Abstract
Many overlapping surveillance and repair mechanisms operate in eukaryotic cells to ensure the stability of the genome. We have screened to isolate yeast mutants exhibiting increased levels of recombination between repeated sequences. Here we characterize one of these mutants, elg1. Strains lacking Elg1p exhibit elevated levels of recombination between homologous and nonhomologous chromosomes, as well as between sister chromatids and direct repeats. These strains also exhibit increased levels of chromosome loss. The Elg1 protein shares sequence homology with the large subunit of the clamp loader replication factor C (RFC) and with the product of two additional genes involved in checkpoint functions and genome maintenance: RAD24 and CTF18. Elg1p forms a complex with the Rfc2–5 subunits of RFC that is distinct from the previously described RFC-like complexes containing Rad24 and Ctf18. Genetic data indicate that the Elg1, Ctf18, and Rad24 RFC-like complexes work in three separate pathways important for maintaining the integrity of the genome and for coping with various genomic stresses.
The processes of DNA replication, repair, and recombination are intimately linked. During DNA replication, the activity of the DNA polymerases may be impaired by the presence of secondary structures, bound proteins, or DNA lesions. This may lead to stalling and even collapse of replication forks. In response, cellular mechanisms are activated that arrest cell cycle progression, induce DNA repair, and restore replication (1).
Replication factor C (RFC), a five-subunit protein complex, associates with the DNA polymerase processivity factor proliferating cell nuclear antigen (PCNA) and loads it onto DNA. PCNA tethers the polymerase to the DNA template and serves as a central platform to load many enzymes involved in the replication, repair, and modification of DNA. Recently, two alternative RFC-like protein complexes (RLCs) have been described in the yeast Saccharomyces cerevisiae and in other organisms, which include the Rfc2–5 subunits but not the Rfc1 protein. In one of these RLCs, the large RFC subunit is replaced by the checkpoint protein Rad24 (Rad17 in Schizosaccharomyces pombe and hRad17 humans) (2, 3). This complex is predicted to load an alternative DNA sliding clamp (4) composed of check-point proteins (5). In the second RLC, the large subunit of RFC is replaced by the Ctf18/Chl12 protein, implicated in sister chromatid cohesion (6–8). It is not yet clear whether this alternative complex interacts with PCNA or with a yet-to-be-found alternative clamp.
The yeast S. cerevisiae has been extensively used to identify and dissect the response to DNA damage. These studies have demonstrated that a network of overlapping pathways operates to maintain genomic stability (9, 10). Checkpoint control failure and elevated levels of genomic instability are a hallmark of cancer cells (1).
Homologous recombination is one of the main mechanisms able to restore replication competence to cells with stalled replication forks. Accordingly, mutations that affect replication proteins, such as DNA ligase, DNA polymerases, topoisomerases, etc., result in increased levels of recombination (reviewed in refs. 11 and 12). On the other hand, under certain circumstances recombination may have deleterious consequences by itself. Recombination between homologous sequences located at nonallelic positions can result in potentially lethal chromosomal aberrations (13). However, recombination between naturally occurring repetitive sequences in yeast is rarely detected, and it is seldom associated with the formation of chromosomal aberrations (13), suggesting the existence of mechanisms that control this type of event.
Here we report the isolation of ELG1, a gene that plays an important role in limiting potentially deleterious recombination events. Its product exhibits sequence similarity to Rfc1p and forms an alternative, RLC. We show that Elg1p, Rad24p, and Ctf18p are components of three distinct RLCs that act in concert to maintain genomic integrity.
Methods
Media, Growth, and General Procedures. Yeast cells were grown at 30°C in yeast extract/peptone/dextrose (YPD) or SD. Canavanine (CAN) medium is SD-complete without arginine, plus 40 μg/ml CAN sulfate (Sigma).
Yeast Strains. All strains used for mutagenesis and to check the genetic requirements for elg1 recombination are isogenic derivatives of strain MK166 (14, 15).
Diploid trains used for measuring allelic mitotic recombination were generated by mating of Sy72 (MATα lys2-1 can1 ura3-1 his1 ade5 trp5-2 leu1-12 ade2-1) and Sy73 (MATa tyr1-1 ura3-13 hom3 met13-c cyh2 trp5-d leu1-c ade6 ade2-1) or their elg1::KanMX derivatives. SBA272 (MATa/MATα CAN1/can1 URA3/ura3-1 HOM3/hom3 HIS1/his1) and its elg1::KanMX/elg1::KanMX derivative were used for crossing over and chromosome loss measurements. Unequal sister chromatid recombination was measured in BLS2 (15) and an elg1::KanMX derivative.
