Abstract
Thiol-containing compounds, such as glutathione and cysteine, react with selenite under specific conditions to form selenotrisulfides. Previous studies have focused on isolation and characterization of intermolecular selenotrisulfides. This study describes the preparation and characterization of intramolecular selenotrisulfide derivatives of lipoic acid and lipoamide. These derivatives, after separation from other reaction products by reverse-phase HPLC, exhibit an absorbance maximum at 288 nm with an extinction coefficient of 1,500 M−1⋅cm−1. The selenotrisulfide derivative of lipoic acid was significantly stable at or below pH 8.0 in contrast to several other previously studied selenotrisulfides. Mass spectral analysis of the lipoic acid and lipoamide derivatives confirmed both the expected molecular weights and also the presence of a single atom of selenium as revealed by its isotopic distribution. The selenotrisulfide derivative of lipoic acid was found to serve as an effective substrate for recombinant human thioredoxin reductase as well as native rat thioredoxin reductase in the presence of NADPH. Likewise, the lipoamide derivative was efficiently reduced by NADH-dependent bovine lipoamide dehydrogenase. The significant in vitro stability of these intramolecular selenotrisulfide derivatives of lipoic acid can serve as an important asset in the study of such selenium adducts as model selenium donor compounds for selenophosphate biosynthesis and as rate enhancement effectors in various redox reactions.
The reaction of sulfhydryl compounds with selenite to yield selenotrisulfides was first described by Painter in a study using cysteine and mercaptoethanol (1). As a result of his initial work, this reaction later became known as the Painter reaction. Ganther (2), using reduced glutathione as the thiol, successfully isolated selenodiglutathione (GSSeSG), which was formed on reacting glutathione with selenite in a molar ratio of 4:1 under acidic conditions, thus corroborating the results of Painter. The first detailed reaction mechanism for the Painter reaction was proposed by Kice et al. (3), who used a butanethiol derivative as a model compound. More recently, a selenotrisulfide derivative of penicillamine, 3-mercaptovaline, has been isolated and characterized (4). Each of these selenotrisulfides is formed most efficiently under acidic conditions. At pH values near neutrality they undergo rapid decomposition, releasing the oxidized dithiol and selenium (5).
GSSeSG formed from selenite and the naturally abundant thiol, glutathione, has been proposed by some investigators to be an intermediate in the reduction of selenium in vivo (6, 7) particularly because this selenotrisulfide was shown to be reduced by NADH-dependent glutathione reductase (5) and also by NADPH-dependent thioredoxin reductase (TrxR) (8). In these reduction reactions the initial products are presumed to be a reduced thiol and a perselenide derivative. The perselenide derivative is unstable and is decomposed rapidly to elemental selenium and the oxidized thiol. Stabilization of the perselenide intermediate by conversion to its carboxymethyl derivative was accomplished by immediate reaction with iodoacetate (5). In vivo, the reaction of selenite with various low molecular weight thiols to form selenotrisulfides followed by enzymatic reductions then might feed into the pathway for selenocysteine biosynthesis and eventually selenophosphate formation (9). Alternatively, a perselenide intermediate could donate selenium directly to a rhodanese-type of enzyme that could act as a selenotransferase to deliver selenium to selenophosphate synthetase. In support of this reduction scheme, Gailer et al. (10) recently reported the isolation of seleno-bis(S-glutathionyl) arsinium ion from the bile of rabbits after injection with selenite and arsenite. Although this isolation of a compound containing glutathione and selenium, apparently stabilized by arsinium ion, is promising, a definitive pathway for the reduction of selenium in vivo under normal conditions remains elusive.
Lipoic acid is the covalently bound dithiol cofactor (11–13) for the α-keto acid dehydrogenase enzyme complexes, i.e., pyruvate dehydrogenase and α-ketoglutarate dehydrogenase, as well as the glycine cleavage system (14). Keto acid dehydrogenases are essential enzymes in plants, animals, and bacteria, and thus lipoic acid is synthesized and present in both eukaryotes and prokaryotes. In the lipoic acid biosynthetic pathway the intermediate octanoic acid is known to be derived from acetyl CoA but the mechanism of introduction of the two thiol groups in positions 6 and 8 is incompletely characterized. The ligation of lipoic acid in amide linkage to the various apoenzymes has been studied best in Escherichia coli (15–17). Lipoamide dehydrogenase (LD) is responsible for reducing the oxidized lipoamide generated during the reaction cycle of the keto acid dehydrogenase complex (18). The role of lipoic acid as a covalently bound cofactor is thought to be its major function in vivo. However, as pointed out by Packer in a recent review (19), free oxidized lipoic acid is taken up rapidly by cells and can be reduced to dihydrolipoic acid by glutathione reductase, TrxR, and LD, thus making it available for action as a low molecular weight thiol in other cellular functions.
