected with empty vector. (B) Cell lysates were immunoprecipitated (IP) with anti-pTyr antibody and blotted with anti-pTyr antibody or anti-p85 antibody or assayed for PI3K activity. (C) CHO-IR cells were transfected with GFP-p85-Flag and with PIP4K IIβ or empty vector (V). Protein lysates were immunoprecipitated with anti-flag antibody and blotted with anti-pTyr or anti-GFP antibodies. Total cell lysates were blotted with anti-PIP4K IIβ antibody. Results are representative of two or more experiments.
To determine whether PIP4K IIβ expression affects PI3K recruitment to the IR complex, we used anti-pTyr antibodies to immunoprecipitate the activated IR and associated proteins. Coimmunoprecipitation of the PI3K present in the total population of cells was measured by Western blot of the regulatory subunit (p85) or a lipid kinase assay (Fig. 2B). PIP4K IIβ expression had no detectable effect on insulin-induced PI3K recruitment to pTyr complexes. To measure the recruitment of PI3K to the receptor complex in transfected cells only, we cotransfected CHO-IR cells with GFP-p85-Flag and PIP4K IIβ or vector control. The cells were stimulated or not with insulin, and the exogenous p85 was immunoprecipitated with anti-Flag antibody. The immunoprecipitates then were blotted with anti-pTyr antibody to detect phospho-IRS associated with p85. PIP4K IIβ did not affect GFP-p85-Flag recruitment to the activated receptor complex in this assay (Fig. 2C). These results show that PIP4K IIβ expression decreased PI-3,4,5-P3 levels without affecting the events upstream of PI3K activation that follow insulin stimulation.
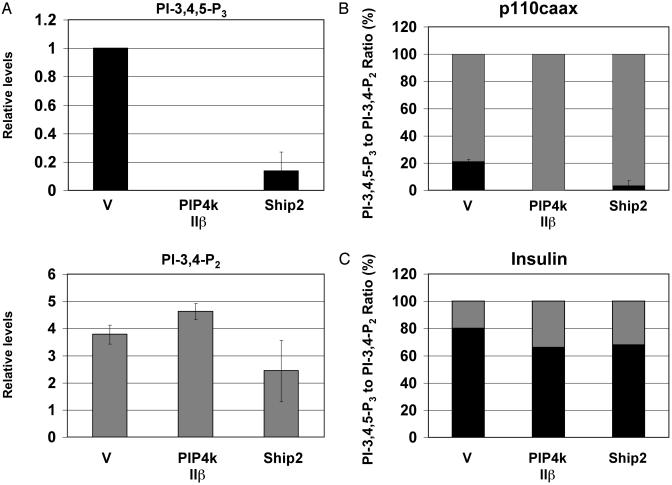
PIP4K IIβ Expression Abolishes PI-3,4,5-P3 Levels in Cells Expressing p110caax. The effect of PIP4K IIβ expression on the levels of PI-3,4,5-P3 induced by constitutively active PI3K was examined by cotransfection of CHO-IR cells with p110caax and either PIP4K IIβ or vector control. The cells were labeled with [3H]inositol for 24 h in the absence of serum. The lipids were extracted, deacylated, and analyzed by HPLC. As shown in Fig. 3A, PIP4K IIβ expression abolished the constitutive levels of PI-3,4,5-P3 in cells expressing p110caax. A dramatic reduction in PI-3,4,5-P3 levels was also observed when we cotransfected COS cells with PIP4K IIβ and p110caax (data not shown). Expression of p110caax was not affected by PIP4K IIβ expression (data not shown).
Fig. 3.
PIP4K IIβ decreases PI-3,4,5-P3, but not PI-3,4-P2 levels. (A) CHO-IR cells were transfected with p110caax and with PIP4K IIβ, Ship2, or empty vector (V) as indicated. Cells were serum-starved and labeled with [3H]inositol for 24 h. The deacylated lipids were analyzed by HPLC, quantified, and normalized as described. The graphs represent the relative levels of PI-3,4,5-P3 (black) or PI-3,4-P2 (gray) when compared with the PI-3,4,5-P3 levels in cells transfected with empty vector. (B and C) The ratio of PI-3,4,5-P3 and PI-3,4-P2 in cells expressing p110caax (B) or cells stimulated with insulin (C). The gray bars represent the percentage of PI-3,4-P2, and the black bars represent the percentage of PI-3,4,5-P3 over the sum of PI-3,4-P2 and PI-3,4,5-P3. The error bars represent the range from two experiments.