The yeast strains used for immunoprecipitation (IP) were constructed as follows: SBA290 (ELG1::3HA-His3MX6), SBA291(ELG1::3Myc-His3MX6), and SBA316 (RAD24:: 3Myc-kanMX6) were created by transformation of MK244 (MATα ade2-1 leu2-3,112 his3-11,13 trp1::hisG arg4::hisG ura3Δ can1). The three alleles were functional, as judged by their ability to complement the recombination phenotype and synthetic lethality of elg1 strains (SBA290 and SBA291) or the UV sensitivity of rad24 strains (SBA316). YJH40.4, YJH62.6, YJH71.1, and YJH72.1 (6) were kindly provided by Forrest A. Spencer (The Johns Hopkins University School of Medicine, Baltimore). Each of them contains a different tagged gene: CTF18::9Myc-klTRP1, RFC1:: 9Myc-klTRP1, RFC4::6HA-KanMX, and RFC5::6HA-KanMX, respectively. Double- and triple-mutant strains were then created by crosses. SBA333 was created by transformation of SBA290 with a RFC4::3Myc-kanMX6 construct.
Mutant Screen. SBA1 is a yeast strain that allows easy scoring of several types of recombination events (Fig. 1A and ref. 14). Its genotype is MATa ade2-1 can1-10 leu2Δ ura3-52 trp1Δ lys2::Ty1Sup his4::TRP1::his4 HIS3::lys2::ura3-x. The suppressible mutations ade2-1 and can1-100 confer adenine auxotrophy and resistance to CAN, respectively. A Ty1 element marked by the insertion of a suppressor tRNA gene (Ty1Sup) renders the cells white (Ade+) and sensitive to CAN (CanS). Gene conversion events between Ty1Sup and other Ty elements in the genome, or recombination between the LTRs of the Ty, yield red, CanR Ade– cells. Forward mutation at the CAN1 gene gives rise to white CanR colonies. In addition, the strain carries a duplication of part of the HIS4 gene; direct-repeat recombination (DRR) between the duplicated segments can give rise to His+ Trp– cells. Strain SBA1 was transformed with a mTn-LEU2/LacZ-mutagenized library (16). Individual Leu+ colonies were patched onto master plates and replica-plated onto CAN plates. Patches showing elevated levels of CANR papillation were retested, first as patches, and later by fluctuation tests.
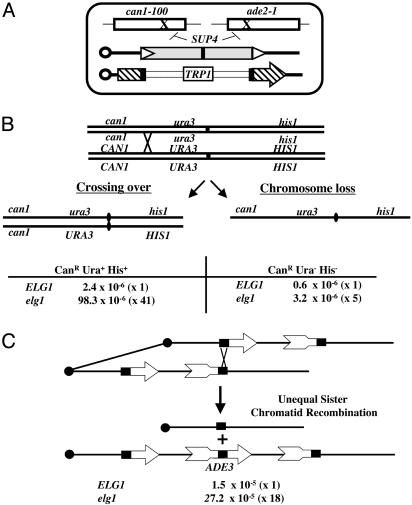
Fig. 1.
(A) Genetic system used to isolate ELG1. The strain SBA1 carries several substrates for recombination. The presence of the SUP4 suppressor tRNA within a Ty element renders the cells white (Ade+) and sensitive to CAN (CanS) by suppressing the mutations can1-100 and ade2-1. Recombination events (gene conversion) between Ty1Sup and other Ty elements in the genome, or interactions between the LTRs of the Ty (triangles), yield CanR Ade– cells, which can be selected on CAN plates. In addition, SBA1 carries a duplication of part of the HIS4 gene, created by the insertion of a TRP1-marked plasmid; DRR between the duplicated segments (black rectangles) gives rise to His+ Trp– cells. (B) Schematic representation of the genetic system used to measure crossing over and chromosome loss. Crossing over between CAN1 and URA3 on chromosome V gives rise to CanR Ura+ His+ colonies, whereas chromosome loss generates CanR Ura– His– colonies. These events can be selected on appropriate plates. Rates of crossing over and chromosome loss in elg1/elg1 and in WT controls are shown. (C) Schematic representation of the genetic system used to measure USCE. Two nonfunctional fragments of the ADE3 gene overlap in a small region (black square). Only unequal sister chromatid recombination after DNA replication generates a functional ADE3 gene. Rates of USCE in elg1 and WT strains are shown. Fold induction appears in parentheses.
Recombination and Mutation Measurement. Ectopic recombination and mutation levels were measured in MK166 derivatives by fluctuation tests as described (14, 15). Standard deviations were usually <15%. Each experiment was repeated at least twice. Crossing over and chromosome loss were measured in strains SBA272 (ELG1/ELG1) and SBA273 (elg1/elg1) by fluctuation test on CAN plates or CAN plus 5-fluoroorotic acid plates. Colonies obtained were replica-plated to SD-His. Unequal sister chromatid recombination was assayed in BLS2 (ELG1) and BLS2elg1 by fluctuation test on SD-His plates.
Synthetic Lethality Screen. The screen was carried out as described (17). A total of 140,000 colonies were screened. Synthetic lethality was verified as plasmid dependence conferring 5-fluoroorotic acid sensitivity. Mutant genes were cloned by complementation from a genomic library.