In addition to being a required cofactor for keto acid dehydrogenases, it has been proposed that lipoic acid may be involved in the redox regulation of transcription factors (20) as well as serving as a scavenger of hydroxyl radicals (21). Although reduced glutathione is present in most cells in higher concentrations than dihydrolipoic acid (DHLA), it has been reported that lipoic acid supplementation can overcome deleterious effects of the redox imbalance created when glutathione biosynthesis is inhibited by buthionine sulfoximine in an animal model (22). Whether lipoate has any role as a redox-sensitive mediator of regulation or in protection from oxidative stress in living systems under normal conditions is still somewhat speculative. However, lipoic acid is one of the relatively few small molecule thiols present in living systems and as such must not be excluded as a possible player in the reduction of selenite, regulation of redox-sensitive transcription factors, and general defense against oxidative stress.
The present study describes the synthesis and characterization of selenotrisulfide derivatives of lipoic acid and lipoamide.
Materials and Methods
Materials.
α-Lipoic acid and lipoamide were purchased from Fluka, and sodium selenite was from Sigma. 75Se-selenite was obtained from the University of Missouri Reactor Center, Columbia. Bovine LD was purchased from Sigma. TrxR isolated from rat liver was a gift of Seung-Rock Lee, National Institutes of Health (23), and human TrxR (hTrxR) expressed in Escherichia coli (23) was a gift of Shoshana Bar-Noy, National Institutes of Health. All other reagents were of the highest grade available.
Methods.
Spectra of reaction mixtures or changes in absorption in enzyme assays were recorded by using a Milton Roy (Rochester, NY) Spectronic 3000 diode-array spectrophotometer. Spectra of purified selenium-derivatives of lipoic acid and lipoamide were recorded by using a diode array spectrophotometer integrated into a series 1050 Hewlett–Packard HPLC system, which was used for all HPLC analyses.
Preparation of DHLA and Dihydrolipoamide (DHLN).
Reduction and isolation of DHLA and DHLN were preformed essentially as reported (24), with the exception that toluene instead of benzene was used to extract DHLA or DHLN after reduction by sodium borohydride. Toluene was evaporated, and the reduced compound subsequently was suspended in a solution of 100% EtOH and stored in the dark at −20°C. The concentrations of DHLA and DHLN were determined spectrophotometrically by using Ellman's reagent (25).
Synthesis and Purification of Selenotrisulfide Derivatives.
A solution of sodium selenite (20 mM) in 0.1 M HCl (pH 1.0) was diluted 1:1 with a 40 mM solution of DHLA or DHLN in EtOH. After 15 min at 25°C the reaction mixture was applied to either a preparative C18 HPLC column (Jones Chromatography, Lakewood, CO) for large-scale separations or an analytical C18 column (Hewlett–Packard) for analytical determination of products. After washing with H2O containing 0.1% trifluoroacetic acid (TFA), adsorbed oxidized lipoic acid (or lipoamide) and selenium containing derivatives were eluted at a flow rate of 2.0 ml/min (0.5 ml/min for an analytical column) using a linear gradient of 0–100% MeOH (30 min) in 0.1% TFA. In each case a major selenium-containing derivative (based on parallel experiments using 75Se-selenite) was eluted at approximately 80% methanol. This product accounted for more than 90% of the total selenium present in the reaction products. The selenium derivatives, stored as isolated in 80% MeOH-TFA at −20°C, were stable for at least 3 months. After an extended period some elemental selenium was detected as a precipitate, but this represented a loss of less than 5% over a 3-month period. Oxidized lipoic acid (or lipoamide) was eluted in the methanol gradient at approximately 72% MeOH.
Enzymatic Reduction of Selenotrisulfide Derivatives.