In contrast to PI-3,4,5-P3, PI-3,4-P2 levels did not decrease with PIP4K IIβ expression but rather increased (Fig. 3A). To normalize for variations in p110caax expression, we plotted the data shown in Fig. 3A as the ratio between PI-3,4,5-P3 and PI-3,4-P2. PIP4K IIβ expression changed the ratio of PI-3,4,5-P3/PI-3,4-P2 from 20:80 to 0:100 in cells expressing constitutively active PI3K (Fig. 3B). The PI-3,4,5-P3/PI-3,4-P2 ratio in cells stimulated with insulin is also shown (Fig. 3C). The effects of PIP4K IIβ on the PI-3,4,5-P3/PI-3,4-P2 ratio were comparable with the effect of Ship2 overexpression in the same cells (Fig. 3).
Together, these results indicate that PIP4K IIβ affects insulin signaling at a step downstream of PI3K activation. Because the levels of PI-3,4-P2 did not decrease with PIP4K II expression, it is unlikely that PIP4K II can directly affect PI3K activity.
Although the main product of PIP4K II is PI-4,5-P2, the levels of this lipid did not increase with PIP4K IIβ expression (data not shown). This observation is not surprising, given that the levels of PI-5-P in cells are low and previous data have suggested that the alternative pathway for PI-4,5-P2 synthesis only contributes to a small fraction of this lipid in cells (4). Notably, the levels of PI-3,4-P2 in serum-starved cells were not increased by PIP4K IIβ expression (data not shown), even though these cells have normal levels of phosphatidylinositol-3-phosphate. This result indicates that PI-3,4-P2 is not a likely product of PIP4K II in vivo.
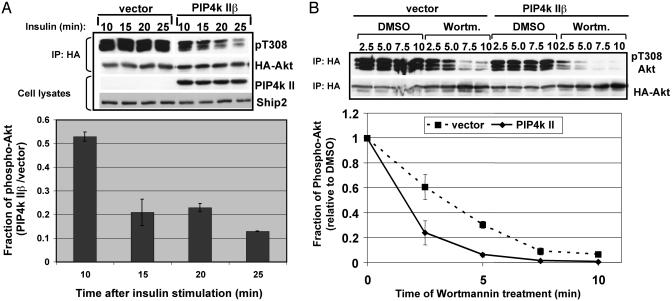
PIP4K IIβ Expression Abbreviates the PI3K Signal. When we examined the kinetics of insulin-induced Akt phosphorylation in CHO-IR cells, we noticed that stimulation with low levels of insulin (1 nM) resulted in a transient activation of Akt in cells expressing PIP4K IIβ, whereas in control cells, insulin-induced activation of Akt was sustained (Fig. 4A). Ten minutes after insulin stimulation, for example, Akt phosphorylation in cells expressing PIP4K IIβ was 52% of control cells, whereas 25 min after stimulation, Akt phosphorylation in PIP4K IIβ-expressing cells was 12%. A possible interpretation for this result is that PIP4K IIβ promotes the termination of the PI-3,4,5-P3 signal that leads to Akt phosphorylation.
Fig. 4.
PIP4K IIβ causes premature termination of the insulin signal. CHO-IR cells were transfected with HA-Akt and with PIP4K IIβ or vector control. Cells were serum-starved for 24 h and stimulated with insulin. Phosphorylation of the transfected HA-Akt was assayed by using phospho-specific antibodies against pT308 of Akt. (A) Cells were stimulated with 1 nM insulin for 10, 15, 20, or 25 min as indicated. Total cell lysates were blotted with antibodies against PIP4K IIβ or endogenous Ship2. The bar graph shows the levels of phospho-Akt in PIP4K IIβ-expressing cells as a fraction of the levels of phospho-Akt in vector-transfected cells. The quantification was performed by using two independent experiments. (B) Cells were stimulated with 10 nM insulin for 5 min and treated with DMSO or wortmannin (Wortm., 100 nM) for 2.5–10 min. The bar graph is a plot of the results obtained from two independent experiments. The data were normalized by calculating the fraction of Akt activation in wortmannin-treated cells relative to DMSO-treated cells. IP, immunoprecipitation.
