Abstract
Novel agents that target the proteasome, a proteolytic complex responsible for the degradation of ubiquitinated proteins, have demonstrated remarkable therapeutic efficacy in multiple myeloma, a plasma cell malignancy. However, the mechanism by which these compounds act remains unknown. A signaling pathway called the unfolded protein response (UPR) allows cells to handle the proper folding of proteins. The transcription factor XBP-1, a regulator of the UPR, is also required for plasma cell differentiation, suggesting a link between the UPR and plasma cell differentiation. Here we show that proteasome inhibitors target XBP-1 and the UPR in myeloma cells. Proteasome inhibitors suppress the activity of the translumenal endoplasmic reticulum endoribonuclease/kinase, IRE1α, to impair the generation of the active, spliced XBP-1 species and simultaneously stabilize the unspliced species that acts as a dominant negative. Myeloma cells rendered functionally deficient in XBP-1 undergo increased apoptosis in response to endoplasmic reticulum stress. Identification of compounds that target the activity of IRE1α/XBP-1 may yield novel therapies for the treatment of multiple myeloma and other malignancies that rely on an intact UPR.
Remarkable regression or stabilization of multiple myeloma (MM) in chemotherapy and stem cell transplant refractory patients has recently been observed with a series of novel drugs that inhibit the proteasome, a highly conserved multienzyme complex that destroys proteins covalently modified by ubiquitin (1–5). One such drug, PS-341, induces apoptosis of MM cells and interferes with their interaction with the stromal microenvironment and subsequent production of the MM survival cytokine IL-6 (6–9). In MM cells, PS-341 decreases levels of several antiapoptotic proteins, resulting in mitochondrial cytochrome c release and activation of caspase-9, jun kinase, and Fas-dependent pathways (9, 10). The molecular switch that initiates these apoptotic cascades, however, has not yet been defined.
Both the normal and malignant plasma cells produce and secrete abundant Igs. This requires a highly developed endoplasmic reticulum and the production of chaperone proteins that effect proper translation and folding. A signaling pathway called the unfolded protein response (UPR), or stress response, ensures that the plasma cells can handle the proper folding of Ig proteins (11). Three signaling pathways responsible for mediating the UPR have been described. Two of them involve the activation of transcription factors XBP-1 and ATF6, whereas the third depends on translational repression mediated by PERK/eIF2α.On sensing unfolded proteins, an endoplasmic reticulum (ER) transmembrane endoribonuclease and kinase called IRE1 oligomerizes, is activated by autophosphorylation, and uses its endoribonuclease activity to excise an intron from yeast Hac1p or mammalian XBP-1 mRNA, resulting in the conversion of a 267-aa unspliced XBP-1 protein to a 371-aa spliced XBP-1 protein (12–20). XBP-1 then translocates into the nucleus where it binds to its target sequence in the regulatory regions of the chaperone genes to induce their transcription.
We recently reported that XBP-1 is required for the generation of plasma cells (21) and that only the spliced XBP-1 species can reconstitute Ig secretion (22). The abundant expression of XBP-1 in myelomas suggested a role for it in perpetuating this malignancy (23) and raised the possibility that it was one molecular target of the novel anticancer compounds that target the proteasome.
Materials and Methods
Western Blot and Pulse–Chase Experiments. Cells were lysed in RIPA buffer (50 mM Tris, pH 7.4/150 mM NaCl/1 mM EDTA/1% Triton X-100/1% sodium deoxycholate/0.1% SDS), and lysates were subjected to SDS/PAGE and transferred to Hybond-P membranes (Amersham Biosciences). Blots were revealed by anti-XBP-1 (Santa Cruz Biotechnology), anti-caspase-12 (gift of J. Yuan, Harvard University, Boston), and anti-IRE1α (24) antibodies by standard procedures. HeLa cells were cotransfected with XBP-1u and His-tagged ubiquitin expression plasmids (pMT107, gift of D. Bohmann, EMBL, Heidelberg, Germany) by using Lipofectamine 2000 reagent (Invitrogen). Cell extracts were purified through Ni-NTA columns as described (25), and ubiquitinated XBP-1u proteins were revealed by Western blot analysis with anti-XBP-1 antiserum. Degradation rates of XBP-1u and -1s proteins were determined by pulse labeling J558 cells with [35S]Met/[35S]Cys for 1 h and chasing for the indicated times. Radiolabeled XBP-1 proteins were immunoprecipitated from total cell extracts, separated on 10% SDS/PAGE, and revealed by autoradiography.