Reaction of the selenotrisulfide derivative of DHLA (LASe) with recombinant hTrxR or rat liver TrxR and NADPH (1.0 mM) was carried out at 25°C in 100 mM sodium phosphate buffer (pH 7.0). Reaction of the selenotrisulfide derivative of DHLN (LNSe) with bovine LD and NADH was performed in sodium phosphate buffer (pH 6.5) at 25°C. Reduction of oxidized lipoic acid by recombinant hTrxR or rat liver TrxR and NADPH (0.2 mM) in 100 mM sodium phosphate buffer was followed after addition of enzyme by monitoring the decrease in absorbance at 340 nm. The reduction of LASe or LNSe was followed by reverse-phase HPLC analysis as described above for the synthetic reactions. The peak area at 288 nm for either LASe or LNSe was recorded and converted into a percentage of the peak area obtained in the absence of enzyme.
Mass Spectral Analysis.
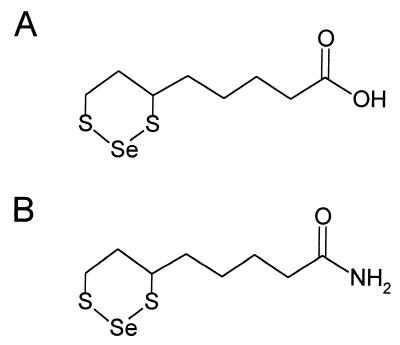
Positive ion electron impact spectra were obtained on a JEOL SX102 mass spectrometer operating at 70 eV. Negative ion chemical ionization spectra were determined on a Finnigan quadrupole mass spectrometer using ammonia as reactant gas. From both methods, a molecular ion at m/z 286 was observed for LASe (see Fig. 4A). The isotopic distribution was consistent with the natural abundance of selenium isotopes, thus indicating the presence of a single selenium atom in a molecular formula of C8H1402S2Se. Similarly, a molecular ion at m/z 285 (C8H15ONS2Se) was revealed for LNSe (see Fig. 4B).
Figure 4.
Proposed structures of selenotrisulfide derivatives. (A) Schematic representation of LASe. (B) Schematic representation of LNSe.
Results
Reaction of Selenite with DHLA and DHLN.
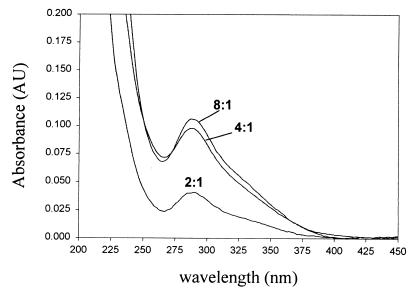
Based on previous findings that acidic conditions were more favorable for preparation and stabilization of selenotrisulfides, initial attempts to prepare selenotrisulfide derivatives of lipoic acid consisted of mixing equal volumes of aqueous 1 mM Na2SeO3 in 0.1 M HCl with various concentrations of DHLA in ethanol. Spectral analysis of the reaction products formed during 15 min at room temperature revealed a strong increase in absorbance at 288 nm (Fig. 1). Oxidized lipoic acid presumably accounted for the shoulder at 333 nm. From these experiments a 4:1 ratio of sulfhydryl equivalents to selenite (Fig. 1) corresponding to a molar ratio of two DHLA to one SeO32− appeared to be optimal for the formation of a compound with significant absorbance at 288 nm. These results are similar to those previously reported for the reaction of SeO32− with other thiol-containing compounds (1, 2, 4), and suggested that a selenotrisulfide having an absorption maximum at 288 nm might be the primary reaction product. No significant changes in absorbance were observed when DHLA was replaced with oxidized lipoic acid.
Figure 1.
Spectral analysis of the reaction of selenite with DHLA. Spectra were recorded after addition of 10 μl of a reaction mixture containing DHLA and selenite in an acidic solution of 50% EtOH (Materials and Methods) to 1 ml of 100% EtOH. Ratios given for each spectrum represent the molar ratio of free sulfhydryl to selenium in the reaction mixture.
Separation of Reaction Products Using Reverse-Phase HPLC.