IP and Western Blot Analysis. IP and Western blot analysis were performed as described (6).
Sensitivity to Methyl Methanesulfonate (MMS) and Hydroxyurea (HU). The WT strain (MK166) and its single-, double-, and triplemutant derivatives were grown to midlogarithmic phase, washed, and resuspended in water. Various concentrations of MMS were added, and after incubation at 30°C for 20 min, the MMS was neutralized with 10% sodium thiosulfate. Cells were washed and plated at various dilutions on YPD plates. Survival was scored after 3 days. HU sensitivity was measured by directly plating on plates containing increasing amounts of HU. Survival was scored similarly.
Results
Isolation of the elg1 Mutant. Ty elements are the largest family of naturally occurring repeated sequences in yeast, comprising ≈3% of the genomic DNA. Despite their abundance, Ty elements spontaneously recombine at remarkably low rates (13). In addition, Ty recombination is not increased by DNA-damaging treatment, such as exposure to MMS or to UV irradiation (18), although it can be induced by directed double-strand breaks (DSBs) (19). These results suggest the existence of mechanisms that prevent recombination between repeated sequences. We screened for mutants exhibiting increased frequencies of Ty recombination. We used strain SBA1, which enables easy monitoring of recombination between Ty elements and between direct repeats (ref. 14; see Fig. 1 A and Methods). Strain SBA1 was subjected to mutagenesis by randomly disrupting genes with a modified transposon (16), and individual colonies showing elevated levels of Ty recombination were isolated. One of the mutants, which exhibited a particularly high level of Ty recombination, carried a transposon insertion at ORF YOR144c. Based on the results presented below, we have named this mutant elg1 (enhanced levels of genome instability). In the original isolate, the transposon was inserted at nucleotide +260 from the start codon of the ELG1 gene. A complete deletion of ELG1 exhibited identical phenotypes. We have therefore used both alleles interchangeably in our studies.
elg1 Mutants Exhibit Increased Levels of DRR and Gene Conversion. Similar to retroviruses, Ty elements are composed of a central element flanked by LTRs. Ty elements can therefore carry out recombination with other Ty elements in the genome, or engage in DRR between LTRs. elg1 mutants exhibited a 30-fold increase in the level of LTR recombination, a 5-fold increase in ectopic recombination between Ty elements, and a 5-fold elevation in DRR at the HIS4 locus. No effect on the level of spontaneous mutagenesis [measured as forward mutations at the can1-100 allele (14, 15)] was observed in the elg1 mutants.
The elevated recombination seen in the absence of Elg1p is not restricted to haploid strains. We also observed an effect of deleting ELG1 on intragenic recombination in diploid cells. In elg1/elg1 diploids carrying heteroallelic markers at three different loci, recombination (gene conversion) was elevated 4- to 6-fold (data not shown).
Deletion of ELG1 Causes Increased Levels of Crossing Over, Chromosome Loss, and Sister Chromatid Recombination. We also found that deletion of ELG1 affects the levels of mitotic reciprocal recombination. Isogenic ELG1 and elg1 diploid strains were generated, heterozygous for the can1, ura3, and his1 mutations on chromosome V. Crossing over events in the interval between CAN1 and the centromere result in colonies resistant to CAN and able to grow on media lacking histidine and uracil (Fig. 1B). By using the same strains it is also possible to estimate the level of chromosome loss, by selecting for CanR Ura– His– cells. In elg1/elg1 diploid cells crossing over was enhanced 41-fold, and chromosome loss rates were elevated 5-fold relative to WT (Fig. 1B).
Deletion of ELG1 also affects the levels of unequal sister chromatid exchange (USCE). In strains that allow monitoring of USCE (ref. 15 and Fig. 1C), elg1 strains exhibited 18-fold more USCE than the isogenic WT.
We thus conclude that mutations in ELG1 result in a genome-wide increase of all forms of recombination (inter- and intragenic, ectopic and allelic, between sister chromatids, and between direct repeats). Both nonreciprocal (gene conversion) and reciprocal recombination (crossing over) are elevated. In addition, chromosome loss is increased.
Genetic Requirements for the Increased Levels of Recombination in elg1 Strains. Repair of DNA damage in yeast is carried out by numerous proteins. These can be assorted into four major epistasis groups: the excision repair, postreplicational repair, nonhomologous end-joining, and recombinational repair pathways. To understand the genetic requirements for the hyperrecombination phenotype observed in elg1 mutants, we deleted ELG1 in conjunction with representative genes of each DNA repair group.
Mutations in the excision repair group and in the nonhomologous end-joining pathways usually do not lead to hyperrecombination. However, in some instances (20, 21), spontaneous lesions may need processing by enzymes from one of these groups to become recombinogenic. Table 1 shows that the high rates of recombination conferred by the elg1 mutation were not affected by mutations in RAD4 (excision repair) or YKU70 (nonhomologous end-joining). This finding indicates that the excision repair and nonhomologous end-joining pathways are not required for the elevated recombination observed in elg1 strains.