Northern Blot and RT-PCR Analysis. Total RNA was prepared by using TRIzol reagent, electrophoresed on 1.2% agarose/6% formaldehyde gels, and then transferred onto Genescreen Plus membrane (NEN). Hybridizations with 32P-radiolabeled probes were performed as demonstrated (22). Probes for ERdj4 and p58IPK were generated from cDNA excised from EST clones (American Type Culture Collection) by using appropriate restriction enzymes (ERdj4, IMAGE:1920927; p58IPK, IMAGE:2646147). The ratio of XBP-1u to -1s mRNA was revealed by RT-PCR analysis with a probe set spanning the spliced-out region as demonstrated (22).
Plasmids and Reporter Assays. Two or three lysine residues in the C terminus of XBP-1u were replaced by arginines to generate XBP-1uKK (K235R, K252R) and XBP-1uKKK (K146R, K235R, K252R) by site-directed mutagenesis (22). dn-XBP contains the N-terminal 188 aa of XBP-1u. NIH 3T3 cells were transfected by using Lipofectamine 2000 reagent as recommended by the manufacturer (Invitrogen) with indicated amount of UPR element (UPRE) reporter (26) and various effector plasmids. Cells were treated for 16 h before harvest in certain experiments. Cells were lysed in passive lysis buffer for dual luciferase assays according to the manufacturer's protocol (Promega).
Production of iXBP-1 and dn-XBP-1 Myeloma Cells. An XBP-1-specific RNAi vector was constructed by inserting two complementary oligonucleotides for 5′-GGGATTCATGAATGGCCCTTA-3′ into the pBS/U6 vector as described (27). To make the SGFΔU3 shuttle retroviral vector for RNAi, a polylinker (PmlI, SalI, BamHI, and MluI) was inserted between the PmlI and BamHI sites of SFG tcLucECT3 (28). The neomycin resistance gene expression cassette was removed by PCR amplification from the pMCSV vector (Invitrogen) and inserted between the BamHI and MluI sites of SGFΔU3 to generate SGFΔU3neo. Lastly, the U6 promoter–iXBP cassette was excised from the pBS/U6-driven vector by SmaI and BamHI digestion and then inserted into SFGΔU3neo between the PmlI and BamHI sites to generate the SFGΔU3neo-iXBP retroviral vector. Retroviral supernatant was prepared and used to transduce J558 cells as described (22). Uninfected cells were removed by culturing cells in the presence of 1 mg/ml G418 for >1 week. Suppression of XBP-1 mRNA and proteins by RNAi was confirmed by Northern and Western blot analysis.
Apoptosis Assays. Cells were stained with annexin V-PE (BD Pharmingen) as recommended and analyzed on a FACScan flow cytometer (BD Biosciences).
Results
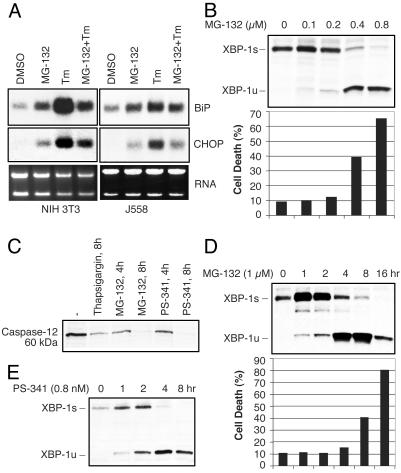
Proteasome Inhibitors (PIs) Induce ER Stress but Suppress the UPR in Myeloma Cells. The maturation and folding of ER membrane and secretory proteins relies on the activity of ER-resident chaperones and folding enzymes. ER proteins that ultimately fail to fold properly are degraded by the 26S proteasome, or ER-associated degradation (ERAD). Suppression of proteasome activity induces the accumulation of ERAD substrates in the ER, thereby inducing ER stress. To test the effect of proteasome dysfunction on UPR activation, NIH 3T3 fibroblasts and J558 myeloma cells were treated with the PI MG-132 in the presence or absence of the ER stress inducer, tunicamycin (Tm), and the expression of UPR target genes was assessed (Fig. 1A). As expected, Tm treatment resulted in the induction of expression of representative UPR target genes such as BiP (Grp78) and CHOP. Treatment of cells with PIs alone also induced UPR gene expression as reported (29, 30) (Fig. 1 A). PIs also induced caspase 12 activation, as evidenced by cleavage of the precursor species (Fig. 1C), confirming that the inhibition of proteasome activity induces ER stress and apoptotic signaling pathways (31). Surprisingly, however, PI treatment blocked rather than further augmented the Tm-induced stress response in both NIH 3T3 and J558 myeloma cells, raising the possibility that PIs might also suppress the UPR (Fig. 1 A).
Fig. 1.