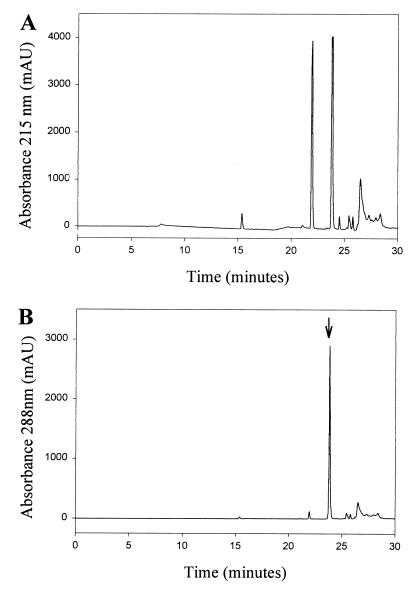
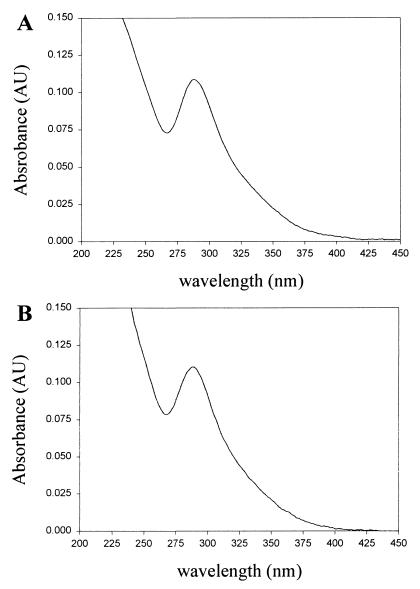
For the initial purification and characterization of the product absorbing at 288 nm a portion (0.1 ml) of a reaction mixture containing a 2:1 molar ratio of DHLA and selenite (20 mM/10 mM) was subjected to HPLC analysis using an analytical C18 column. Hydrophobic compounds bound to the column were eluted in a linear methanol gradient (0–100%) in 0.1% trifluoroacetic acid. The elution pattern monitored at 215 nm showed two major peaks (Fig. 2A). A peak at 22 min exhibited the characteristic spectrum of oxidized lipoic acid (maximum at 333 nm) and a second peak at 24 min (Fig. 2B) had an absorbance maximum at 288 nm (Fig. 3A). From this analysis it was clear that the major compound produced in the reaction of selenite with DHLA had a peak absorbance of 288 nm and was slightly more hydrophobic than oxidized lipoic acid. These results supported an intramolecular selenotrisulfide derivative of DHLA (Fig. 4A) as the reaction product. Using 75Se-labeled SeO32− and higher concentrations of SeO32− (10 mM) and DHLA (20 mM), the putative selenotrisulfide formed in the reaction mixture was separated chromatographically as above and examined for the presence of radioactive selenium. More than 90% of the 75Se eluted in the methanol gradient was present in the major peak fraction that absorbed at 288 nm (data not shown). This clearly demonstrated that selenium had been incorporated into the lipoic acid derivative, presumably forming a selenotrisulfide. A stoichiometry of one selenium per lipoic acid molecule was confirmed by mass spectral analysis of the isolated product. As expected, a mass of m/z 286 was determined for the Se-derivative of DHLA, corresponding to the structure shown in Fig. 4A (data not shown). The characteristic isotopes of selenium also were revealed by this analysis, thus confirming the identification of an intramolecular selenotrisulfide derivative of lipoic acid. With the knowledge that one atom of selenium is present in each derivative, a molar extinction coefficient of 1,500 at 288 nm was calculated for the selenium derivative of lipoic acid based on the activity of the 75Se-selenite used in the synthesis of the derivative.
Figure 2.
Reverse-phase HPLC analysis of the products formed by reaction of selenite with DHLA. Spectrophotometric analysis of reaction products monitored at (A) 215 nm and (B) 288 nm. Arrow in B indicates the peak with absorption maximum at 288 nm, which corresponds to the selenium-containing derivative of DHLA. No compounds with significant absorbance at 288 nm were detected in the elution profile before the application of the MeOH gradient. Chromatographic details are described in Materials and Methods.
Figure 3.
Absorption spectra of selenotrisulfide derivatives. (A) Selenium-containing derivative of DHLA. (B) Selenium-containing derivative of DHLN. Spectra were recorded during HPLC analysis using an integrated Diode array spectrophotometer in Hewlett–Packard series 1050 HPLC system.
To prepare the corresponding selenotrisulfide of the amide derivative of lipoic acid, DHLN and selenite were reacted under acidic conditions, and the products were analyzed as described in Materials and Methods. The HPLC elution pattern of a reaction mixture containing DHLN and selenite was similar to the analogous mixture containing DHLA. A compound formed during this reaction, which also had an absorption maximum at 288 nm (Fig. 3B), was purified and analyzed by mass spectrometry. As expected a mass of m/z 285, identical to that predicted for the structure shown in Fig. 4B, was determined. The molar absorption coefficient calculated as above for the lipoic acid derivative was also found to be approximately 1,500 at 288 nm. These compounds are termed LASe (Fig. 4A) and LNSe (Fig. 4B).
Stability of LASe at Varying pH Values.