Table 1. Genetic interactions between elg1 and mutations in DNA repair genes.
| Strain | His+ DRR | Ty DRR |
|---|---|---|
| MK 166 | (6.0 × 10-6) × 1 | (1.0 × 10-6) × 1 |
| elg1 | ×5.1 | ×30 |
| rad4 | ×1.0 | ×1.3 |
| elg1 rad4 | ×5.6 | ×29.7 |
| rad18 | ×5.7 | ×20 |
| elg1 rad18 | ×11 | ×68 |
| yku70 | ×1.0 | ×1.3 |
| elg1 yku70 | ×4.8 | ×29.5 |
| rad52 | ×0.11 | ×0.09 |
| elg1 rad52 | ×0.07 | <×0.1 |
Fold induction levels, compared to the WT control (MK 166), are shown.
Mutations in genes of the postreplicational repair group have been shown to cause spontaneous hyperrecombination (15). An elg1 rad18 double mutant exhibited higher levels of recombination than either single mutant (Table 1). This additive relationship indicates that Elg1p is not a component of the postreplicational repair group.
In contrast, the hyperrecombination phenotype was totally eliminated by mutations in the RAD52 gene. This central gene of the recombinational repair group is essential for the repair of DSBs (22). In addition, elg1 rad52 double mutants exhibited very slow growth and extremely reduced viability, resulting in a nearly lethal phenotype. A similar effect, albeit to a lesser degree, was observed in elg1 strains deleted for the additional recombinational repair genes RAD50, RAD51, RAD54, RAD55, and RAD57. Microscopic examination (>300 cells counted for each strain) revealed that a very large proportion of elg1 rad52 cells were large-budded, consistent with a cell cycle arrest at the G2/M phase (88% in elg1 rad52, vs. 56% in rad52, 31% in elg1, and 11% in WT cells; all comparisons with P < 0.01). These results suggest that Elg1p-deficient cells probably suffer a large number of DSBs that, in the absence of Rad52p, are not repaired, resulting in cell cycle arrest and cell death.
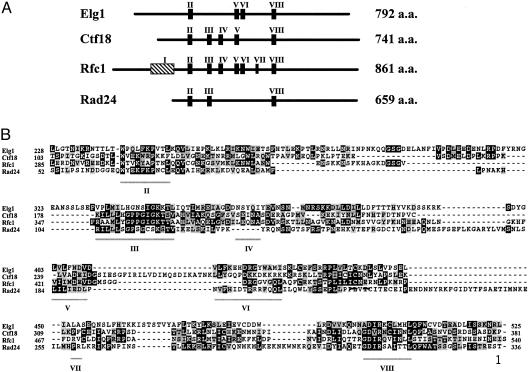
Elg1p Shows Similarity to the Large Subunit of RFC. The ELG1 gene encodes a 792-aa long protein. Database searches detected sequence similarity between Elg1p and Rfc1p, the large subunit of the RFC complex (Fig. 2). The five RFC subunits share seven conserved domains, termed boxes II–VIII (23). Elg1p possesses clearly identifiable domains II, V, VI, and VIII (Fig. 2B). Two other proteins, Rad24p and Ctf18p/Chl12p, also share similarities with Rfc1p and Elg1p. Rad24p plays an important role in the DNA damage checkpoint: mutations in RAD24 cause sensitivity to genotoxic agents and abrogate damage-dependent cell-cycle arrest (24). CTF18 was isolated in a screen for yeast mutants that exhibit elevated rates of mitotic chromosome loss (25). Mutations in this gene also cause increased levels of mitotic recombination (26). Elg1p exhibits low similarity to Rad24p and Ctf18p throughout most of its length (Fig. 2B). Potential Elg1p orthologs are present in Schizosaccharomyces pombe (AL021837.2/Q43086), Candida albicans (IPF625), Caenorhabditis elegans (C08B6.2), and Drosophila melanogaster (Q9VUZ2). In addition, several mammalian ESTs share low levels of similarity with both Elg1p and Ctf18p.
Fig. 2.
Elg1p shares similarities with Rfc1p, Rad24p, and Ctf18p. (A) Schematic representation of the four proteins. Boxes I–VIII represent motives conserved in RFC proteins (23). The overall identity/similarity between Elg1p and the other three proteins are as follows: Rfc1p, 20/13.3%; Ctf18p, 15.4/15.4%; and Rad24p, 17.2/12.8%. (B) Detailed comparison between the four proteins. Boxes II–VIII are underscored. Black squares represent identical residues and gray squares mark conserved amino acids. The comparison was generated with clustalw and matchbox programs.