To test this possibility, we transfected CHO-IR cells with PIP4K IIβ or vector control and stimulated them with insulin. After 5 min, de novo PI-3,4,5-P3 synthesis was inhibited by the addition of the PI3K inhibitor wortmannin. The duration of the insulin signal was measured by determining the level of phosphorylated Akt over time. In the absence of wortmannin, PIP4K IIβ caused a 40–50% inhibition of insulin-induced Akt phosphorylation as compared with control cells as previously shown (Figs. 1 and 4B). In control cells treated with wortmannin, the level of phosphorylated Akt decreased steadily as expected. However, in cells expressing PIP4K IIβ the levels of phosphorylated Akt dropped dramatically (75%) after 2.5 min of wortmannin treatment (Fig. 4B). These results indicate that PIP4K IIβ attenuates the insulin signal that leads to Akt phosphorylation by accelerating the degradation of PI-3,4,5-P3. This effect cannot be attributed to changes in the expression of the PI-3,4,5-P3 phosphatases PTEN and Ship2 (Figs. 1 and 4A).
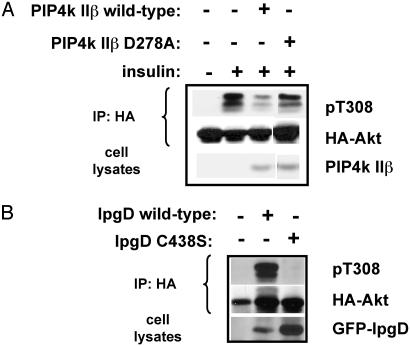
Catalytic Activities of PIP4K II and IpgD Are Important for Their Function in Regulating the PI3K Signal. To understand whether the kinase activity of PIP4K II is necessary for its effect on decreasing the insulin signal that leads to Akt phosphorylation, CHO-IR cells were cotransfected with HA-Akt and with either wild-type PIP4K IIβ or kinase-impaired D278A mutant. Mutation of this aspartic acid present in the catalytic site of PIP4K IIβ reduced but did not completely abolish the PI-5-P kinase activity of this enzyme (5% activity was retained) as determined by an in vitro kinase assay of recombinant PIP4K IIβ expressed in bacteria (data not shown). Fig. 5A shows that expression of wild-type PIP4K IIβ significantly inhibited the insulin-induced Akt phosphorylation, whereas the D278A mutant did not. When expressed at high levels (5- to 10-fold higher than wild-type), the PIP4K IIβ D278A mutant was able to inhibit insulin-induced Akt phosphorylation (data not shown), possibly because of its residual kinase activity. Like the IIβ isoform, expression of PIP4K IIα and PIP4K IIγ in CHO-IR cells reduced the insulin-stimulated Akt phosphorylation (data not shown). Thus, the ability of the PI-5-P pathway to regulate insulin signaling cannot be explained solely by a scaffolding effect of PIP4K IIβ but rather depends on the conversion of PI-5-P to PI-4,5-P2, catalyzed by PIP4K II.
Fig. 5.
Inactivation of PIP4K II and IpgD impairs their ability to regulate Akt phosphorylation. CHO-IR cells were transfected with HA-Akt and with wild-type or D278A PIP4K IIβ (A) or wild-type or C438S GFP-IpgD (B). Cells were serum-starved for 24 h and stimulated with insulin (10 nM) for 10 min (A) or left untreated (B). Lysates were prepared, immunoprecipitated (IP) with anti-HA, and assayed for phosphorylated Akt by using anti-Akt pT308 antibody. The result shown is representative of two or more separate experiments.
To understand whether the phosphatase activity of IpgD is required for its effect on Akt phosphorylation, CHO-IR cells were cotransfected with HA-Akt and with either wild-type IpgD or phosphatase-dead C438S mutant. Fig. 5B shows that the phosphatase activity of IpgD is essential for its effect on PI3K signaling, which leads to Akt phosphorylation. These results indicate that production of PI-5-P and/or consumption of PI-4,5-P2 by IpgD are positive regulators of PI3K signaling.
