Abstract
Long-range enhancer–promoter interactions are commonly seen in complex genetic loci such as Hox genes and globin genes. In the case of the Drosophila Antennapedia complex, the T1 enhancer bypasses the neighboring ftz gene and interacts with the distant Scr promoter to activate expression in posterior head segments. Previous studies identified a 450-bp promoter-proximal sequence, the tethering element, which is essential for T1–Scr interactions. To obtain a more comprehensive view of how individual enhancers selectively interact with appropriate target genes, we used bioinformatic methods to identify new cis-regulatory DNAs in the ≈50-kb Scr-Antp interval. Three previously uncharacterized regulatory elements were identified: a distal T1 tethering sequence mapping >40 kb from the proximal tethering sequence, a repressor element that excludes activation of Scr by inappropriate enhancers, and a new ftz enhancer that directs expression within the limits of stripes 1 and 5. Many of the regulatory DNAs in the Scr-Antp interval are transcribed, including the proximal and distal tethering elements. We suggest that homotypic interactions between the tethering elements stabilize long-range T1–Scr interactions during development.
Enhancers direct localized stripes, bands, and tissue-specific patterns of gene expression in the early Drosophila embryo (1). They are typically 300 bp to 1 kb in length and contain clustered binding sites for both transcriptional activators and repressors. Enhancers usually activate nearby target genes, although there are examples where they ignore the most proximal promoters and interact with distantly linked genes. Examples include the 3′ enhancers of the dpp gene and the T1 enhancer of Scr.
The dpp enhancers fail to activate the neighboring slh and oaf genes but instead activate the expression of the distal dpp gene in imaginal disks (2). The selective regulation of dpp expression appears to depend on promoter specificity (3, 4). The oaf and slh promoters are incompatible for activation by the dpp enhancers, despite the fact that they map much closer than does the preferred dpp promoter. Similarly, the distal T1 enhancer jumps over the intervening ftz gene to activate Scr in posterior head segments (5). The failure of the T1 enhancer to activate ftz might also depend on promoter specificity. The T1 enhancer only weakly activates a minimal ftz-lacZ fusion gene, despite the fact that it contains a strong TATA element. However, the possible incompatibility between T1 and the ftz promoter is not sufficient to account for selective T1–Scr interactions, because T1 also fails to activate a Scr-lacZ fusion gene containing the minimal Scr core promoter. We have previously identified a 450-bp tethering element that maps immediately 5′ of the Scr core promoter (6). This element is essential for T1–Scr interactions and is sufficient to mediate long-range T1–ftz interactions when placed immediately 5′ of the ftz promoter (Fig. 1).
Fig. 1.
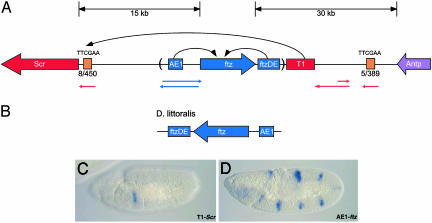
Summary of the Scr-Antp region of the Antennapedia complex. The Scr-Antp interval of the Antennapedia complex contains two homeotic selector genes, Scr and Antp, and the ftz segmentation gene. (A) Scr and ftz are divergently transcribed and separated by a 15-kb intergenic spacer. The AE1 enhancer, which drives localized expression of ftz in seven pairrule stripes (D), is positioned upstream of the ftz promoter within this spacer. A 30-kb intergenic spacer separates ftz from the homeotic selector gene Antp.A newly identified ftz enhancer, ftzDE, is contained within this spacer, just downstream of the ftz coding region. ftzDE directs expression specifically in ftz stripes 1 and 5. The T1 enhancer directs Scr expression in the labial head segment and anterior compartment of the first thoracic segment (C). The Scr promoter-proximal tethering element is required for the proper targeting of T1 to the Scr promoter (A). The 450-bp tethering element is positioned immediately upstream of the Scr core promoter and contains eight copies of a palindromic motif, TTCGAA. Another cluster of this motif is located within 3 kb downstream of the Antp coding region. This “distal” tethering cluster contains five copies of the palindromic motif. Colored arrows beneath the diagram indicate regions within the intergenic spacer that are transcribed. Red arrows represent regions transcribed in patterns overlapping the endogenous Scr expression domain. Blue arrows represent intergenic regions transcribed in the ftz pair-rule pattern. A 14-kb region containing the ftz gene and associated enhancers has undergone a chromosomal inversion in related Drosophila species, such as D. littoralis (B).
In the present study, we have conducted a systematic analysis of cis-regulatory DNAs in the 50-kb interval that separates Scr and Antp within the Antennapedia complex (ANT-C). An ftz enhancer is identified that maps 3′ of the ftz transcription unit (ftzDE; Fig. 1 A). This enhancer initiates gene expression within the limits of ftz stripes 1 and 5. The previously identified Scr tethering element contains eight copies of a simple palindromic sequence, TTCGAA. Four tandem copies of this motif are sufficient to mediate T1–ftz interactions in transgenic embryos. A whole-genome survey of high-density clusters of the TTCGAA motif identifies a 389-bp sequence located just 3′ of the Antp transcription unit (Fig. 1 A, 5/389 cluster). This cluster can function as a tethering element when attached to the minimal ftz promoter. It also diminishes the position effects observed for T1–Scr interactions in transgenic strains. We propose a model whereby proteins that bind the TTCGAA motif in the proximal tethering element and distal cluster mediate the formation of a transcription loop, which stabilizes T1–Scr interactions. The putative loop might depend on the transcription of the cis-regulatory DNAs within the ANT-C, including the tethering element and distal cluster themselves.