Elg1p Operates in Parallel to Rad24p and Ctf18p with Respect to DNA Damage Sensitivity and Recombination. The sequence homology between ELG1, CTF18, and RAD24 and the partial phenotypic similarity between the mutants encouraged us to test for genetic interactions between the genes. We reasoned that if these proteins play overlapping functions, double-mutant strains should show a more severe phenotype than each single mutant. Alternatively, the three proteins (or two of them) may work together in a single pathway; in that case no additive behavior of the double mutants is expected. We therefore generated all of the double mutant combinations and the triple elg1 ctf18 rad24 mutant, which was viable but exhibited an extreme slow growth phenotype and reduced viability.
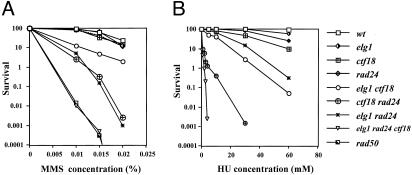
We tested the different strains for sensitivity to the radiomimetic drug MMS, or to the inhibitor of DNA replication HU. Although individual mutants showed no significant sensitivity to DNA damage by MMS, the double mutants were sensitive (Fig. 3A). The triple mutant was extremely sensitive; survival in the presence of very low concentrations of MMS was comparable to that of rad50 or rad52 strains, which lack DSB repair activity. A similar additive relationship could be seen with respect to HU sensitivity. Single mutants were mildly sensitive, and double mutants exhibited an intermediate level of sensitivity. Remarkably, the elg1 ctf18 rad24 strain failed to grow in the presence of even minute amounts of the drug (Fig. 3B).
Fig. 3.
elg1, ctf18, and rad24 are additive in their sensitivity to MMS and HU. (A) Midlogarithmic cells were treated for 20 min at 30°C with various concentrations of MMS, washed, and plated on YPD plates. (B) Mid-logarithmic cells were plated on plates containing different concentrations of HU.
As with MMS and HU sensitivity, additive or synergistic relationships between elg1, ctf18, and rad24 were observed for the hyperrecombination phenotype. Triple mutants exhibited the highest elevation of recombination rates: DRR was elevated 50-fold at the HIS4 locus and almost 100-fold for Ty elements (Table 2). Taken together, our results suggest that Elg1p, Rad24p, and Ctf18p operate in three redundant pathways. They may participate in alternative responses to DNA damage; alternatively, their absence may lead to replication defects and endogenous damage that renders the cells more sensitive to exogenous insults.
Table 2. elg1 is additive to ctf18 and rad24 for hyper-recombination.
| Strain | His+ DRR | Ty DRR |
|---|---|---|
| MK 166 | (6 × 10-6) × 1 | (1.0 × 10-6) × 1 |
| elg1 | ×5.1 | ×30 |
| ctf18 | ×4.2 | ×4.9 |
| rad24 | ×1.1 | ×2.2 |
| elg1 ctf18 | ×16 | ×37 |
| elg1 rad24 | ×21 | ×36 |
| ctf18 rad24 | ×6.9 | ×47 |
| ctf18 rad24 elg1 | ×49 | ×96 |
Fold induction levels, compared to the WT control (MK166), are shown.
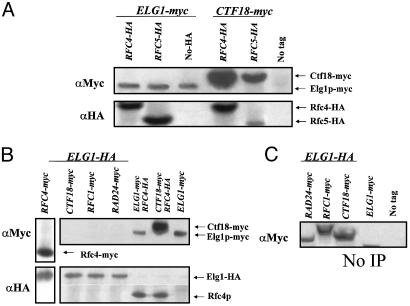
Elg1p, Rad24p, and Ctf18p Form Three Alternative RLCs. Recent work has shown that Rad24p forms a RLC, in which it replaces Rfc1p and interacts with Rfc2–5p (2). A similar complex in which Rfc1p is replaced by Ctf18p has also been described (6–8). We tested whether Elg1p interacts with the small RFC subunits by IP. We generated strains in which the genomic ELG1 gene is replaced by a myc-tagged allele and which carry a hemagglutinin (HA) epitope-tagged version of either the RFC4 or the RFC5 genes. Extracts were prepared from cells and subjected to IP with an anti-myc antibody. The immunoprecipitates were separated on denaturing SDS/PAGE and analyzed by immunoblotting with antibodies against the HA and myc epitopes. As a control we used a strain carrying a myc-tagged CTF18 allele and a HA-tagged RFC4 allele; these proteins have been shown to coimmunoprecipitate (6–8). Indeed, Rfc4-HA and Rfc5-HA can be efficiently coimmunoprecipitated with Elg1-myc, indicating that Rfc4p and Rfc5p interact with Elg1p (Fig. 4A). This finding is consistent with recent reports from genome-wide affinity purification studies (27, 28), which identified physical interactions between Elg1p and the Rfc2, Rfc4, and Rfc5 proteins.
Fig. 4.
