Abstract
A coordinated cellular response to oxidative stress occurs in part through transcriptional regulation via a cis-acting sequence known as the antioxidant response element (ARE). NF-E2-related factor 2 (Nrf2), a member of the Cap'n'Collar family of basic region-leucine zipper (bZIP) transcription factors, has been implicated as an essential component of an ARE-binding transcriptional complex, but the signaling pathway leading to its activation has remained unclear. Using a reporter gene assay, we found that ARE-directed transcription was activated by phorbol 12-myristate 13-acetate (PMA), but completely suppressed by staurosporine and Ro-32–0432, selective inhibitors of protein kinase C (PKC). Immunocytochemistry and subcellular fractionation revealed that PMA, like tert-butylhydroquinone (tBHQ), promoted the nuclear localization of Nrf2, a process that was blocked by staurosporine or Ro-32–0432. We showed that Nrf2, a previously unidentified kinase target, was phosphorylated in HepG2 cells. PMA transiently activated Nrf2 phosphorylation, whereas the addition of tBHQ or β-naphthoflavone (βNF) led to a persistent stimulation, which was abolished by staurosporine, but not by U0126 and SB203580, respective inhibitors of MEK and p38 kinases. Purified Nrf2 was phosphorylated in vitro by the catalytic subunit of PKC, or by PKC immunoprecipitated from cell lysates. Significantly, PKC precipitated from tBHQ- or βNF-treated cells showed enhanced activity against Nrf2. These findings indicate an important role of the PKC pathway in the ARE-mediated gene expression, and suggest that PKC-directed phosphorylation of Nrf2 may be a critical event for the nuclear translocation of this transcription factor in response to oxidative stress.
Elevated intracellular levels of reactive oxygen species (ROS), or oxidative stress, can result from cellular exposure to a variety of chemical and physical sources including ionizing radiation, heavy metals, inflammatory cytokines, and xenobiotic agents (1, 2). ROS are highly cytotoxic, causing damage to DNA, lipids, and proteins, and consequently lead to numerous pathological states such as cancer, neurodegenerative disease, atherosclerosis, and aging (3–6). To protect against such ROS-induced damages, cells have developed a defense mechanism involving the coordinated induction of a number of genes; central among them are those encoding phase II detoxifying enzymes, which convert reactive electrophiles to less toxic and more readily excretable products (7, 8). Extensive biochemical analyses on the promoter region of the rat glutathione S-transferase (GST) A2 and mouse GST A1 genes (9, 10) and of the rat and human NAD(P)H:quinone oxidoreductase (QR or NQO1) genes (11, 12) have established that their constitutive and xenobiotic-inducible expression is primarily mediated by a cis-acting regulatory sequence known as the antioxidant response element (ARE) or electrophile response element (10, 13).
The consensus ARE core sequence, 5′-TGACNNNGC-3′, shows remarkable similarity to the binding sequence of the erythroid transcription factor NF-E2 (14). Accumulated evidence has now implicated NF-E2-related factor 2 (Nrf2), a member of the Cap'n'Collar subfamily of basic region-leucine zipper (bZIP) transcription factors (15–17), as part of a heterodimer with small Maf proteins forming an ARE-binding transcriptional complex (18–21). Nrf2 appears critical for ARE-mediated gene expression, as nrf2-null mice showed either severe impairment in the induction of several phase II metabolizing enzymes in their liver and intestine (18), or a much reduced basal expression of such enzymes (22). In Nrf2-deficient macrophages, a number of anti-oxidative stress genes could no longer be induced by electrophilic or ROS-generating agents (23). A recent study suggested that Nrf2 activity is normally repressed through its localization in the cytoplasm by binding to the cytoskeleton-associated protein Keap1 (24). Immunocytochemistry experiments involving cotransfection of Nrf2 and Keap1 showed that electrophilic agents released Nrf2 from Keap1, allowing the transcription factor to traverse to the nucleus to activate ARE-dependent gene expression.
Whereas the Nrf2-Keap1 interaction may constitute a cytoplasmic sensor system for oxidative stress as has been proposed (24), little is understood of precisely how that signal is transduced. One likely mechanism involves regulation by phosphorylation (25). A role for mitogen-activated protein kinase (MAPK) pathways has been suggested by results from transfection and specific kinase inhibitor studies, which revealed that extracellular signal-regulated kinase 2 positively (26), whereas p38 negatively (27), regulated ARE-mediated transcription. In earlier studies, we and others had also shown that phorbol esters, potent tumor promoters that activate protein kinase C (PKC) (28), induced ARE-driven expression of the rat GST A1 (29), rat QR (30), and mouse GST A2 (31) genes. To date, however, there has been no report of any specific cellular components serving as kinase targets in the signaling cascade leading to ARE activation. We therefore undertook the present study to determine whether Nrf2 itself is phosphorylated, and to examine whether PKC plays a role in Nrf2 phosphorylation and in its nuclear translocation in response to oxidative stress.
Materials and Methods
Cell Culture, Plasmids, Transfection, and Reporter Assays.