Materials and Methods
Bioinformatics. The cis-analyst program (http://rana.lbl.gov/cis-analyst) was used to scan a 10-kb region flanking the ftz coding region for high-density clusters of binding sites for transcription factors involved in early A-P axis patterning. Of the three identified clusters, two correspond to known ftz regulatory elements. PCR primers were used to isolate a genomic DNA fragment encompassing the third cluster (5′-GACGGGCACGAAATTTTAC; 5′-AGTTGTTTTGCAAACATCAACC).
The distal tethering element was identified through a genomewide screen for high-density TTCGAA clusters. The fly enhancer web server (www.flyenhancer.org) was used to scan the Drosophila genome for clusters of five or more TTCGAA hexamers within 500 bp. The distal tether was selected on the basis of its location within the Scr-Antp domain. Primers were designed to amplify the 389-bp distal cluster: 5′-TTCGAAACTCGGGGGCGGAGCCATC; 5′-TTCGAAAATCATTCGAAATTCGAAAATTCG.
P Transformations. yw67 flies were used for all P transformation assays. Fusion genes were introduced into the Drosophila germ line by using standard methods (7). Multiple transformants were generated for each construct, and at least three independent lines were examined by in situ hybridization. Embryos were prepared and hybridized with digoxigenin-labeled CAT and lacZ probes, as described (8, 9).
Preparation of Cis Elements. Cloning of the AE1 and T1 enhancers has been described (6). The 389-bp distal TTCGAA cluster was amplified from the genome as a SmaI fragment and subcloned into p-Bluescript. It was also subcloned into a p-Bluescript vector containing the 3.8-kb T1 enhancer (HindIII fragment) for the constructs in Fig. 3.
Fig. 3.
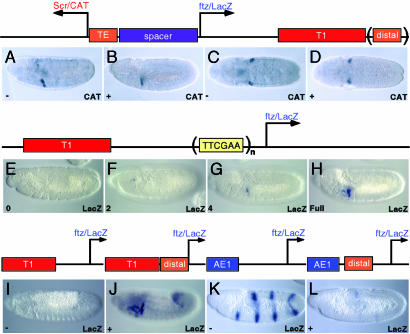
The distal cluster mediates tethering. (A–D) A P element expression vector was used to determine the activities of the newly identified distal cluster, which contains five copies of the TTCGAA motif in a 389-bp DNA fragment located downstream of Antp (see Fig. 1 A legend). The distal cluster was placed downstream of the 3′ T1 enhancer (top). Divergently transcribed CAT and lacZ reporter genes were placed under the control of the Scr and ftz promoters, respectively. The 450-bp proximal tethering element was positioned immediately 5′ of Scr and separated from the divergently transcribed ftz-lacZ fusion gene with a 1.6-kb spacer sequence from bacteriophage λ. The T1 enhancer activates Scr-CAT expression in the posterior head segment (A–D). However, in the absence of the distal cluster (“distal,” top), CAT exhibits variable background staining (A and C). With the distal cluster, background staining is eliminated, and CAT expression is restricted to the posterior head segment (B and D). One hundred stained embryos undergoing germband retraction were scored for CAT expression outside of the T1 expression domain. This was done for three separate lines for both transgenes. In lines containing the distal cluster, only 5% of the embryos displayed any ectopic staining. In contrast, more than half of the embryos lacking the distal cluster express lacZ outside of the T1 expression domain. (E–H) Multimers of the TTCGAA palindrome were inserted immediately upstream of a ftz-lacZ reporter to test whether it is sufficient to mediate tethering activity (center). The T1 enhancer was placed upstream of the modified reporter, and T1 activity was assayed by in situ hybridization. Copies of the palindrome were separated by 10 bp or one helical turn. None, 2, 4, and 6 copies of the palindrome were stained in parallel with a ftz-lacZ reporter modified with the full-length Scr proximal tethering (E–G). Weak activation of the lacZ reporter gene is detected on insertion of four tandem copies of the TTCGAA motif (G). An additional two copies of the palindrome do not significantly alter lacZ expression (not shown). Stronger activation of lacZ expression is obtained with either the proximal tethering element (H; “full”) or distal cluster (not shown). (I–L) The distal cluster was inserted between the 5′ T1 enhancer and ftz-lacZ fusion gene, or between AE1 and ftz-lacZ. The distal cluster mediates strong activation by T1 (J; compare with I), but diminishes activation by the AE1 enhancer (L; compare with K).