Discussion
The data presented here show that activation of the PI-5-P pathway for PI-4,5-P2 synthesis by PIP4K IIβ expression negatively regulates insulin signaling by decreasing the cellular levels of PI-3,4,5-P3. This result was unexpected given that PI-4,5-P2 is the intermediate for PI-3,4,5-P3 synthesis that leads to Akt activation. Activation of PI3K is not impaired by PIP4K IIβ expression, because PI-3,4-P2 levels did not decrease in cells expressing PIP4K IIβ. Instead, we found that PIP4K IIβ expression abbreviated the PI3K signal. Thus, our data suggest that PIP4K IIβ expression causes the removal of PI-3,4,5-P3 from cells rather than inhibition of its synthesis. We measured the effect of PIP4K II on inositol polyphosphate levels (data not shown) and found no evidence that the reduced levels of PI-3,4,5-P3 in PIP4K IIβ-expressing cells could be due to phospholipases. Instead, our data suggest that PIP4K IIβ expression activates a PI-3,4,5-P3-specific phosphatase as illustrated in Fig. 6. Ship1, Ship2, and PTEN are well characterized lipid phosphatases known to dephosphorylate PI-3,4,5-P3 (10, 11). More recently, SKIP was identified as a PI-3,4,5-P3-specific phosphatase as well (12). Ship and SKIP dephosphorylate PI-3,4,5-P3 at the 5′ position of the inositol ring to generate PI-3,4-P2. PTEN, on the other hand, dephosphorylates PI-3,4,5-P3 and PI-3,4-P2 at the 3′ position of the inositol ring to generate PI-4,5-P2 and PI-4-P. Because PIP4K IIβ expression did not decrease PI-3,4-P2 levels, it is unlikely that the effect of PIP4K IIβ on PI-3,4,5-P3 is due to activation of PTEN. Our results are consistent with PIP4K IIβ activating a PI-3,4,5-P3-specific 5-phosphatase. In fact, we observed that in cells expressing PIP4K IIβ there is an increase in the number of counts present in the PI-3,4-P2 peak, which is equivalent to the number of counts lost from the PI-3,4,5-P3 peak (Fig. 3A). Nonetheless, in some experiments, the increase in PI-3,4-P2 in PIP4K IIβ-expressing (Fig. 2 A) or Ship2-expressing (Fig. 3A) cells does not account for the total loss in PI-3,4,5-P3, indicating the presence of a PI-3,4-P2-specific phosphatase in cells. Other groups have also expressed Ship2 in cells and reported similar findings (13). Interestingly, the effect of PIP4K IIβ expression on phosphorylation of Akt at T308 was more dramatic than the effect on S473, when cells were treated with insulin for 10 min (Fig. 1C). This could be due to the fact that dephosphorylation of pT308 is faster than dephosphorylation of pS473. In addition, Scheid et al. (14) recently reported that high levels of PI-3,4-P2 correlate with phosphorylation of Akt at S473, whereas high levels of PI-3,4,5-P3 correlate with phosphorylation at T308. Our results are consistent with this report.
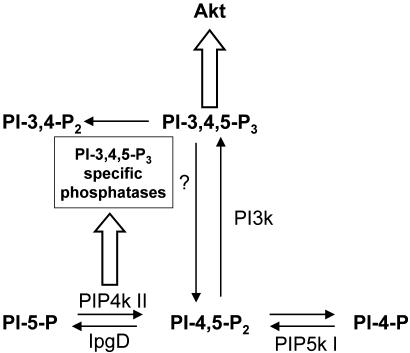
Fig. 6.
Schematic diagram showing our current model for the role of PIP4K II in PI-3,4,5-P3 regulation. PIP4K II expression in cells stimulates PI-3,4,5-P3 degradation. Our results are consistent with PIP4K II activating a PI-3,4,5-P3-specific 5-phosphatase. However, we have not eliminated the possibility that a PI-3,4,5-P3-specific 3-phosphatase is also activated by PIP4K II. The thick arrows indicate a pathway, and the thin arrows indicate a biochemical reaction.
It has been reported previously that the translocation of IpgD into mammalian cells during bacterial invasion leads to rapid changes in phosphoinositide levels in the host cell including a dramatic increase in cellular PI-3,4-P2 and PI-3,4,5-P3 (8). Here we show that IpgD phosphatase activity is essential for activating the PI3K signal that leads to Akt phosphorylation, indicating that the conversion of PI-4,5-P2 to PI-5-P catalyzed by IpgD regulates PI3K signaling. Furthermore, the effect of IpgD on Akt phosphorylation can be partially reversed by PIP4K IIβ expression. The mechanism by which IpgD affects PI3K signaling is likely to be complex, given the massive changes in phosphoinositide metabolism caused by its activity. Therefore, it is possible that the effect of IpgD on PI3K signaling involves concomitant activation of stress signals that stimulate PI-3,4,5-P3 synthesis and inhibition of phosphoinositide phosphatases.
Consistent with our data, mice lacking PIP4K IIβ have increased insulin sensitivity that potentially could be explained by increased stability of the insulin-dependent PI3K signal (K.A.L., O. M. Peroni, L.E.R., B. B. Kahn, and L.C.C., unpublished data). Interestingly, this phenotype resembles the phenotype observed in mice heterozygous for disruption of Ship2 (15).