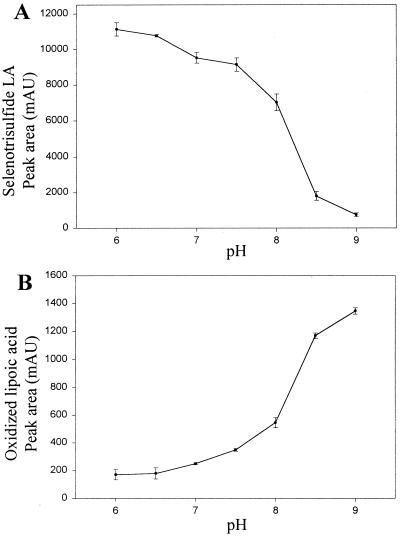
Previously isolated selenotrisulfides, such as GSSeSG, are relatively unstable at physiological (neutral) pH (2, 3). The stability of LASe, 0.27 mM, in aqueous buffers of varying pH value during 15 min at 25°C is shown in Fig. 5A. Under these conditions significant decomposition is observed at pH 7.5–8.0 and above and, in contrast to other isolated selenotrisulfides, LASe is remarkably stable at neutral pH. Loss of LASe as a function of increasing pH was accompanied by concomitant increases in the level of oxidized lipoic acid (Fig. 5B).
Figure 5.
pH stability of LASe. (A) Total area units of the peak representing LASe recorded during HPLC analysis are plotted as a function of the pH of the buffer in which LASe was incubated. (B) Total area units of the peak representing oxidized lipoic acid released from LASe are plotted as a function of the pH of the buffer in which LASe was incubated. The area units given in both plots are the average of at least three independent experiments, and the error bars represent one standard deviation from the mean. For LASe, the absorbance was followed at 288 nm, and for oxidized lipoic acid at 333 nm. Other experimental details are described in Materials and Methods.
The peak area (arbitrary units) of oxidized lipoic acid (measured at 333 nm) produced during base decomposition of LASe was approximately 9-fold lower than the peak area corresponding to LASe. The molar extinction coefficient for oxidized lipoic acid measured at 333 nm is 150 (26). Thus approximately 1 mole (0.9) of oxidized lipoic acid is released on incubation of LASe in aqueous buffer above pH 8.0. The fact that this ratio of peak area units is almost 10-fold higher also corroborates the calculated extinction coefficients determined for LASe.
Enzymatic Reduction of LASe by Recombinant hTrxR.
GSSeSG previously has been shown to be reduced by glutathione reductase and TrxR (5, 8). TrxR also has been shown to reduce oxidized lipoic acid (27). To test the ability of TrxR preparations to reduce LASe, reaction mixtures containing 100 mM sodium phosphate buffer (pH 7), 0.5 mM NADPH, and 0.3 mM LASe were supplemented with either E. coli expressed hTrxR or native rat liver TrxR. Within 5 min the solutions became bright yellow in color and within 15 min a red color developed. In the absence either of NADPH or enzyme there was no color change, indicating the ability of TrxR to catalyze the reduction of LASe. However, when the reactions were monitored spectrophotometrically both an overall increase at 333 nm and a decrease at 340 nm was observed, hampering any attempt to quantitate the rate of reduction by following the loss of NADPH at 340 nm. Light scattering also caused an increase in absorbance presumably because of the formation of elemental selenium, which was observed to precipitate at the end of the reaction.
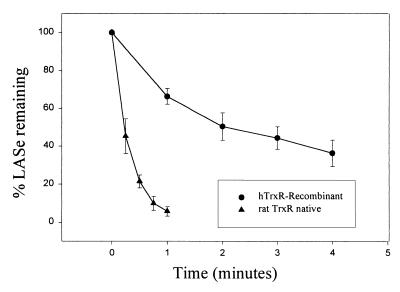
Because these color and light scattering changes complicated the spectrophotometric determination of enzyme activity based on oxidation of NADPH, reverse-phase HPLC analysis was used to measure the reduction of LASe by TrxR. After incubation of enzymes with NADPH and substrate in sodium phosphate buffer for various times, aliquots were withdrawn and immediately made to 200 mM with iodoacetic acid. Under these conditions there was immediate termination of the reaction and stabilization of reaction products for HPLC analysis. Shown in Fig. 6 are the rates of reduction of LASe by recombinant hTrxR (1 μg) and by native rat liver TrxR (1 μg) determined by HPLC analysis. The much more rapid rate of reduction of LASe by the native rat liver enzyme, about 500 μmol⋅min−1⋅mg−1 protein, as compared with the rate with the E. coli expressed hTrxR, about 115 μmol⋅min−1⋅mg−1 protein, is comparable to the relative differences in rates of reduction of a standard disulfide substrate, 5,5′ dithiobis(2-nitrobenzoic acid), observed with these two enzyme preparations (4 and 42 μmol⋅min−1⋅mg−1 protein for hTrxR and rat TrxR, respectively). In similar experiments using hTrxR it was found that with increasing concentrations of LASe as substrate there was a linear increase in activity between 100 μM and 1 mM (data not shown), indicating that the apparent maximum velocity for the reaction had not been reached. From these experiments it is clear that the selenium adduct of lipoic acid serves as an excellent substrate for TrxR.