Most of the Scr and ftz promoters used in this study have been characterized (6). For the extended 520-bp tethering element, a hybrid oligonucleotide was used to insert the element (–555 to –34 upstream of Scr +1) upstream of the ftz core promoter.
The ftz promoters containing multimers of the TTCGAA hexamer were created by inserting copies of the palindrome into a unique SbfI site engineered upstream of the minimal ftz core promoter. All manipulations were performed in a p-Bluescript vector modified with AscI sites flanking the polylinker. For the two-copy multimer, an oligo containing the sequence 5′-CCTGCAGGTTCGAAGATCCGTTAGTTCGAAGTTACCTGCAGG was used. For the four-copy vector, an extended oligo was used: 5′-GGAGTCGGACTTCGAAGATCCGTTACTTCGAAGTTACCTGCATTCGAATGGCATGGGCTTCGAACCGTCCTGCA. In both cases, individual TTCGAA motifs are separated by 10-bp spacer sequences.
Construction of P Element Transposons. The CAT/lacZ P transformation vector used for all of the experiments presented in this study is a modification of pCasPer, which contains divergently transcribed white and lacZ reporter genes (7). It was modified by insertion of a CAT reporter gene between white and lacZ (10). Construction of P element injection vectors has been described (6).
For the distal tether constructs in Fig. 3, a 1.6-kb spacer from bacteriophage λ was isolated as an AscI fragment and cloned into the unique AscI site in pCasPer. The T1 enhancer and distal tether were isolated from p-Bluescript as a single SbfI-flanked fragment and cloned into the unique PstI site located downstream of the lacZ reporter gene.
Noncoding Cis Element Transcription. Transcription of cis elements in the Scr-ftz and ftz-Antp intergenic spacers was assayed by in situ hybridization. The intergenic spacers were amplified from the genome in 2-kb fragments by PCR. Amplified products were cloned into the pGEM-T cloning vector. Digoxigenin probes were synthesized from the T7 and SP6 polymerase sites in the pGEM-T.
Results
Previous studies have identified three cis-regulatory DNAs in the 50-kb interval that separate the Scr and Antp genes: the T1 and AE1 enhancers and a 450-bp tethering element located immediately 5′ of the Scr core promoter (Fig. 1 A). The tethering element is required for long-range T1–Scr interactions and localized expression in the posterior head segment (Fig. 1C). AE1 maintains the seven stripes of ftz expression in the germband of elongating embryos (Fig. 1D). To identify new cis-regulatory DNAs, different genomic DNA fragments from the Scr-Antp interval were assayed in transgenic embryos by using a variety of P element expression vectors.
Identification of a 3′ ftz Enhancer. Using the cis-analyst search algorithm, a new ftz enhancer was identified by scanning the Antp-Scr interval for clusters of cis-regulatory elements that are recognized by transcription factors encoded by maternal (bicoid and caudal), gap (hb, Kr, kni), and pair-rule (ftz) genes (11). A total of three clusters were identified (Fig. 2A). Two of the clusters correspond to previously identified cis-regulatory DNAs, the AE1 enhancer, and the ftz zebra element, which initiates ftz expression in early embryos (12–14). A third cluster was also identified that maps just downstream of the ftz transcription unit (cluster 3, see Fig. 2 A). A 1.25-kb genomic DNA fragment that encompasses this cluster was inserted into a P element expression vector containing divergently transcribed CAT and lacZ reporter genes (Fig. 2B). CAT is under the control of the Scr promoter region, whereas lacZ contains the ftz promoter region. Transgenic embryos that contain this reporter gene were collected and hybridized with CAT and lacZ antisense RNA probes. Cluster 3 selectively activates the lacZ reporter gene but fails to induce CAT expression (Fig. 2D; compare with Fig. 2C). ftz-lacZ expression is detected in two stripes in cellularizing embryos (Fig. 2D). Double-staining experiments using a probe that visualizes the endogenous ftz stripes indicates that the newly identified enhancer directs expression in stripes 1 and 5 (arrows, Fig. 2F). ftz stripes 1 and 5 flank the expression domain of the gap repressor Krüppel (Kr), suggesting Kr might repress expression in the center of the embryo. In mutant embryos homozygous for a null mutation in the Kr gene, these stripes are expanded into a broad band (Kr1, Fig. 2E).
Fig. 2.
Identification of an ftz enhancer. The ftz regulatory region was scanned for clusters of cis-regulatory elements that are recognized by transcription factors encoded by maternal (bicoid and caudal), gap (hb, Kr, kni), and pair-rule (ftz) genes. Three clusters were found in the 10-kb region flanking the ftz coding region. The first two clusters correspond to previously identified cis-regulatory elements: AE1 and the zebra element, respectively (A). The third cluster is located between the ftz coding region and the T1 enhancer (B). A 1.25-kb genomic DNA fragment that encompasses this cluster was inserted into a P element expression vector that contains divergently transcribed CAT and lacZ reporter genes (B). CAT is under the control of the Scr promoter region, whereas lacZ contains the ftz promoter region. Cluster 3 selectively activates the lacZ reporter gene but fails to induce CAT expression (C). ftz-lacZ expression is detected in two stripes in cellularizing embryos (D). These stripes are expanded into a broad band in mutant embryos homozygous for a null mutation in the Kr gene (E). Double-staining experiments using a probe that visualizes the endogenous ftz stripes indicate the newly identified enhancer directs expression in stripes 1 and 5 (F).