HepG2 and H4IIEC3 cells were obtained from the American Type Culture Collection, and were maintained as previously described (19). All media and supplements were from Life Technologies (Grand Island, NY). All reagents were from Sigma unless noted otherwise. H4IIEC3 cells stably transfected with an expression plasmid containing rat QR ARE (5′-TCTAGAGTCACAGTGACTTGGCAAAATCTGA-3′) linked to chloramphenicol acetyltransferase (CAT) reporter gene (30) were obtained by following the manufacturer's instructions for Lipofectamine Plus Reagent (Life Technologies). HepG2 cells stably transfected with the rat QR ARE-CAT construct were a kind gift of Leonard Favreau (Schering–Plough Research Institute). After incubation in tert-butylhydroquinone (tBHQ) or phorbol 12-myristate 13-acetate (PMA) for 18 h, in the presence or absence of Ro-32–0432 (Calbiochem) or staurosporine (Boehringer Mannheim), cells were harvested in M-PER Mammalian Protein Extraction Reagent (Pierce), and cell lysates were assayed for CAT activity as described (19). A high-level expression plasmid of rat Nrf2 gene linked at its N terminus to a 6xHis tag was obtained by cloning the rat Nrf2 cDNA (GenBank accession no. AF037350) into the pQE-30 vector (Qiagen, Chatsworth, CA). After expression in Escherichia coli M15 cells, Nrf2 protein was purified under native conditions to near homogeneity by Ni-NTA metal chelate affinity chromatography essentially according to the manufacturer's protocols (Qiagen).
Immunocytochemistry and Subcellular Fractionation.
HepG2 or H4IIEC3 cells were grown on coverslips and treated with tBHQ or PMA for 4 h, or where indicated, pretreated with staurosporine for 1 h before exposure to tBHQ. Immunocytochemistry using an affinity-purified rabbit polyclonal anti-Nrf2 antibody (sc-722; Santa Cruz Biotechnology), followed by a FITC-conjugated anti-rabbit IgG antibody (62–6111; Zymed), was performed as described (24). Propidium iodide (PI) counterstaining verified the location and integrity of the nuclei. Fluorescence was monitored with a confocal laser scanning microscope (DM IRBE; Leica, Deerfield, IL). To confirm the subcellular distributions of Nrf2, HepG2 cells exposed to tBHQ or PMA for 1 h, in the presence or absence of staurosporine or Ro-32–0432, were fractionated into cytosolic and nuclear fractions as described (32). Fractions were resolved by SDS/12% PAGE, transferred to poly(vinylidene difluoride) membrane, and the relative amounts of Nrf2 protein present were determined by immunoblotting with an anti-Nrf2 antibody followed by enhanced chemiluminescence detection (Amersham Pharmacia).
Metabolic Labeling and Immunoprecipitation.
HepG2 cells were cultured for 1 h in phosphate-free MEM supplemented with dialyzed FBS, and then metabolically labeled for 3.5 h with [32P]orthophosphate (1 mCi per 60-mm dish; 1 Ci = 37 GBq). After washes in ice-cold phosphate-free MEM, cells were lysed in lysis/IP buffer [50 mM Tris (pH 8.0)/150 mM NaCl/1% Nonidet P-40/0.5% DOC/0.1% SDS/50 mM NaF/1 mM Na3VO4/20 mM β-glycerophosphate/1 μM okadaic acid] on ice for 30 min, then centrifuged at 14,000 rpm for 15 min in a microcentrifuge. Before lysis, cells were exposed to tBHQ, β-naphthoflavone (βNF), or PMA for various times as indicated. In experiments with kinase inhibitors, cells were pretreated with staurosporine, U0126 (Calbiochem), or SB203580 (Calbiochem) for 1 h, followed by the addition of tBHQ for 30 min before lysis. 32P-labeled Nrf2 was immunoprecipitated from cell lysates by incubating with an anti-Nrf2 antibody on ice overnight, followed by the addition of Protein A-Trisacryl beads (Pierce). The mixture was rotated at 4°C for 1 h, and washed extensively in lysis/IP buffer with 0.4 M NaCl. Precipitates were resolved by SDS/PAGE and analyzed by autoradiography.
Kinase Assays.
E. coli-expressed rat Nrf2 protein was purified as described above and used as substrate in in vitro kinase assays. In assays using catalytic subunits of rat brain PKC (Calbiochem), 50-μl reactions were carried out for various times at 30°C in a buffer containing 25 mM Hepes (pH 7.5), 10 mM MgCl2, 200 mM ATP, and 2 mCi of [γ-33P]ATP. Where indicated, assays were performed for 30 min at 30°C after preincubation of PKC with staurosporine or Ro-32–0432 for 10 min at room temperature. Phosphorylation of Nrf2 was also assayed by using endogenous PKC immunoprecipitated from HepG2 cell lysates. A monoclonal anti-PKC antibody conjugated to agarose (sc-80AC; Santa Cruz Biotechnology) was used to precipitate PKC from lysates of HepG2 cells treated with tBHQ, βNF, or PMA for various times, according to the immunoprecipitation protocol described above. The anti-PKC immunocomplexes were then incubated with purified Nrf2 and [γ-33P]ATP for 30 min at 30°C in a reaction buffer from the GIBCO/BRL Protein Kinase C Assay System (Life Technologies). Kinase reactions were stopped by the addition of SDS/PAGE sample buffer, the products were resolved by SDS/PAGE, and the level of [γ-33P]ATP incorporation was determined by a phosphor imager (FLA-2000; Fuji).