Elg1p coimmunoprecipitates with Rfc4p and Rfc5p, but not with Ctf18p, Rfc1p, or Rad24p. (A) Extracts were prepared from strains carrying combinations of the following alleles: ELG1::3myc, CTF18::9myc, RFC4::6HA, and RFC5::6HA. After IP with anti-myc antibody, immunocomplexes were separated by SDS/PAGE and immunoblotted with anti-myc and anti-HA antibodies. Rfc4p and Rfc5p coimmunoprecipitated with both Elg1p and Ctf18p. MK244, carrying no tags, served as a negative control. (B) Proteins were extracted from strains in which the genomic ELG1 gene was HA-tagged and carried myc-tagged versions of CTF18, RFC1, RAD24, or, as a positive control, RFC4. Anti-HA antibody was used to precipitate Elg1-3HA. As expected, Rfc4-myc was detected in the immunocomplexes, whereas no coimmunoprecipitation of Elg1p with Rfc1p, Ctf18p, or Rad24p could be detected. As additional controls, we show that Rfc4-HA is easily detected after IP of Elg1-myc or Ctf18-myc. (C) Samples treated similarly, but without IP, are shown to indicate the expected electrophoretic mobility of the various tagged proteins. MK244, carrying no tags, served as a negative control.
The genetic data suggest that Elg1p, Rad24p, and Ctf18p act in separate pathways. To test whether Elg1p belongs to one of the known complexes, or whether it is part of an alternative RLC, we carried out IP experiments with strains in which the genomic ELG1 gene was replaced by an HA-tagged allele, and carried myc-tagged versions of RFC1, CTF18, RAD24, or, as a positive control, RFC4. Fig. 4B shows that although Rfc4p readily coimmunoprecipitated with Elg1p, the Rfc1, Ctf18, or Rad24 proteins were not detected after IP of Elg1p. In extracts not immunoprecipitated, the three proteins are readily detected (Fig. 4C). These results imply that Elg1p forms in vivo a complex with the Rfc4 and Rfc5 proteins (and by inference Rfc2p and Rfc3p) but not with Rfc1p, Rad24p, or Ctf18p. The IP experiments and genetic data are consistent and suggest that the Elg1p complex comprises a RLC that acts independently of the two previously characterized RLCs.
elg1 Is Synthetically Lethal with ctf4. To further characterize the function of ELG1, we screened to identify mutants unable to survive in the absence of Elg1p (see Methods). We found that ELG1-deleted cells require a functional CTF4 gene for survival. Tetrad analysis confirmed that the double mutant elg1 ctf4 is inviable. Ctf4p was found to bind DNA polymerase α (pol α) (29) and was independently isolated, along with ctf18, in a screen for mutants with increased chromosome instability (25, 30). The double mutant ctf4 ctf18 is not viable (31). In addition, ctf4 mutants exhibit synthetic lethality with mutations in many genes whose products participate in DNA replication, such as RFC1, RAD27 (encoding the flap endonuclease), and DNA2 (encoding a replication helicase) (31). The synthetic lethality with mutations in CTF4 further supports a role for Elg1p during DNA replication.
Discussion
Stalling or collapse of replication forks can lead to aneuploidy and chromosomal rearrangements (9). Such events may occur spontaneously, or may be induced by DNA damage or interference with the replication machinery. Under these circumstances, recombination can provide a mechanism to increase cell survival, reforming functional replication forks (32).
The results presented here indicate that, in the absence of Elg1p, increased rates of spontaneous recombination are observed. Allelic and ectopic intra- and inter-chromosomal recombination, as well as recombination between sister chromatids, are elevated in elg1 mutants. A high rate of chromosome loss is also detected (Fig. 1). Recently, elg1 was identified in a screen for yeast mutants exhibiting elongated telomeres (S. Smolikov and A. Krauskopf, personal communication). In addition, a mutation in ELG1 (termed rtt110) was also found to increase the rate of transposition of Ty elements by a posttranscriptional mechanism (33). In summary, hyperrecombination, chromosome loss, hypertransposition, and elongated telomeres are all phenotypes of elg1 mutants. Elg1p function is thus clearly required for maintaining genome stability during normal growth, and its absence has severe genetic consequences.
The Role of Elg1p in DNA Replication and Damage-Associated Replication. We suggest that the spontaneous genomic instability in elg1 cells is associated with DNA replication defects. Absence of Elg1p results in the accumulation of lesions that are repaired mainly through a recombination mechanism. The vast majority of the cell population in the elg1 rad52 strains is composed of checkpoint-arrested cells. This finding suggests that in elg1 strains replication may not be complete, and that the remaining single-strand DNA gaps or DSBs must be processed by a recombination mechanism. This phenotype is reminiscent of that observed in cells defective in PCNA, DNA ligase, flap endonuclease, and other replication proteins that accumulate discontinuities in their DNA (11, 12).
Further linkage of ELG1 to replication is provided by the presence in its promoter of an MluI site. This sequence is part of the binding site for the MBF (MluI-box binding factor) transcription factor and is present in the regulatory regions of many DNA replication genes expressed during the G1/S transition of the cell cycle (34). Accordingly, ELG1 mRNA levels fluctuate during the cell cycle, with a pattern similar to that of CDC9, RAD27, POL12, and other DNA replication genes that contain MluI boxes (ref. 35; data not shown).