The newly identified enhancer (cluster 3) is adjacent to the T1 enhancer (see Fig. 1 A), which regulates Scr expression in the labial head segment and anterior compartment of the first thoracic segment (5). Despite its proximity to T1, the new enhancer appears to regulate ftz expression, not Scr. First, the enhancer selectively activates the ftz-lacZ gene and fails to stimulate expression from the Scr promoter, even though the leftward CAT reporter gene contains both the Scr core promoter and the adjacent tethering sequence (Fig. 2C). In contrast, the T1 enhancer exhibits the opposite regulatory specificity; it selectively activates Scr-CAT and not ftz-lacZ. Another argument that the new enhancer is a component of the ftz locus is the observation that other Drosophila species, such as Drosophila littoralis, contain an inversion that inverts the ftz transcription unit (see Fig. 1B). This inversion includes the 5′ zebra element and AE1 enhancer (15). It also includes the newly identified enhancer. The “rightward” chromosomal breakpoint maps between the new enhancer and T1. We hereafter refer to the new enhancer as the ftz distal enhancer (ftzDE) and suggest it is a remnant of the homeotic function seen for Ftz in other insects, such as the flour beetle (see Discussion).
Identification of a Distal Tethering Element. We recently identified a promoter-proximal regulatory element located immediately 5′ of the Scr core promoter (6). This tethering element is required for specific T1–Scr interactions. When positioned upstream of a ftz-lacZ fusion gene, the T1 enhancer now activates transcription from the heterologous ftz promoter. The 450-bp tethering element contains an overrepresented hexamer motif, TTCGAA (Fig. 1 A). A survey of the entire Drosophila genome using the flyenhancer search engine (16) identified a relatively small number of short DNA segments (<400 bp) that contain at least five perfect copies of this motif. One of the clusters maps within the Antp-Scr interval, just downstream of the Antp gene (Fig. 1 A). This newly identified distal cluster is also able to function as a tethering element and recruit the T1 enhancer when placed 5′ of the ftz core promoter (Fig. 3J; compare with Fig. 3I).
The newly identified distal cluster maps >40 kb from the Scr promoter. To determine whether it might play a role in the normal regulation of Scr expression, CAT/lacZ fusion genes were created that contain an authentic arrangement of cis-regulatory elements (diagram, Fig. 3 Top). The tethering element was placed 5′ of the leftward Scr-CAT reporter gene, whereas the T1 enhancer was placed 3′ of the ftz-lacZ reporter gene. The distal cluster (distal, Fig. 3) was inserted just downstream of the T1 enhancer. Thus, as seen for the normal organization of Scr regulatory elements, the tethering element and distal cluster bracket the remote T1 enhancer (see Fig. 1 A).
As expected, only the Scr-CAT reporter gene is activated by the T1 enhancer (Fig. 3A). CAT staining is restricted to a groove of cells located between the labial head and first thoracic segments. The ftz-lacZ gene is silent and does not exhibit expression (data not shown). In the absence of the distal cluster, variable background staining is produced by the Scr-CAT reporter gene (Fig. 3 A and C). However, extraneous staining is lost in each of the individual lines that contain the distal cluster in the 3′ position (Fig. 3 B and D; compare with Fig. 3 A and C). The addition of the distal cluster does not augment T1–Scr interactions. The same levels of CAT staining are observed in the labial-T1 region with or without the distal cluster (e.g., Fig. 3 C and D). The addition of the distal cluster serves to eliminate background staining and to produce a more precise pattern of expression in the labial-T1 region. One interpretation of these results is that proteins bind to the TTCGAA motif in the proximal tethering element and distal cluster and mediate a long-range chromatin loop, which stabilizes T1-Scr.
The TTCGAA motif is the most obvious component of the proximal tethering element and distal cluster. To determine whether it is sufficient to recruit the T1 enhancer, different multiples of the motif were placed immediately 5′ of the ftz-lacZ reporter gene (diagram above Fig. 3 E–H). In the complete absence of the motif, there is no activation of ftz-lacZ expression by the T1 enhancer (Fig. 3E). There is a similar absence of expression when two copies of the TTCGAA motif were placed 5′ of the ftz promoter (Fig. 3F). However, four tandem copies of the motif led to weak but consistent activation of the ftz-lacZ reporter gene in the labial-T1 region of transgenic embryos (Fig. 3G). Similar staining was obtained with a fusion gene that contains six copies of the TTCGAA motif (data not shown). Stronger ftz-lacZ expression was obtained when either the proximal tethering element (Fig. 3H) or distal cluster (data not shown) was placed 5′ of the ftz promoter. These observations suggest that the TTCGAA motif is an important component of the regulatory activities of the tethering element and distal cluster, but additional sequence elements and DNA-binding proteins are required for long-range T1–Scr interactions (see Discussion).