Results
Inhibitors of PKC Suppress Transcriptional Activation of the ARE.
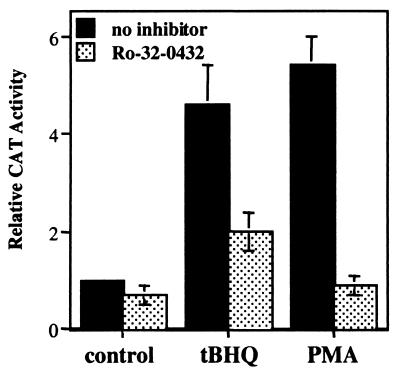
In our prior studies using transient transfection of ARE-linked CAT reporter gene contructs into human hepatoma HepG2 cells, we observed that the phorbol ester PMA, like the phenolic antioxidant tBHQ, caused a 3- to 4-fold increase in CAT activity directed by the rat QR ARE (30). A similar PMA induction of CAT activity was also seen when a CAT construct linked to the ARE from the rat GST A2 gene was stably transfected into HepG2 cells (29). We show here that PMA (100 nM) treatment of HepG2 cells stably transfected with a rat QR ARE-CAT expression plasmid resulted in an ≈5-fold induction of CAT activity, comparable to that caused by tBHQ (100 μM) (Fig. 1). Significant induction was seen at PMA concentrations as low as 10 nM (data not shown). However, in the presence of Ro-32–0432 (2 μm), a bisindolylmaleimide derivative of staurosporine that has been shown to be highly a selective inhibitor of PKC, Ro-32–0432 (0.2 μM), PMA induction was abolished, while tBHQ activation was also reduced by more than 50% (Fig. 1). Similar results were also obtained in rat hepatoma H4IIEC3 cells stably transfected with the rat QR ARE-CAT construct, or when staurosporine (15 nM), another potent inhibitor of PKC, was used (data not shown). Thus, activation of PKC by PMA enhanced ARE-mediated gene expression, and the ARE activation in response to antioxidants appeared to require, at least in part, active PKC.
Figure 1.
A PKC inhibitor suppresses transcriptional activation of the ARE. HepG2 cells were stably transfected with the rat QR ARE-CAT reporter construct. After incubation in dimethyl sulfoxide (solvent control), tBHQ (100 mM), or PMA (100 nM) for 18 h, in the presence or absence of Ro-32–0432 (0.2 mM), cells were harvested, and cell lysates were ssayed for CAT activity. Quantitation of the results was performed by a phosphor imager; the level of CAT activity from solvent-treated cells was set at 1. The data shown are means of three independent experiments.
Involvement of PKC in Nrf2 Nuclear Localization.
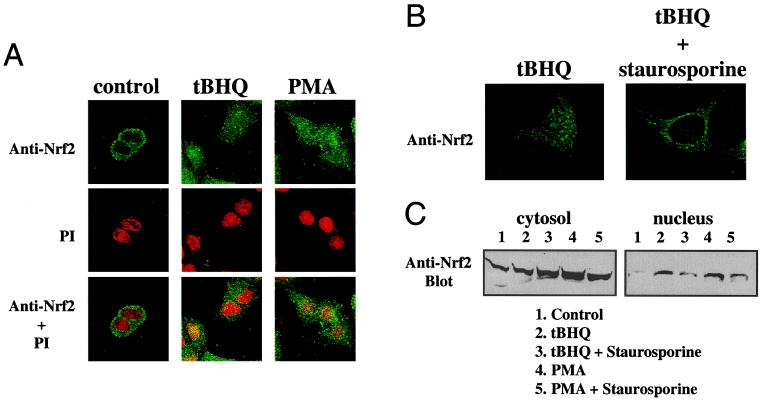
Immunocytochemical studies of 293T cells overexpressed with Nrf2 and Keap1 plasmids led Itoh et al. (24) to propose that Nrf2 is normally retained in the cytoplasm through its association with the cytoskeleton-binding Keap1, and is liberated to translocate into the nucleus upon induction by electrophilic agents. We sought to determine whether antioxidants and phorbol esters that activate the ARE can cause a similar subcellular redistribution of endogenous Nrf2 in hepatoma cells. HepG2 cells were treated with either 100 μM tBHQ or 100 nM PMA for 4 h before they were fixed and permeabilized. Nrf2 localization was visualized by incubation with an anti-Nrf2 antibody, followed by a FITC-conjugated secondary antibody. PI staining of the same cells verified the location and integrity of the nuclei. As shown in Fig. 2A, Nrf2 was found predominantly in the cytoplasm in solvent-treated control cells, but appeared uniformly distributed in both the nucleus and the cytoplasm after PMA or tBHQ treatment. These results involving endogenous Nrf2 confirmed those from previous overexpression studies (24), and demonstrated that activation of PKC by PMA induced nuclear localization of Nrf2. Immunocytochemistry of rat H4IIEC3 cells revealed similar PMA- and tBHQ-induced Nrf2 nuclear localization (data not shown). We also performed subcellular fractionation followed by Western blot analysis. As expected, the nuclear fractions of HepG2 cells exposed to tBHQ or PMA for 1 h showed elevated amounts of Nrf2 compared with the solvent-treated control (Fig. 2C).