Figure 6.
Reduction of LASe by recombinant hTrxR and rat liver TrxR. Reaction mixtures (1 ml), containing 0.1 M sodium phosphate buffer (pH 7.0), 1.0 mM, and 300 μM LASe were incubated with recombinant hTrxR [1 μg, 4 units of activity with 5,5′ dithiobis(2-nitrobenzoic acid) as substrate] or native rat TrxR [1 μg, 42 units of activity with 5,5′ dithiobis(2-nitrobenzoic acid) as substrate]. Samples were taken at the indicated time intervals and stabilized for HPLC analysis by reaction with 200 mM iodoacetic acid. The area of the peak representing LASe determined for each time point by HPLC analysis was converted to a percentage of the initial LASe peak area of a sample taken before enzyme addition. The area units given in both plots are the average of at least three independent experiments, and the error bars represent one standard deviation from the mean.
For comparison the rates of reduction of oxidized lipoic acid by the two TrxR preparations were determined under comparable conditions. With reaction mixtures containing 100 mM sodium phosphate buffer (pH 7.0), 1.0 mM oxidized lipoic acid and 0.2 mM NADPH the apparent specific activities of the recombinant hTrxR and the native rat liver TrxR were 57 and 150 nmol⋅min−1⋅mg−1 protein, respectively. Thus LASe is reduced by native rat liver TrxR at a 3,300-fold higher rate in comparison to oxidized lipoic acid, and recombinant hTrxR reduced LASe at a 2,000-fold higher rate. These results clearly demonstrate that the reduction of LASe is significantly more efficient than the reduction of oxidized lipoic acid by TrxR, and that the presence of a S—Se bond within the selenotrisulfide apparently causes a significant rate enhancement with respect to the activity of TrxR.
Reduction of LASe by Bovine LD.
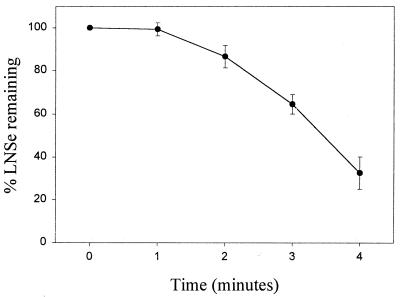
LD also used LNSe as substrate as determined by HPLC analysis similar to that described for the analysis of the reduction of LASe in the previous section. Visually the LNSe solution turned yellow on reduction with LD in the presence of NADH, and a precipitate of red elemental selenium was formed. Fig. 7 illustrates the loss of the LNSe compound observed over time when incubated with of 33 ng of LD and 1.0 mM NADH. A 1- to 2-min lag period was followed by a rapid rate of reduction. The amount of LD enzyme used, 33 ng, corresponds to 0.003 unit of activity on lipoamide or a rate of reduction of 3 nmol/min. During an approximately linear rate of reduction of LNSe between 3 and 4 min (Fig. 7) the amount of substrate decreased from 180 to 90 nmol. Thus a rate of 90 nmol/min with LNSe as substrate can be compared with a rate of 3 nmol/min with lipoamide under similar conditions, or a 33-fold rate enhancement when LNSe is used as substrate. It may be noted that LD, unlike TrxR, does not contain selenocysteine. The lower rate enhancement observed with LD using LNSe as a substrate may reflect the greater efficiency of selenium as a catalyst. This would explain the 100-fold increase in the rate enhancement when both enzyme and substrate contain selenium (3,300-fold increase with rat TrxR reducing LASe) as compared with only one molecule containing selenium (33-fold increase with LD reducing LNSe).
Figure 7.