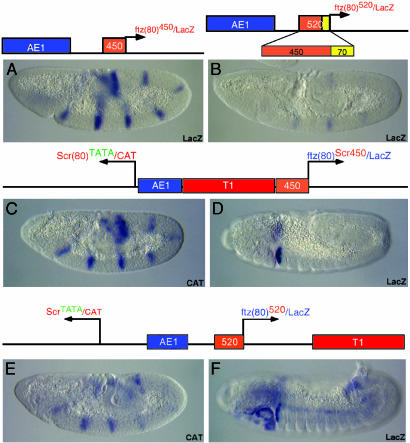
A Complete Regulatory Swap in the Scr-ftz Interval. We have previously shown that creating a TATA element in the minimal Scr promoter and inserting the tethering element 5′ of the minimal ftz promoter were sufficient to swap the regulatory activities of the T1 and AE1 enhancers (6). When placed between divergently transcribed Scr-CAT and ftz-lacZ reporter genes, T1 now activates ftz-lacZ expression in the labial head segment (Fig. 4D), and AE1 activates Scr-CAT in seven stripes along the germ band (Fig. 4C). A limitation of this earlier experiment, however, is that the arrangement of cis-regulatory DNAs does not reflect the in vivo organization seen in the ANT-C. Moreover, the AE1 enhancer retains the capacity to activate ftz-lacZ expression when the minimal 450-bp tethering element is placed 5′ of the ftz promoter (not shown). This residual AE1-ftz activity was diminished by placing AE1 5′ of the 3.8-kb T1 enhancer (see diagram above Fig. 4 C and D). The intervening T1 enhancer somehow attenuates AE1, either through weak enhancer blocking activity or by simply increasing the distance separating AE1 from the ftz promoter.
Fig. 4.
A negative element upstream of the Scr core promoter interferes with AE1–Scr interactions. (A and B) P element expression vectors contain the AE1 enhancer positioned 5′ of the ftz-lacZ fusion gene. The minimal 450-bp proximal tethering element does not block activation of lacZ expression by the AE1 enhancer (A). However, a larger 520-bp tethering element, containing 70 bp of flanking sequence in the Scr promoter-proximal region, attenuates AE1–ftz interactions (B). (C and D) A regulatory swap in the activities of the AE1 and T1 enhancers was obtained by inserting a TATA element in the minimal Scr promoter and placing the 450-bp proximal tethering element 5′ of the ftz promoter (6). AE1 now activates the Scr-CAT reporter gene in seven stripes (C), whereas T1 activates ftz-lacZ in the posterior head segment (D). The ftz-lacZ fusion gene is weakly activated by AE1, but these interactions are attenuated by the 3.8-kb T1 (not shown). A more complete regulatory swap was obtained in the following experiment. (E and F) The AE1 enhancer was positioned between the divergently transcribed CAT and lacZ reporter genes, whereas the T1 enhancer was downstream of lacZ. The “full-length” 520-bp proximal tethering element was positioned 5′ of the ftz-lacZ fusion gene. It contains the negative elements that impede AE1–Scr interactions. This tethering sequence completely blocks AE1–ftz interactions, but AE1 is able to activate the TATA-containing Scr promoter and direct seven stripes of CAT expression (E). Conversely, the 3′ T1 enhancer strongly activates the ftz-lacZ fusion gene in posterior head segments and the anterior compartment of the first thoracic segment.
The further characterization of promoter-proximal sequences located just 5′ of the core Scr promoter identified a negative element that diminishes AE1–Scr interactions. This negative element is located within a 70-bp sequence adjacent to the previously identified 450-bp tethering element (diagram above Fig. 4B). The larger 520-bp tethering sequence diminishes activation of the ftz-lacZ fusion gene by the 5′ AE1 enhancer (Fig. 4B). In contrast, AE1 can activate the same fusion gene when the minimal 450-bp tethering element is used in place of the 520-bp fragment (Fig. 4A; compare with Fig. 4B). The distal tether also contains a negative activity that interferes with AE1–ftz interactions (Fig. 3L; compare with Fig. 3K), reaffirming the similarities between the two tethering elements.
The use of the 520-bp tethering sequence permitted a clean reversal in the regulatory activities of the AE1 and T1 enhancers (Fig. 4 E and F). In this experiment, the AE1 enhancer was placed between the divergently transcribed Scr-CAT and ftz-lacZ reporter genes. The Scr promoter contains a synthetic TATA element, whereas the minimal ftz promoter was attached to the full-length 520-bp tethering element. As observed in the ANT-C, the T1 enhancer was placed in a 3′ position, in this case downstream of the ftz-lacZ reporter gene. The Scr-CAT fusion gene exhibits seven stripes of expression, indicating activation by AE1 (Fig. 4E). In contrast, the authentic Scr promoter (and associated tethering element) is not activated by AE1 but is expressed in the labial head segment due to activation by T1 (see Fig. 1C). The ftz-lacZ fusion gene exhibits only residual activation by AE1, due to the use of the full-length 520-bp tethering sequence containing the negative elements that repel AE1. Instead, ftz-lacZ exhibits robust expression in the posterior head segments and first thoracic segment due to activation by the 3′ T1 enhancer (Fig. 4F). The normal ftz-lacZ fusion gene is not regulated by T1 but instead exhibits seven stripes of expression due to activation by AE1 (see Fig. 1D).