Figure 2.
Involvement of PKC in the nuclear localization of Nrf2. (A) HepG2 cells were treated with DMSO (control), tBHQ (100 μM), or PMA (100 nM) for 4 h. Nrf2 localization was detected by immunocytochemistry using an anti-Nrf2 antibody followed by FITC-conjugated secondary antibody. To confirm the location of the nuclei, PI counterstaining of the same fields was shown both separately and overlaid with Nrf2 immunofluorescence. (B) HepG2 cells were preincubated with solvent or staurosporine (15 nM) for 1 h before their exposure to tBHQ (100 μM) for 4 h. Nrf2 immunofluorescence was monitored as in A. (C) Subcellular fractionation of HepG2 cells was carried out after treatment of tBHQ (100 μM) or PMA (100 nM) for 1 h, in the presence or absence of staurosporine (15 nM). Eighty micrograms of proteins of the cytosolic fraction and 20 μg of the nuclear fraction were resolved by SDS/PAGE. The relative amounts of Nrf2 protein present in each were determined by Western blotting using an anti-Nrf2 antibody followed by enhanced chemiluminescence detection.
To probe further the involvement of PKC in antioxidant-induced nuclear translocation of Nrf2, we preincubated HepG2 cells with 15 nM staurosporine for 1 h before their exposure to tBHQ, and performed immunocytochemical analysis as before. Fig. 2B demonstrates that this potent PKC inhibitor clearly abrogated the tBHQ-induced nuclear localization of Nrf2. Western blot analysis of subcellular fractions confirmed that staurosporine prevented the tBHQ- and PMA-induced accumulation of Nrf2 in the nucleus (Fig. 2C). Pretreatment with Ro-32–0432 (0.2 μM) had similar inhibitory effects on Nrf2 nuclear localization as staurosporine (data not shown). Together, these findings suggest a critical role for active PKC in the nuclear translocation of Nrf2 in response to oxidative stress.
Phosphorylation of Nrf2 in Vivo.
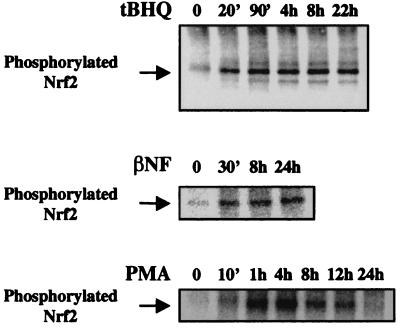
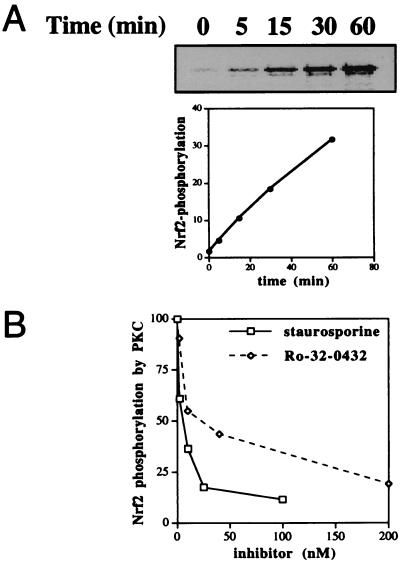
Previous studies implicating the involvement of various kinase pathways in ARE-mediated gene expression have failed to identify any cellular kinase targets (26, 27, 31). To ascertain whether Nrf2 can be phosphorylated in vivo, we performed metabolic labeling of HepG2 cells with [32P]orthophosphate, followed by immunoprecipitation with an anti-Nrf2 antibody of the cell lysates. In untreated cells, a weak phosphorylation of Nrf2 was observed, but upon exposure to antioxidants tBHQ (100 μM) or βNF (50 μM), an approximately 2-fold stimulation of Nrf2 phosphorylation appeared by 20 min and persisted for at least 24 h (Fig. 3). PMA (100 nM) also activated Nrf2 phosphorylation, but in contrast to tBHQ and βNF, its effect began to diminish after 4 h, and by 24 h Nrf2 phosphorylation had returned to near basal level (Fig. 3), consistent with the well-established time course of PMA-induced transient stimulation and subsequent down-regulation of PKC activity (33). Indeed, after incubation in the presence of PMA for 24 h, addition of tBHQ for 20 min no longer resulted in stimulation of Nrf2 phosphorylation (data not shown), suggesting a requirement for active PKC. Transfection of an Nrf2 expression vector into HepG2 cells resulted in recovery of a phosphoprotein containing higher amounts of incorporated label with identical mobility as endogenous Nrf2, whereas mock experiments using normal rabbit IgG did not recover any phosphoproteins under identical conditions (data not shown). We have thus identified Nrf2 as a substrate for intracellular phosphorylation, a process stimulated by antioxidants that activate the ARE.