Additional support for a role of Elg1p in replication is inferred from the synthetic lethality between mutations in the CTF4 and ELG1 genes. Ctf4 is a pol α-binding protein (29). ctf4 mutations are lethal in combination with mutations in genes required for lagging-strand DNA synthesis, including POL1, RAD27, and RFC1 (31).
In summary, in the absence of Elg1p, single-strand DNA gaps or DSBs may remain after the bulk of replication is completed. The RAD52 recombinational repair pathway acts as a fail-safe mechanism that allows the cells to bypass these potentially lethal lesions. Repair of the damaged DNA by single-strand annealing or by ectopic recombination accounts for the elevated levels of recombination in elg1 mutants. When damage cannot be repaired, chromosome loss ensues.
Elg1p Forms a RLC. We have shown that ELG1 encodes a protein with homology to Rfc1p, Ctf18p, and Rad24p (Fig. 2) that forms a RLC (Fig. 4 and refs. 27 and 28). The Elg1p RLC is distinct from the other three protein complexes, as Rfc1p, Ctf18p, Rad24p, and Elg1p do not coimmunoprecipitate (Fig. 4).
What might be the role of the four different complexes? Of the four unique large components, only Rfc1p is essential for viability. The RFC complex is thus absolutely required to carry out regular DNA replication. The role of the other three complexes is not yet clear. Although rad24, ctf18, and elg1 were isolated as checkpoint, chromosome segregation, and hyperrecombination mutants, respectively, it is clear that the function of these genes overlaps considerably. Deletion of all three genes results in reduced viability and growth rates, indicating that they function redundantly in some crucial function that is required during normal growth. The existence of a fourth RLC on which the survival of the triple mutant depends cannot be ruled out at this point.
The MMS and HU survival curves show, however, that the three RLCs analyzed do not play identical roles in coping with different insults. For example, ctf18 rad24 mutants, with a functional ELG1 gene, are significantly more HU-sensitive than elg1 ctf18 or elg1 rad24 strains. This increased sensitivity implies that in the absence of the other two RLCs, the Elg1 complex is relatively inefficient in conferring HU resistance. Similarly, elg1 ctf18 double mutants are more resistant to MMS than any of the double mutants lacking Rad24p. Still, the triple mutant is much more sensitive to both HU and MMS than the double mutants, implying that the three proteins contribute to survival after DNA damage. Thus, although carrying out overlapping functions, it seems likely that each RLC may differentially contribute to survival under specific circumstances.
An attractive hypothesis is that the RLCs may be required for the loading of specific DNA polymerases. Several polymerases have been described that play various roles related to genome stability. They include error-prone and error-free translesion polymerases, including pol ζ, pol η, Rev1, and pol σ (reviewed in ref. 36). The RLCs may act similarly to RFC, loading PCNA-related complexes that act as clamps for specific polymerases. Consistent with this idea, it has recently been shown that SpRad17, the Schizosaccharomyces pombe homologue of Rad24p, is required for the loading of the DinB (pol κ) translesion polymerase (37).
The identification of an additional RLC requires a reevaluation of the function of the related complexes. ctf18 and elg1 mutants share many phenotypes, including an enhanced level of chromosome loss. Our results are consistent with a role for both RLCs in DNA replication-associated repair. Moreover, mutations in both ELG1 and CTF18 cause lethality in the absence of Ctf4p, as do mutations in genes encoding well characterized replication factors (31). We favor a model by which Ctf18p and Elg1p function in the loading or unloading of different DNA polymerases during DNA replication, according to particular cellular needs. Regarding Rad24p, its crucial function appears to reside in its role in DNA processing, rather than in its effect on cell cycle progression control (38). Similarly, the Ctf18 and Elg1 RLCs may affect still-to-be-defined global cellular processes, perhaps through signals that convey information on sister chromatid cohesion or chromatin status.
Further functional characterization of the different RLCs will undoubtedly be highly informative about the mechanisms that link and control DNA replication, recombination, and repair, and will shed light on their influence on cell cycle control and genome stability.
Acknowledgments
We thank Forrest Spencer for the generous gift of strains, Yael Aylon for excellent ideas and help, Anat Krauskopf and Sarit Smolikov for sharing unpublished information, and all of the members of the Kupiec laboratory for helpful comments and support. This work was supported by a grant from the Israel Science Foundation (to M.K.).
Abbreviations: RFC, replication factor C; RLC, RFC-like complex; PCNA, proliferating cell nuclear antigen; CAN, canavanine; CanS, CAN-sensitive; CanR, CAN-resistant; MMS, methyl methanesulfonate; HU, hydroxyurea; DSB, double-strand break; USCE, unequal sister chromatid exchange; HA, hemagglutinin; IP, immunoprecipitation; pol X, DNA polymerase X; DRR, direct-repeat recombination.