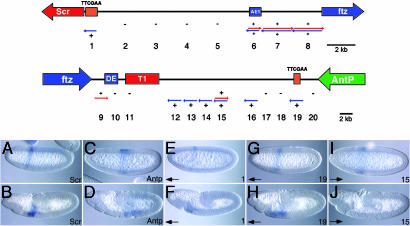
Transcription of Cis-Regulatory DNAs in the Antp-Scr Interval. Previous studies have shown that cis-regulatory DNAs in the Bithorax complex are transcribed (17–19). To date, there is no evidence for the transcription of cis-regulatory DNAs in the Antennapedia complex. To investigate this issue, a series of short DNA fragments from the Antp-Scr interval were used to prepare digoxigenin-labeled RNA probes for in situ hybridization (Fig. 5). There is extensive transcription in this region, beginning with probe 19, which maps just 3 kb downstream of the Antp transcription unit. Antp is expressed in a broad band in central regions of early embryos (20). Staining encompasses the presumptive middle thorax and is centered on parasegment 4, which is composed of the posterior compartment of T1 and the anterior compartment of T2 (Fig. 5 C and D). Despite its proximity to Antp, probe 19 is transcribed in a domain that is distinct from the Antp pattern. Staining is detected in parasegments 2 and 3 (Fig. 5 G and H). The anterior component of this expression pattern overlaps the normal Scr profile in parasegment 2 (Fig. 5 A and B). Thus, the newly identified distal cluster, which is contained within probe 19, appears to be transcribed in the Scr domain and in parasegment 3, which lacks both Scr and Antp expression in early embryos. This probe 19 pattern persists during germband elongation (Fig. 5H).
Fig. 5.
Transcription of intergenic DNA in the Scr-Antp interval. A series of 2-kb DNA fragments covering the Antp-Scr interval were used to prepare 20 digoxigenin-labeled RNA probes for in situ hybridization (diagram). Colored arrows indicate the orientation of the transcripts detected by each probe. Minus signs denote regions where no transcription is detected. Extensive transcription is seen throughout the ftz-Antp intergenic spacer (probes 10–20). Staining is detected in parasegments 2 and 3 (G–J). The anterior component of this expression pattern overlaps the normal Scr profile in parasegment 2 (A and B). Staining does not overlap the endogenous Antp pattern, which encompasses the presumptive middle thorax and is centered on parasegment 4 (C and D). Staining persists during germband elongation for probe 19 (H), which encompasses the distal TTCGAA cluster, but expression does not persist for the other detectable transcripts in the ftz-Antp spacer (e.g., J). One region, 15, exhibits transcription from both DNA strands. Double-stranded transcription is also detected throughout the AE1-ftz interval (6–8). The staining pattern observed is similar to the endogenous ftz pattern and consists of seven pair-rule stripes (data not shown). This same pattern of transcription is seen for probe 9. There is little or no transcription in the interval between AE1 and the proximal tethering element (1–5), although the tethering element (probe 1) is itself transcribed (E and F). Like transcription from the distal cluster (probe 19), staining persists during germband elongation (F).
Similar staining patterns, expression in parasegments 2 and 3, are observed for the other probes in the 20-kb interval separating the distal TTCGAA cluster and the T1 enhancer. However, in contrast to probe 19, the other sequences in this interval exhibit transient expression in early embryos; staining is lost by the onset of elongation (e.g., Fig. 5 I and J). One region, probe 15, exhibits transcription from both DNA strands. There is also doublestranded transcription in the AE1-ftz interval. The staining pattern that is observed is similar to the endogenous ftz pattern and consists of seven pair-rule stripes (data not shown). There is little or no transcription in the interval between AE1 and the proximal tethering element, although the tethering element (probe 1) is itself transcribed (Fig. 5 E and F). Like transcription from the distal cluster (probe 19), staining persists during germband elongation (Fig. 5F). However, unlike probe 19, the proximal tethering element exhibits a narrower stripe of transcription within parasegment 2, similar or identical to the normal Scr expression pattern (Fig. 5 A and B). The transcription of the proximal and distal tethering elements might render them in an open conformation during the time when the distal T1 enhancer activates Scr expression (see Discussion).
Discussion
The preceding analysis identified three new cis-regulatory DNAs in the Scr-Antp interval of the ANT-C: a 3′ ftz enhancer, a distal cluster of TTCGAA elements, and negative elements that inhibit AE1–Scr interactions adjacent to the originally defined Scr tethering sequence. The tethering sequence and newly identified distal cluster are themselves transcribed and exhibit similar patterns of transcription even though they map quite far from one another (40 kb). This transcription might promote the formation of a long-range chromatin loop domain that stabilizes T1–Scr interactions.