Figure 3.
Phosphorylation of Nrf2 in HepG2 cells. HepG2 cells were metabolically labeled with [32P]orthophosphate, followed by immunoprecipitation of cell lysates with an anti-Nrf2 antibody. The immunoprecipitates were analyzed by SDS/PAGE and autoradiography. Cells were exposed to tBHQ (100 μM), βNF (50 mM), or PMA (100 nM) for the indicated times before lysis. Mock immunoprecipitations using normal rabbit IgG did not recover any phosphoproteins.
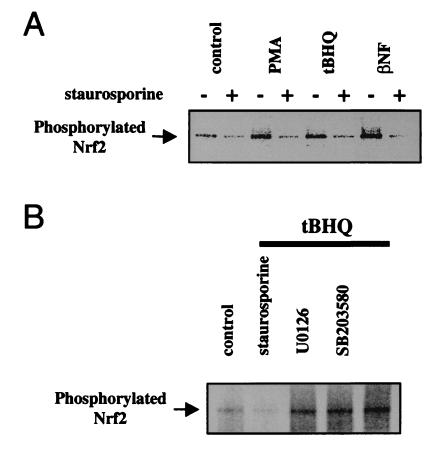
To confirm a role for PKC in Nrf2 phosphorylation, we performed 32P-metabolic labeling and immunoprecipitation of Nrf2 in the presence of 15 nM staurosporine. Results in Fig. 4A revealed that this PKC inhibitor effectively blocked the phosphorylation of Nrf2 in untreated, or tBHQ-, βNF-, and PMA-treated cells. Because the MAPK pathways have been shown to influence ARE-mediated transcription (26, 27), we also tested whether selective inhibitors of the extracellular signal-regulated kinase 2 and p38 pathways might affect Nrf2 phosphorylation. In contrast to staurosporine, however, U0126 (10 μM), which targets the MEK kinases (34), and SB203580 (10 μM), a selective inhibitor of the p38 kinase (35), were both ineffective in preventing the tBHQ-induced stimulation of Nrf2 phosphorylation (Fig. 4B). These results establish a specific requirement for active PKC in normal and antioxidant-induced phosphorylation of Nrf2.
Figure 4.
Involvement of PKC in Nrf2 phosphorylation in vivo. 32P-labeling of HepG2 cells was followed by immunoprecipitation of Nrf2 as in Fig. 3. (A) Cells were treated with tBHQ (100 μM), βNF (50 μM), or PMA (100 nM) for 30 min, after preincubation for 1 h in solvent or in 15 nM staurosporine. (B) Cells were exposed to tBHQ (100 μM) for 30 min, after preincubation for 1 h in solvent, staurosporine (15 nM), U0126 (10 mM), or SB203580 (10 μM).
In Vitro Phosphorylation of Nrf2 by PKC.
While we have shown that PKC appears to play an essential role in Nrf2 phosphorylation in vivo, it remained unclear whether PKC can directly phosphorylate Nrf2 itself, which contains seven putative phosphorylation sites for PKC. We therefore constructed a high-level expression plasmid of rat Nrf2 gene linked at its N terminus to a 6xHis tag, and purified to near homogeneity by metal chelate affinity chromatography E. coli-expressed Nrf2 proteins (data not shown). In vitro kinase assays were performed by using purified Nrf2 and commercially available catalytic subunits of rat brain PKC. Fig. 5A reveals Nrf2 to be an excellent substrate of PKC in vitro, comparable to other standard PKC substrates tested (data not shown). As expected, staurosporine (Ki ≈ 2.5 nM) and Ro-32–0432 (Ki ≈ 25 nM) both effectively inhibited PKC-mediated phosphorylation of Nrf2 (Fig. 5B).
Figure 5.
In vitro phosphorylation of Nrf2 by PKC. In vitro kinase assays were performed by using purified Nrf2 and catalytic subunits of rat brain PKC. (A) Nrf2 (2 mM) was incubated with 5 mM PKC and 2 mCi of [γ-33P]ATP for the indicated times at 30°C. The reaction products were analyzed by SDS/PAGE and autoradiography. [γ-33P]ATP incorporation was quantitated by a phosphor imager. (B) Identical kinase assays were carried out for 30 min at 30°C in the presence of various concentrations of staurosporine or Ro-32–0432.