Reduction of LNSe by bovine LD. Reaction mixtures contained 0.1 M sodium phosphate buffer (pH 6.5), 1.0 mM NADPH, 300 μM LNSe, and 33 ng LD. After removal of an initial aliquot before addition of enzyme, samples were taken at 1-min intervals up to 4 min. The area of the peak representing LNSe determined for each time point by HPLC analysis was converted to a percentage of the initial LNSe peak. The area units given are the average of at least three independent experiments, and the error bars represent one standard deviation from the mean.
Discussion
The intramolecular selenotrisulfide containing the 6,8 dithiol groups of lipoic acid described in the present study represents a type of selenium adduct involving a widely distributed small molecular mass biological thiol. Normally, in vivo levels of free lipoic acid are very low and most of the cofactor is bound in amide linkage to lysine residues in certain enzymes. However, numerous current studies of Lester Packer and associates (19–22) and other investigators on the efficacy of lipoic acid as an antioxidant when added to cultured mammalian cells or administered to test animals indicates a possible clinical use. Under such conditions significant levels of the free dithiol would be available to react with selenium compounds also commonly administered as antioxidants. Both selenium and lipoic acid are now being marketed to consumers as over-the-counter antioxidants, and thus the possibility that selenotrisulfide compounds such as LASe or GSSeSG could form in vivo and what effects these compounds might have on biological systems should be investigated.
The synthesis and characterization of LASe and LNSe in this report documents the isolation of intramolecular selenotrisulfide compounds. These derivatives of lipoic acid are significantly more stable than other previously isolated selenotrisulfides, perhaps owing to both the hydrophobic nature of lipoic acid as well as the stability of the six-membered ring formed during the formation of the selenotrisulfide. As a model compound, LASe may provide clues necessary for attainment of more stable perselenide derivatives for study in vitro. This type of perselenide is postulated as part of the active site of native selenium-dependent molybdenum hydroxylase enzymes such as xanthine dehydrogenase and purine hydroxylase from Clostridia species (28, 29). Part of the reasoning for this type of compound comes from information gained during the recent structural analysis of CO dehydrogenase (30), in which a required modified cysteine, termed a selanylcysteine, was identified. Future investigations may uncover a variety of new enzymes that require selenium in a form other than a selenocysteine residue.
Likewise, the study of the mechanism of redox-active enzymes like TrxR and glutathione peroxidase, which are critical in the defense of oxidative stress in mammalian systems, can be facilitated by the use of alternative substrates like LASe in vitro. The reduction and oxidation of S—S and S—Se bonds have been found to be of great significance with respect to the overall redox state of the cell and will continue to be the focus of the study of selenium-dependent enzymes well into the future. Equally as important is the elucidation of the in vivo reduction of selenite for entrance into the pathways for incorporation into proteins in the form of selenocysteine or labile selenium cofactors as well as modification of tRNAs. Although not demonstrated definitively in vitro, it has been presumed by many investigators that the Painter reaction is involved in the reduction of selenium in vivo. Therefore detailed analysis of the reaction of biologically relevant thiol-containing compounds with selenite may prove fruitful with respect to the many questions still to be answered regarding utilization of selenium in biological systems.
Acknowledgments
We thank Dr. Noel F. Whittaker, Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health for mass spectroscopic determinations. We thank Dr. Shoshana Bar-Noy for providing the recombinant hTrxR and Dr. Seung-Rock Lee for providing the rat liver TrxR.