PIs induce ER stress and caspase-12 activation, but suppress the UPR. (A) BiP and CHOP mRNA induction in NIH 3T3 or J558 myeloma cells treated with MG-132 (20 μM), Tm (10 μg/ml), or both. Cells were pretreated with MG-132 for 1 h and then further treated with Tm for 4 h. Ethidium bromide staining of the gel is shown at the bottom. (B) Alteration in the ratio of XBP-1 protein species in J558 cells treated with increasing amounts of MG-132 for 16 h. Cells undergoing apoptosis were counted by annexin V staining. (C) Inhibition of caspase-12 processing by PIs. Processing of full-length caspase-12 was examined by Western blotting in J558 myeloma cells treated with thap-sigargin (1 μM) or PIs (20 μM) during the indicated time periods. (D) Time course of the XBP-1s to -1u shift. Cells were treated with MG-132 (1 μM) for the indicated times, and XBP-1u and -1s protein levels and cell death were determined. (E) Alteration in the ratio of XBP-1 protein species in the MM.1s human myeloma cell line. Cells were treated with PS-341 (8 nM) in a time course analysis, and XBP-1 protein species were quantified.
PIs Prevent IRE1α-Mediated XBP-1 mRNA Splicing. Treatment of J558 cells, which express high levels of the active spliced form of XBP-1 (XBP-1s), with MG132 or PS-341 (data not shown) resulted in a striking accumulation of XBP-1u and a concomitant decrease in XBP-1s proteins at concentrations of MG-132 between 0.2 and 0.4 μM (Fig. 1B). A time course of the kinetics of induction and loss of the two XBP-1 species by MG-132 revealed that XBP-1s was induced at early time points but rapidly declined after 4 h of treatment and was barely detectable by 16 h (Fig. 1D). Conversely, XBP-1u levels were increased from as early as 1 h after treatment and were sustained throughout the experiment, peaking at 8 h (Fig. 1D). Of note, we found that MG-132 and PS-341 (not shown) induced apoptosis in J558 cells (Fig. 1 B and D). A close correlation between the dose dependency of the XBP-1s to -1u shift and apoptosis was observed, with the most marked increase in both occurring between 0.2 and 0.4 μM MG-132 (Fig. 1B). Similarly, the kinetics of XBP-1u accumulation and XBP-1s loss mirrored the kinetics of MG-132-induced apoptosis of these cells (Fig. 1D). PS-341 induced the same marked shift in the ratio of XBP-1s to -1u in the human MM cell line MM.1s (Fig. 1E) and in primary MM cells derived from patient bone marrow (not shown).
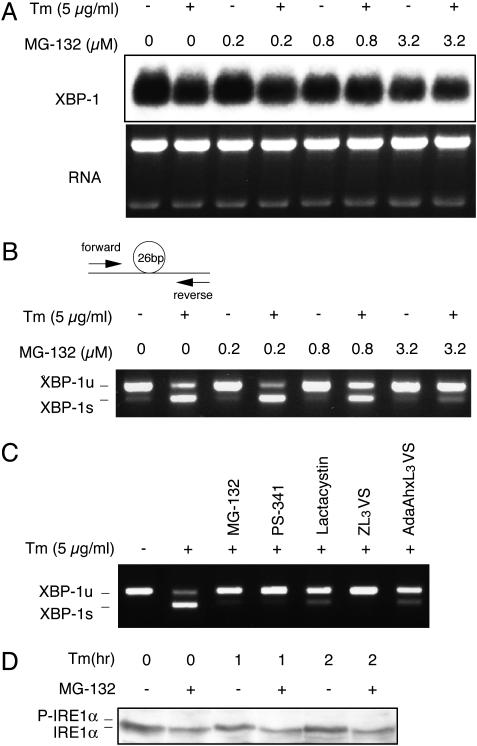
The disappearance of the spliced XBP-1s species suggested that PIs suppressed IRE1α-mediated XBP-1 mRNA splicing. Overall levels of XBP-1 RNA were not significantly altered by either Tm or MG-132 in J558 cells by Northern blot analysis, which does not distinguish between XBP-1u and -1s transcripts (Fig. 2A). Relative amounts of XBP-1u and -1s transcripts were measured by RT-PCR with a primer set that amplified 145 and 119 bp of XBP-1u and -1s mRNA, respectively (Fig. 2B). As expected, Tm treatment markedly induced XBP-1 mRNA splicing (Fig. 2B, first two lanes). MG-132 alone did not induce any XBP-1mRNA splicing, even after prolonged treatment up to 8 h at high concentrations (Fig. 2B and data not shown). Interestingly, however, Tm-induced XBP-1 mRNA splicing was suppressed by MG-132 in a dose-dependent manner, as reflected by a decrease of the ratio of XBP-1s to -1u forms (Fig. 2B). We conclude that the marked decrease in XBP-1s protein after MG-132 treatment results from suppression of IRE1-mediated XBP-1 mRNA splicing. To confirm that MG-132 inhibited XBP-1 splicing by targeting the proteasome, we tested a panel of compounds known to specifically inhibit proteasomal activity. PS-341, a reversible inhibitor of chymotryptic activity of the 20S proteasome complex, ZL3VS, and AdaAhx3L3VS, both of which efficiently target all β subunits of the proteasome (32), all suppressed XBP-1 mRNA splicing as efficiently as MG-132 (Fig. 2C), confirming that MG-132 inhibited XBP-1 splicing by targeting the proteasome.