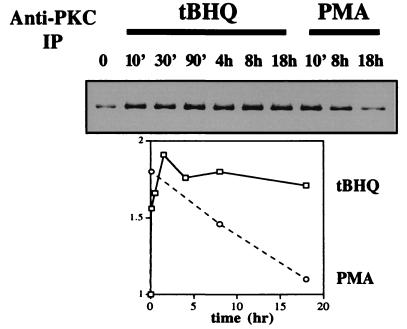
Because Nrf2 phosphorylation in HepG2 cells appeared to require PKC as shown above, we asked whether the activation in Nrf2 phosphorylation upon antioxidant treatment could be accounted for by an increase in cellular PKC activity. In Fig. 6, PKC was immunoprecipitated from lysates of HepG2 cells treated with 100 μM tBHQ or 100 nM PMA for the indicated times, and subsequently used in in vitro kinase assays with purified Nrf2 as substrate. Both tBHQ and PMA led to a near 2-fold enhancement of PKC activity against Nrf2—the former persistently, the latter transiently. Exposure to βNF (50 μM) resulted in a sustained up-regulation of PKC activity similar to tBHQ (data not shown). No phosphorylation of Nrf2 was seen when such an immunocomplex kinase assay was carried out with normal IgG (data not shown). These findings, taken together with those in Fig. 3, lead us to conclude that the antioxidant-induced phosphorylation of Nrf2 in HepG2 cells is most likely catalyzed by activated PKC that directly targets Nrf2.
Figure 6.
Antioxidant-induced increase in PKC activity against Nrf2. PKC was immunoprecipitated by an anti-PKC antibody from lysates of HepG2 cells treated with tBHQ (100 mM) or PMA (100 nM) for the indicated times. PKC activity was assayed by incubating the anti-PKC immunocomplexes with purified Nrf2 and [γ-33P]ATP for 30 min at 30°C. Quantitation was performed by a PhosphorImager. The results shown are typical of three separate experiments. No phosphorylation of Nrf2 was observed in a mock immunocomplex kinase assay using normal rabbit IgG.
Discussion
We have demonstrated the in vivo and in vitro phosphorylation of Nrf2, a transcription factor responsible for ARE-mediated gene expression. Our results indicate the critical involvement of PKC in the signaling pathway leading to ARE activation, and suggest a mechanism in which PKC-directed phosphorylation of Nrf2 triggers its nuclear translocation to potentiate the cellular response against oxidative stress.
Phosphorylation has long been established as a major posttranslational mechanism for the regulation of gene expression in eukaryotic cells. Almost every eukaryotic transcription factor analyzed has been shown to undergo phosphorylation. One of the principal paradigms that has emerged is a phosphorylation-regulated cytoplasmic–nuclear shuttling of transcription factors (36). A well-studied example is the Rel-related family member NF-κB, which translocates into the nucleus upon phosphorylation of its IκB regulatory subunit in response to various stimuli, including phorbol esters (37). Oxidative stress has been known to activate specific protein kinase signaling pathways that trigger nuclear import of other transcription factors. In response to cytokine stimulation, STATs (signal transducers and activators of transcription) are phosphorylated by activated Janus kinase family of protein tyrosine kinases and translocated into the nucleus (38). Antioxidants such as pyrrolidine dithiocarbamate induce protein kinase A-mediated serine phosphorylation and subsequent nuclear translocation of CCAAT/enhancer-binding protein β (39). We have presented evidence here that suggests a similar regulatory mechanism of PKC-mediated phosphorylation of Nrf2 signaling its nuclear import in response to antioxidants. Our findings demonstrate phosphorylation of a member of the Cap'n'Collar family of basic region-leucine zipper (bZIP) transcription factors, which also includes NF-E2, Nrf1, the recently identified Nrf3, and the Bach family (14, 40–42). It will be of interest to investigate whether these other transcription factors can also undergo phosphorylation, and what role it might play in their regulation.
Our results indicating the oxidative stress-inducible release of endogenous Nrf2 into the nucleus confirm a recently proposed model based on cotransfection of Nrf2 and Keap1 expression plasmids (24). Because Nrf2 has been shown to interact with the cytoskeleton-associated Keap1 through its N-terminal domain Neh2 (24), it seems conceivable that PKC may phosphorylate Nrf2 at a site within this domain, resulting in conformational changes that trigger the dissociation of the transcription factor from its cytoplasmic anchor. The Keap1 protein also contains several putative sites for PKC phosphorylation, as do the small Maf proteins which heterodimerize with Nrf2 to form high-affinity ARE-binding complexes (18). Possibly, PKC may target these Nrf2-associated partners as well, as part of a coordinated response to oxidative stress.