The ftz gene was first cloned 20 years ago (21), and the AE1 enhancer and zebra element were identified just a few years later (13). Here we have identified a third enhancer by using a computer program to scan the Drosophila genome for clusters of binding sites recognized by segmentation regulatory factors, particularly the gap repressor Kr (11). The newly identified ftz enhancer has the properties of a primary pair-rule stripe enhancer in that it directs the expression of just two stripes. The 3′ enhancer, although conserved in Drosophila species containing an inversion at the ftz locus (Fig. 1), is dispensable for ftz gene function. Previous studies have shown that a ftz minigene lacking 3′ regulatory sequences is nonetheless able to complement ftz-mutant embryos (22).
The ftz gene has acquired distinct activities in different insects (23, 24). In short germband insects such as Tribolium, ftz appears to function in both segmentation and homeosis (25, 26). The Tribolium Ftz protein contains two peptide motifs, LRALL and YPWM, that mediate interactions with FtzF1 (segmentation) and Exd (homeosis), respectively. When misexpressed in fly embryos, the Tribolium Ftz protein produces both segmentation and homeotic defects. In contrast, the Drosophila Ftz protein contains only the LRALL motif and thereby functions solely in segmentation. It does not produce homeotic defects when misexpressed in transgenic embryos. Ancestral forms of Ftz functioned in both segmentation and homeosis in primitive insects, but the homeotic function has been lost in more modern insects, such as the Diptera. Perhaps the newly identified ftz enhancer is a remnant of the homeotic functions seen in other insects.
We previously showed that the 450-bp tethering sequence in the promoter-proximal region of the Scr gene is essential for activation by the remote T1 enhancer (6). The further analysis of this tethering sequence identified multiple copies of a simple palindromic sequence motif, TTCGAA. There are eight copies of this motif in the 450-bp tethering sequence, and the fly enhancer program was used to identify additional high-density clusters. One such cluster is also located in the Scr-Antp interval, just downstream of the Antp transcription unit (see Fig. 1 A). This newly identified distal cluster can function as a tethering sequence and augment T1–Scr interactions. It also eliminates position effects when placed downstream of the T1 enhancer (see Fig. 3). Multiple copies of the TTCGAA motif are sufficient to mediate weak T1–ftz interactions in transgenic embryos. This activation is not as robust as that observed for the native tethering sequence. Thus, TTCGAA may be an essential component of the tethering sequence, but additional regulatory elements are likely to play an important role in mediating T1–Scr interactions.
We propose that a common set of proteins bind to both the tethering sequence and distal cluster and form homotypic complexes, which stabilize long-range T1–Scr interactions. It is possible that a chromatin loop forms between the tethering sequence and distal cluster. Alternatively, according to a scanning model for enhancer–promoter interactions, interactions between the tethering sequence and distal cluster might lock the T1 enhancer onto the Scr promoter, after the two encounter one another. In addition to the proposed homotypic interactions between the distal cluster and tethering element, it is conceivable that heterotypic interactions are important for the recruitment of the T1 enhancer to the Scr promoter. The tethering element is sufficient to recruit T1 to either the Scr or ftz promoters in the absence of the distal cluster. These interactions might depend on different classes of proteins. Given that the two tethering elements interfere with activation by AE1, these elements might also serve to isolate the ftz segmentation enhancers away from neighboring homeotic genes. Improper activation of homeotic promoters by segmentation enhancers would be lethal for the developing embryo (27, 28).
Regulatory proteins that bind to promoter-proximal sequences, such as the Scr tethering element, might not interact with the basal transcription complex and function as classical activators. Instead, they might regulate gene expression by recruiting distal enhancers. A number of mammalian promoterproximal regulatory proteins might work through this type of mechanism (29–31). For example, Sp1 has been shown to mediate the formation of DNA loops when bound to both proximal and distal recognition sequences (32).
Previous studies have documented the occurrence of extensive intergenic transcription in the Drosophila Bithorax complex. Many of these transcripts are associated with a number of defined cis-regulatory DNAs, including the Fab-8 insulator and IAB5 enhancer in the extended 3′ regulatory region of the Abd-B gene (18, 19). It has been suggested that this transcription serves to maintain these critical regulatory elements in an open chromatin conformation during Drosophila development (18, 33). For example, the Rox RNAs (dosage compensation) serve as docking sites for histone acetyltransferase complexes that are thought to open the chromatin on the male X chromosome and thereby augment gene expression (34).
The present study provides evidence for intergenic transcription in the Scr-Antp interval of the ANT-C. Interestingly, some of this transcription occurs in the tissues of parasegment (PS) 3, between the major sites of Scr and Antp expression in PS2 and PS4, respectively. Both homeotic genes are activated in PS3 in older embryos, and it is conceivable that intergenic transcription is required for this expression by maintaining the genes in an open conformation. The transcription of the tethering sequence and distal cluster might help ensure the maintenance of T1–Scr interactions during development.
Acknowledgments
We thank Naoe Harafuji for helpful discussion and Freyja Knapp for technical assistance. This work was funded by National Institutes of Health Grant GM 34431.
Abbreviations: ANT-C, Antennapedia complex; Kr, Krüppel.