Abbreviations
- GSSeSG
selenodiglutathione
- DHLA
dihydrolipoic acid
- DHLN
dihydrolipoamide
- TrxR
thioredoxin reductase
- hTrxR
human TrxR
- LD
lipoamide dehydrogenase
- LASe
selenotrisulfide derivative of lipoic acid
- LNSe
selenotrisulfide derivative of lipoamide
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220426897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220426897
References
- 1.Painter H E. Chem Rev. 1941;28:179–213. [Google Scholar]
- 2.Ganther H E. Biochemistry. 1968;7:2898–2905. doi: 10.1021/bi00848a029. [DOI] [PubMed] [Google Scholar]
- 3.Kice J L, Lee T W S, Pan S. J Amer Chem Soc. 1980;102:4448–4455. [Google Scholar]
- 4.Nakagawa T, Aoyama E, Kobayashi N, Tanaka H, Chikuma M, Sakurai H, Nakayama M. Biochem Biophys Res Commun. 1988;150:1149–1154. doi: 10.1016/0006-291x(88)90749-8. [DOI] [PubMed] [Google Scholar]
- 5.Ganther H E. Biochemistry. 1971;10:4089–4098. doi: 10.1021/bi00798a013. [DOI] [PubMed] [Google Scholar]
- 6.Ganther H E. Carcinogenesis. 1999;20:1657–1666. doi: 10.1093/carcin/20.9.1657. [DOI] [PubMed] [Google Scholar]
- 7.Turner R J, Weiner J H, Taylor D E. Biometals. 1998;11:223–227. doi: 10.1023/a:1009290213301. [DOI] [PubMed] [Google Scholar]
- 8.Bjornstedt M, Kumar S, Holmgren A. J Biol Chem. 1992;267:8030–8034. [PubMed] [Google Scholar]
- 9.Lacourciere G M. Biofactors. 1999;10:237–244. doi: 10.1002/biof.5520100222. [DOI] [PubMed] [Google Scholar]
- 10.Gailer J, George G N, Pickering I J, Prince R C, Ringwald S C, Pemberton J E, Glass R S, Younis H S, DeYoung D W, Aposhian H V. J Am Chem Soc. 2000;122:4637–4639. [Google Scholar]
- 11.Reed L J, DeBusk B G, Gunsalus I C, Hornberger C S., Jr Science. 1951;114:93–94. doi: 10.1126/science.114.2952.93. [DOI] [PubMed] [Google Scholar]
- 12.Reed L J, DeBusk B G, Gunsalus I C, Schnakenberg G H F. J Am Chem Soc. 1951;73:5920. [Google Scholar]
- 13.Reed L J. In: The Enzymes. 2nd Ed. Boyer P D, Lardy H, Myerback K, editors. Vol. 3. New York: Academic; 1960. pp. 195–223. [Google Scholar]
- 14.Robinson J R, Sigrid M K, Sagers R D. J Biol Chem. 1973;248:5319–5323. [PubMed] [Google Scholar]
- 15.Jordan S W, Cronan J E., Jr J Biol Chem. 1997;272:17903–17906. doi: 10.1074/jbc.272.29.17903. [DOI] [PubMed] [Google Scholar]
- 16.Morris T W, Reed K E, Cronan J E., Jr J Bacteriol. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green D E, Morris T W, Green J, Cronan J E, Jr, Guest J R. Biochem J. 1995;309:853–862. doi: 10.1042/bj3090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reed L J, Cox D J. Annu Rev Biochem. 1966;35:57–84. [Google Scholar]
- 19.Packer L. Drug Metab Rev. 1998;30:245–275. doi: 10.3109/03602539808996311. [DOI] [PubMed] [Google Scholar]
- 20.Sen C K, Tirosh O, Roy S, Kobayashi M S, Packer L. Biochem Biophys Res Commun. 1998;247:223–228. doi: 10.1006/bbrc.1998.8764. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki Y J, Tsuchiya M, Packer L. Free Radical Res Commun. 1991;15:255–263. doi: 10.3109/10715769109105221. [DOI] [PubMed] [Google Scholar]
- 22.Maitra I, Serbinova E, Tritschler H J, Packer L. Biochem Biophys Res Commun. 1996;221:422–429. doi: 10.1006/bbrc.1996.0611. [DOI] [PubMed] [Google Scholar]
- 23.Lee S R, Bar-Noy S, Kwon J, Levine R L, Stadtman T C, Rhee S G. Proc Natl Acad Sci USA. 2000;97:2521–2526. doi: 10.1073/pnas.050579797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunsalus I C, Barton L S, Gruber W. J Am Chem Soc. 1956;78:1763–1766. [Google Scholar]
- 25.Ellman G L. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 26.Berltrop J A, Hayes P M, Calvin M. J Am Chem Soc. 1954;76:4348–4367. [Google Scholar]
- 27.Arner E S, Nordberg J, Holmgren A. Biochem Biophys Res Commun. 1996;225:268–274. doi: 10.1006/bbrc.1996.1165. [DOI] [PubMed] [Google Scholar]
- 28.Schrader T, Rienhofer A, Andreesen J R. Eur J Biochem. 1999;264:862–871. doi: 10.1046/j.1432-1327.1999.00678.x. [DOI] [PubMed] [Google Scholar]
- 29.Self W T, Stadtman T C. Proc Natl Acad Sci USA. 2000;97:7208–7213. doi: 10.1073/pnas.97.13.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobbek H, Gremer L, Meyer O, Huber R. Proc Natl Acad Sci USA. 1999;96:8884–8889. doi: 10.1073/pnas.96.16.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]