ROS have been known to activate members of the PKC family, which consists of at least 11 isoforms with conserved catalytic domains (28, 43). PKCs contain structural features that make them susceptible to direct redox modifications, which alter their activity against cellular targets. A recent study showed that H2O2-induced cellular apoptotic response involves the mutual phosphorylation and activation of PKC δ and the cytoplasmic form of protein tyrosine kinase c-Abl (44). PKC δ was also shown to target the STAT3 transcription factor in response to interleukin-6 stimulation (45). Earlier studies had shown that polyphenolic agents that generate oxidants, such as catechol and hydroquinone, can activate PKC (46). Our results suggest a mechanism in which the PKC phosphorylation of Nrf2 leads to its activation of the ARE. Because PKC δ has been implicated in various cellular responses to oxidative stress, it is tempting to speculate that it may be the isoform that targets Nrf2. Establishing the spectrum of cellular substrates preferentially phosphorylated by the various PKC isozymes following oxidative stress will help dissect the intricate regulatory mechanisms of ROS-induced PKC activation.
Interestingly, several reports have implicated PKC in the xenobiotic-inducible transcription of the cytochrome P450 (CYP) 1A1 gene mediated through the aryl hydrocarbon receptor (AhR) (49). Binding of AhR to ligands such as TCDD (2,3,7,8-tetrachlorodibenzo-p-dioxin) triggers translocation of the transcription factor into the nucleus where it dimerizes with the ARNT protein and activates transcription of CYP 1A1 (7, 52). Metabolites of certain CYP 1A1-inducing planar aromatic compounds (e.g., βNF) can also function as inducers of the GST A2 and QR genes via the ARE (9). In contrast to our findings with Nrf2-mediated ARE activation, however, PKC was not shown to play a role in the nuclear transport of the AhR in response to TCDD treatment (49, 50). Because ARE-responsive gene expression does not require functional AhR (9), and TCDD cannot directly activate the ARE, the molecular mechanisms of PKC requirement in the AhR- and Nrf2-mediated signal transduction pathways appear to be distinct.
Previous studies have demonstrated the involvement of MAPK pathways in ARE-mediated transcription (26, 27). While it is well established that PKC can activate the MAPK pathways (47), our finding that specific inhibitors of ERK and p38 did not affect Nrf2 phosphorylation appears to rule out the possibility that the role of PKC in Nrf2 phosphorylation is through its indirect action on these downstream kinases. Indeed the identity of the specific MAPK-regulated components in the signaling cascade toward ARE activation is an important mystery that remains to be uncovered. It is likely that kinases other than PKC are also critical in ARE-mediated gene expression, perhaps through regulating transcriptional activation events in addition to Nrf2 nuclear translocation.
It is also noteworthy that PKC inhibitors were only partially effective in blocking tBHQ-induced ARE-driven CAT activity, even though tBHQ-stimulated Nrf2 phosphorylation and nuclear accumulation appeared to be abolished. Our data revealed that even though Nrf2 is predominantly localized to the cytoplasm under unstressed conditions, it is not entirely absent from the nucleus. It is possible that the constitutive levels of Nrf2 in the nucleus account for the residual CAT activity seen in the presence of PKC inhibitors. Indeed, we and others have previously reported the important role of Nrf2 in mediating the constitutive ARE-mediated gene expression (19, 22). Furthermore, inhibition of PKC did not reduce the CAT activity in untreated or PMA-stimulated cells to a level significantly below the basal level. One possibility is that the constitutive expression of ARE-responsive genes is regulated in a PKC-independent manner. Finally, while we have shown a role for PKC acting in the cytoplasm to regulate Nrf2 nuclear import, it may well have additional functions that directly influence Nrf2 activity in the nucleus, similar to its effects on AhR-mediated transcription (49, 50). Future studies addressing the multiple levels of regulation, involving overlapping kinase pathways, should contribute to the elucidation of the coordinated cellular response to oxidative stress.
Acknowledgments
We thank Dr. Leonard Favreau for providing HepG2 cells stably transfected with an ARE-CAT reporter construct, Dr. Lydia Armstrong for invaluable help in immunofluorescence studies, and Drs. W. Robert Bishop, Paul Kirschmeier, Jonathan Pachter, Jessie English, and Ahmed Samatar for useful advice and stimulating discussions.