It is still unclear whether the effect of PIP4K II on PI-3,4,5-P3 levels reported here is due to PI-5-P consumption and/or PI-4,5-P2 synthesis. We favor a model in which PI-5-P functions as an inhibitor of a PI-3,4,5-P3-specific 5-phosphatase, and PIP4K II regulates PI3K signaling by removing PI-5-P from cells.
Acknowledgments
This work was funded by Department of Defense (U.S. Army) Grant DAMD17-01-1-0155, National Institutes of Health Grant GM36624, and Public Health Service Research Grant 5 P30 DK36836-15 of the National Institute of Diabetes and Digestive and Kidney Diseases.
Abbreviations: PI-3,4,5-P3, phosphatidylinositol-3,4,5-trisphosphate; PI3K, phosphoinositide 3-kinase; PI-4,5-P2, phosphatidylinositol-4,5-bisphosphate; PI-5-P, phosphatidylinositol-5-phosphate; PI-4-P, phosphatidylinositol-4-phosphate; PIPK, phosphatidylinositol phosphate kinase; PIP4K, PI-5-P 4-kinase; CHO, Chinese hamster ovary; IR, insulin receptor; HA, hemagglutinin; pTyr, phosphotyrosine.
References
- 1.Tengholm, A. & Meyer, T. (2002) Curr. Biol. 12, 1871–1876. [DOI] [PubMed] [Google Scholar]
- 2.Toker, A. (1998) Curr. Opin. Cell Biol. 10, 254–261. [DOI] [PubMed] [Google Scholar]
- 3.Fruman, D. A., Meyers, R. E. & Cantley, L. C. (1998) Annu. Rev. Biochem. 67, 481–507. [DOI] [PubMed] [Google Scholar]
- 4.Whiteford, C. C., Brearley, C. A. & Ulug, E. T. (1997) Biochem. J. 323, 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishihara, H., Shibasaki, Y., Kizuki, N., Wada, T., Yazaki, Y., Asano, T. & Oka, Y. (1998) J. Biol. Chem. 273, 8741–8748. [DOI] [PubMed] [Google Scholar]
- 6.Rameh, L. E., Tolias, K. F., Duckworth, B. C. & Cantley, L. C. (1997) Nature 390, 192–196. [DOI] [PubMed] [Google Scholar]
- 7.Morris, J. B., Hinchliffe, K. A., Ciruela, A., Letcher, A. J. & Irvine, R. F. (2000) FEBS Lett. 475, 57–60. [DOI] [PubMed] [Google Scholar]
- 8.Niebuhr, K., Giuriato, S., Pedron, T., Philpott, D. J., Gaits, F., Sable, J., Sheetz, M. P., Parsot, C., Sansonetti, P. J. & Payrastre, B. (2002) EMBO J. 21, 5069–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serunian, L. A., Auger, K. R. & Cantley, L. C. (1991) Methods Enzymol. 198, 78–87. [DOI] [PubMed] [Google Scholar]
- 10.Krystal, G., Damen, J. E., Helgason, C. D., Huber, M., Hughes, M. R., Kalesnikoff, J., Lam, V., Rosten, P., Ware, M. D., Yew, S. & Humphries, R. K. (1999) Int. J. Biochem. Cell Biol. 31, 1007–1010. [DOI] [PubMed] [Google Scholar]
- 11.Maehama, T. & Dixon, J. E. (1998) J. Biol. Chem. 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- 12.Ijuin, T. & Takenawa, T. (2003) Mol. Cell. Biol. 23, 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor, V., Wong, M., Brandts, C., Reilly, L., Dean, N. M., Cowsert, L. M., Moodie, S. & Stokoe, D. (2000) Mol. Cell. Biol. 20, 6860–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheid, M. P., Huber, M., Damen, J. E., Hughes, M., Kang, V., Neilsen, P., Prestwich, G. D., Krystal, G. & Duronio, V. (2002) J. Biol. Chem. 277, 9027–9035. [DOI] [PubMed] [Google Scholar]
- 15.Clement, S., Krause, U., Desmedt, F., Tanti, J. F., Behrends, J., Pesesse, X., Sasaki, T., Penninger, J., Doherty, M., Malaisse, W., et al. (2001) Nature 409, 92–97. [DOI] [PubMed] [Google Scholar]