References
- 1.Blackwood, E. M. & Kadonaga, J. T. (1998) Science 281, 61–63. [DOI] [PubMed] [Google Scholar]
- 2.Merli, C., Bergstrom, D. E., Cygan, J. A. & Blackman, R. K. (1996) Genes Dev. 10, 1260–1270. [DOI] [PubMed] [Google Scholar]
- 3.Butler, J. E. & Kadonaga, J. T. (2002) Genes Dev. 16, 2583–2592. [DOI] [PubMed] [Google Scholar]
- 4.Li, X. & Noll, M. (1994) EMBO J. 13, 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gindhart, J. G., Jr., King, A. N. & Kaufman, T. C. (1995) Genetics 139, 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calhoun, V. C., Stathopoulos, A. & Levine, M. (2002) Proc. Natl. Acad. Sci. USA 99, 9243–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Small, S., Blair, A. & Levine, M. (1992) EMBO J. 11, 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tautz, D. & Pfeifle, C. (1989) Chromosoma 98, 81–85. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, J., Kosman, D., Ip, Y. T. & Levine, M. (1991) Genes Dev. 5, 1881–1891. [DOI] [PubMed] [Google Scholar]
- 10.Ohtsuki, S., Levine, M. & Cai, H. N. (1998) Genes Dev. 12, 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman, B. P., Nibu, Y., Pfeiffer, B. D., Tomancak, P., Celniker, S. E., Levine, M., Rubin, G. M. & Eisen, M. B. (2002) Proc. Natl. Acad. Sci. USA 99, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dearolf, C. R., Topol, J. & Parker, C. S. (1989) Genes Dev. 3, 384–398. [DOI] [PubMed] [Google Scholar]
- 13.Hiromi, Y., Kuroiwa, A. & Gehring, W. J. (1985) Cell 43, 603–613. [DOI] [PubMed] [Google Scholar]
- 14.Pick, L., Schier, A., Affolter, M., Schmidt-Glenewinkel, T. & Gehring, W. J. (1990) Genes Dev. 4, 1224–1239. [DOI] [PubMed] [Google Scholar]
- 15.Maier, D., Preiss, A. & Powell, J. R. (1990) EMBO J. 9, 3957–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markstein, M., Markstein, P., Markstein, V. & Levine, M. S. (2002) Proc. Natl. Acad. Sci. USA 99, 763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez-Herrero, E. & Akam, M. (1989) Development (Cambridge, U.K.) 107, 321–329. [DOI] [PubMed] [Google Scholar]
- 18.Zhou, J., Ashe, H., Burks, C. & Levine, M. (1999) Development (Cambridge, U.K.) 126, 3057–3065. [DOI] [PubMed] [Google Scholar]
- 19.Bae, E., Calhoun, V. C., Levine, M., Lewis, E. B. & Drewell, R. A. (2002) Proc. Natl. Acad. Sci. USA 99, 16847–16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine, M., Hafen, E., Garber, R. L. & Gehring, W. J. (1983) EMBO J. 2, 2037–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroiwa, A., Hafen, E. & Gehring, W. J. (1984) Cell 37, 825–831. [DOI] [PubMed] [Google Scholar]
- 22.Hiromi, Y. & Gehring, W. J. (1987) Cell 50, 963–974. [DOI] [PubMed] [Google Scholar]
- 23.Lohr, U., Yussa, M. & Pick, L. (2001) Curr. Biol. 11, 1403–1412. [DOI] [PubMed] [Google Scholar]
- 24.Damen, W. G. (2002) BioEssays 24, 992–995. [DOI] [PubMed] [Google Scholar]
- 25.Brown, S. J., Hilgenfeld, R. B. & Denell, R. E. (1994) Proc. Natl. Acad. Sci. USA 91, 12922–12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Telford, M. J. (2000) Curr. Biol. 10, 349–352. [DOI] [PubMed] [Google Scholar]
- 27.Gibson, G., Schier, A., LeMotte, P. & Gehring, W. J. (1990) Cell 62, 1087–1103. [DOI] [PubMed] [Google Scholar]
- 28.Struhl, G. (1985) Nature 318, 677–680. [DOI] [PubMed] [Google Scholar]
- 29.Mastrangelo, I. A., Courey, A. J., Wall, J. S., Jackson, S. P. & Hough, P. V. (1991) Proc. Natl. Acad. Sci. USA 88, 5670–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong, Q. & Dean, A. (1993) Mol. Cell. Biol. 13, 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gourdon, G., Morle, F., Roche, J., Tourneur, N., Joulain, V. & Godet, J. (1992) Acta Haematol. 87, 136–144. [DOI] [PubMed] [Google Scholar]
- 32.Su, W., Jackson, S., Tjian, R. & Echols, H. (1991) Genes Dev. 5, 820–826. [DOI] [PubMed] [Google Scholar]
- 33.Hogga, I. & Karch, F. (2002) Development (Cambridge, U.K.) 129, 4915–4922. [DOI] [PubMed] [Google Scholar]
- 34.Franke, A. & Baker, B. S. (2000) Curr. Opin. Cell Biol. 12, 351–354. [DOI] [PubMed] [Google Scholar]