References
- 1.Hartwell, L. H. & Kastan, M. B. (1994) Science 266, 1821–1828. [DOI] [PubMed] [Google Scholar]
- 2.Green, C. M., Erdjument-Bromage, H., Tempst, P. & Lowndes, N. F. (2000) Curr. Biol. 10, 39–42. [DOI] [PubMed] [Google Scholar]
- 3.Lindsey-Boltz, L. A., Bermudez, V. P., Hurwitz, J. & Sancar, A. (2001) Proc. Natl. Acad. Sci. USA 98, 11236–11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venclovas, C. & Thelen, M. P. (2000) Nucleic Acids Res. 28, 2481–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burtelow, M. A., Roos-Mattjus, P. M., Rauen, M., Babendure, J. R. & Karnitz, L. M. (2001) J. Biol. Chem. 276, 25903–25909. [DOI] [PubMed] [Google Scholar]
- 6.Hanna, J. S., Kroll, E. S., Lundblad, V. & Spencer, F. A. (2001) Mol. Cell. Biol. 21, 3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer, M. L., Gygi, S. P., Aebersold, R. & Hieter, P. (2001) Mol. Cell 7, 959–970. [DOI] [PubMed] [Google Scholar]
- 8.Naiki, T., Kondo, T., Nakada, D., Matsumoto, K. & Sugimoto, K. (2001) Mol. Cell. Biol. 21, 5838–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolodner, R. D., Putnam, C. D. & Myung, K. (2002) Science 297, 552–557. [DOI] [PubMed] [Google Scholar]
- 10.Klein, H. L. (2001) Genetics 159, 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Symington, L. S. (1998) Nucleic Acids Res. 26, 5589–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debrauwere, H., Loeillet, S., Lin, W., Lopes, J. & Nicolas, A. (2001) Proc. Natl. Acad. Sci. USA 98, 8263–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kupiec, M. & Petes, T. D. (1988) Genetics 119, 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liefshitz, B., Parket, A., Maya, R. & Kupiec, M. (1995) Genetics 140, 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liefshitz, B., Steinlauf, R., Friedl, A., Eckardt-Schupp, F. & Kupiec, M. (1998) Mutat. Res. 407, 135–145. [DOI] [PubMed] [Google Scholar]
- 16.Ross-Macdonald, P., Coelho, P. S., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K. H., Sheehan, A., Symoniatis, D., Umansky, L., et al. (1999) Nature 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 17.Koren, A., Ben-Aroya, S., Steinlauf, R. & Kupiec, M. (2003) Curr. Genet. 43, 62–69. [DOI] [PubMed] [Google Scholar]
- 18.Parket, A. & Kupiec, M. (1992) Mol. Cell. Biol. 12, 4441–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parket, A., Inbar, O. & Kupiec, M. (1995) Genetics 140, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montelone, B. A. & Liang-Chong, B. C. (1993) Curr. Genet. 24, 481–486. [DOI] [PubMed] [Google Scholar]
- 21.Manthey, G. M. & Bailis, A. M. (2002) Mol. Cell. Biol. 22, 5347–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sung, P., Trujillo, K. M. & Van Komen, S. (2000) Mutat. Res. 451, 257–275. [DOI] [PubMed] [Google Scholar]
- 23.Cullmann, G., Fien, K., Kobayashi, R. & Stillman, B. (1995) Mol. Cell. Biol. 15, 4661–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinert, T. A., Kiser, G. L. & Hartwell, L. H. (1994) Genes Dev. 8, 652–665. [DOI] [PubMed] [Google Scholar]
- 25.Spencer, F., Gerring, S. L., Connelly, C. & Hieter, P. (1990) Genetics 124, 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouprina, N., Kroll, E., Kirillov, A., Bannikov, V., Zakharyev, V. & Larionov, V. (1994) Genetics 138, 1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 28.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 29.Miles, J. & Formosa, T. (1992) Mol. Cell. Biol. 12, 5724–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouprina, N., Kroll, E., Bannikov, V., Bliskovsky, V., Gizatullin, R., Kirillov, A., Shestopalov, B., Zakharyev, V., Hieter, P., Spencer, F., et al. (1992) Mol. Cell. Biol. 12, 5736–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Formosa, T. & Nittis, T. (1999) Genetics 151, 1459–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuzminov, A. (2001) Proc. Natl. Acad. Sci. USA 98, 8461–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholes, D. T., Banerjee, M., Bowen, B. & Curcio, M. J. (2001) Genetics 159, 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowndes, N. F., Johnson, A. L., Breeden, L. & Johnston, L. H. (1992) Nature 357, 505–508. [DOI] [PubMed] [Google Scholar]
- 35.Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedberg, E. C., Wagner, R. & Radman, M. (2002) Science 296, 1627–1630. [DOI] [PubMed] [Google Scholar]
- 37.Kai, M. & Wang, T. S. (2003) Genes Dev. 17, 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aylon, Y. & Kupiec, M. (2003) Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]