Abbreviations
- Nrf2
NF-E2-related factor 2
- ARE
antioxidant response element
- tBHQ
tert-butylhydroquinone
- CAT
chloramphenicol acetyltransferase
- βNF
β-naphthoflavone
- PMA
phorbol 12-myristate 13-acetate
- PKC
protein kinase C
- QR
NAD(P)H:quinone oxidoreductase
- GST
glutathione S-transferase
- ROS
reactive oxygen species
- MAPK
mitogen-activated protein kinase
- PI
propidium iodide
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220418997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220418997
References
- 1.Scandalios J G, editor. Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 2.Sies H. Eur J Biochem. 1993;215:213–219. doi: 10.1111/j.1432-1033.1993.tb18025.x. [DOI] [PubMed] [Google Scholar]
- 3.Berlett B S, Stadtman E R. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 4.Jacobson M D. Trends Biochem Sci. 1996;21:83–86. [PubMed] [Google Scholar]
- 5.Cerutti P A. Science. 1985;227:375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 6.Ames B N, Shigenaga M K, Hagen T M. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes J D, McLellan L I. Free Radical Res. 1999;31:273–300. doi: 10.1080/10715769900300851. [DOI] [PubMed] [Google Scholar]
- 8.Primiano T, Sutter T R, Kensler T W. Adv Pharmacol. 1997;38:293–328. doi: 10.1016/s1054-3589(08)60989-8. [DOI] [PubMed] [Google Scholar]
- 9.Rushmore T H, Pickett C B. J Biol Chem. 1990;265:14648–14653. [PubMed] [Google Scholar]
- 10.Friling R S, Bensimon A, Tichauer Y, Daniel V. Proc Natl Acad Sci USA. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favreau L V, Pickett C B. J Biol Chem. 1991;266:4556–4561. [PubMed] [Google Scholar]
- 12.Li Y, Jaiswal A K. J Biol Chem. 1992;267:15097–15104. [PubMed] [Google Scholar]
- 13.Rushmore T H, Morton M R, Pickett C B. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 14.Andrews N C, Erdjument-Bromage H, Davidson M B, Tempst P, Orkin S H. Nature (London) 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 15.Moi P, Chan K, Asunis I, Cao A, Kan Y W. Proc Natl Acad Sci USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Nature (London) 1994;367:568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- 17.Motohashi H, Shavit J A, Igarashi K, Yamamoto M, Engel J D. Nucleic Acids Res. 1997;25:2953–2959. doi: 10.1093/nar/25.15.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen T, Huang H-C, Pickett C B. J Biol Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 20.Wild A C, Moinova H R, Mulcahy R T. J Biol Chem. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 21.Alam J, Stewart D, Touchard C, Boinapally S, Choi A M K, Cook J L. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 22.Hayes J D, Chanas S A, Henderson C J, McMahon M, Sun C, Moffat G J, Wolf C R, Yamamoto M. Biochem Soc Trans. 2000;28:33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- 23.Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, Bannai S, Yamamoto M. J Biol Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 24.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel J D, Yamamoto M. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter T, Karin M. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 26.Yu R, Lei W, Mandlekar S, Weber M J, Der C J, Wu J, Kong A-N T. J Biol Chem. 1999;274:27545–27552. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- 27.Yu R, Mandlekar S, Lei W, Fahl W E, Tan T-H, Kong A-N T. J Biol Chem. 2000;275:2322–2327. doi: 10.1074/jbc.275.4.2322. [DOI] [PubMed] [Google Scholar]
- 28.Nishizuka Y. FASEB J. 1995;9:484–496. [PubMed] [Google Scholar]
- 29.Nguyen T, Rushmore T H, Pickett C B. J Biol Chem. 1994;269:13656–13662. [PubMed] [Google Scholar]
- 30.Favreau L V, Pickett C B. J Biol Chem. 1993;268:19875–19881. [PubMed] [Google Scholar]
- 31.Ainbinder E, Bergelson S, Pinkus R, Daniel V. Eur J Biochem. 1997;243:49–57. doi: 10.1111/j.1432-1033.1997.0049a.x. [DOI] [PubMed] [Google Scholar]
- 32.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chida K, Kato N, Kuroki T. J Biol Chem. 1986;261:13013–13018. [PubMed] [Google Scholar]
- 34.Favata M F, Horiuchi K Y, Manos E J, Daulerio A J, Stradley D A, Feeser W S, Van Dyk D E, Pitts W J, Earl R A, Hobbs F, et al. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 35.Cuenda A, Rouse J, Doza Y N, Meier R, Cohen P, Gallagher T F, Young P R, Lee J C. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 36.Karin M. Curr Opin Cell Biol. 1994;6:415–424. doi: 10.1016/0955-0674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 37.Baeuerle P A, Baltimore D. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- 38.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 39.Chinery R, Brockman J A, Dransfield D T, Coffey R J. J Biol Chem. 1997;272:30356–30361. doi: 10.1074/jbc.272.48.30356. [DOI] [PubMed] [Google Scholar]
- 40.Chan J X, Han X-L, Kan Y W. Proc Natl Acad Sci USA. 1993;90:11366–11370. doi: 10.1073/pnas.90.23.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M. J Biol Chem. 1999;274:6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- 42.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newton A C. J Biol Chem. 1995;270:28495–28498. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- 44.Sun X, Wu F, Datta R, Kharbanda S, Kufe D. J Biol Chem. 2000;275:7470–7473. doi: 10.1074/jbc.275.11.7470. [DOI] [PubMed] [Google Scholar]
- 45.Jain N, Zhang T, Kee W H, Li W, Cao X. J Biol Chem. 1999;274:24392–24400. doi: 10.1074/jbc.274.34.24392. [DOI] [PubMed] [Google Scholar]
- 46.Gopalakrishna R, Chen Z-H, Gundimeda U. Proc Natl Acad Sci USA. 1994;91:12233–12237. doi: 10.1073/pnas.91.25.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buchner K. J Cancer Res Clin Oncol. 2000;126:1–11. doi: 10.1007/PL00008458. [DOI] [PMC free article] [PubMed] [Google Scholar]