Fig. 2.
Effect of PIs on IRE1α-mediated XBP-1 mRNA splicing. (A) XBP-1 mRNA levels in ER-stressed J558 cells treated with Tm for4hinthe absence or presence of MG-132. Cells were pretreated with MG-132 for 1 h before adding Tm. (B) The ratio of XBP-1u to -1s mRNA as revealed by RT-PCR analysis with a probe set spanning the spliced-out region as demonstrated (22). (C) Effect of a panel of PIs on XBP-1 splicing. Cells were treated with Tm for 4 h in the absence or presence of MG-132 (10 μM), PS-341 (10 μM), lactacystin (10 μM), ZL3VS (50 μM), or AdaAhxL3VS (50 μM). (D) IRE1α phosphorylation in NIH 3T3 cells treated with Tm as indicated after 2 h of pretreatment with MG-132 (10 μM).
On sensing misfolded proteins in the ER lumen, IRE1 proteins become activated by oligomerization and autophosphorylation. To determine the step at which PIs interfered with IRE1α function, the integrity of IRE1α phosphorylation was assessed. Western blot analysis of extracts prepared from untreated and Tm-treated cells revealed the increase in phosphorylated (slower mobility) and the decrease in unphosphorylated IRE1α species previously observed (Fig. 2D). Notably, MG-132 completely blocked the phosphorylation of IRE1α by Tm (Fig. 2D). We conclude that the initial steps of IRE1α activation are disrupted in the presence of PIs, resulting in impaired oligomerization and autophosphorylation.
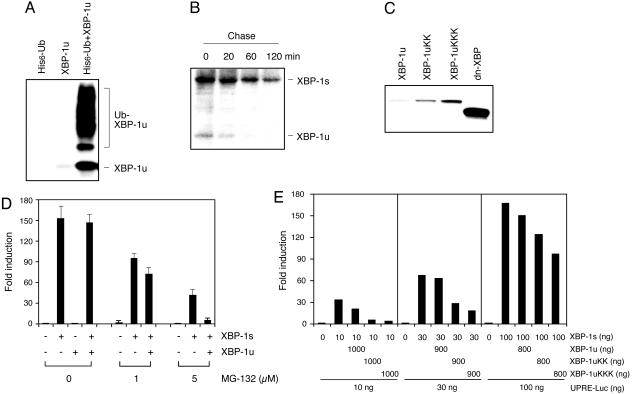
The Stabilized XBP-1u Protein Acts as an Inhibitor of the Spliced Species. PIs could act at multiple stages to alter the balance between XBP-1u and -1s species. Two potential mechanisms were suppression of the splicing event itself or preferential stabilization of XBP-1u protein. Under normal conditions, XBP-1u protein is barely detectable in J558 cells, despite the presence of abundant XBP-1u transcripts, indicating its poor stability (14). Indeed, XBP-1u protein is highly ubiquitinated in vivo (Fig. 3A) and rapidly degraded in myeloma cells with a half-life of ≈10 min (Fig. 3B). Thus, XBP-1u protein is rapidly degraded through the ubiquitin–proteasome pathway and is stabilized and accumulates in the presence of PIs. XBP-1s protein, although also unstable, has a longer half-life of ≈1 h. In conclusion, the initial accumulation of XBP-1s protein at early time points and the rapid increase in XBP-1u protein after MG-132 treatment reflects their stabilization by PIs, and the rapid decline of XBP-1s protein level thereafter is explained by suppression of IRE1α-dependent XBP-1 splicing.
Fig. 3.
PIs stabilize XBP-1u protein to act as a dominant-negative inhibitor of XBP-1s activity. (A) Ubiquitination of XBP-1 in HeLa cells cotransfected with XBP-1u and His-tagged ubiquitin expression plasmids. (B) Degradation rates of XBP-1u and -1s proteins as determined by pulse-labeling J558 cells with [35S]Met/[35S]Cys for 1 h and chasing for the indicated times. (C) Generation and expression of lysine to arginine XBP-1u mutants. Two or three lysine residues in the C terminus of XBP-1u were replaced by arginine to generate XBP-1uKK (235, 252) and XBP-1uKKK (146, 235, 252) by site-directed mutagenesis. dn-XBP contains the N-terminal 188 aa of XBP-1u. Western blot analysis was performed with NIH 3T3 extracts transfected with the indicated plasmids. (D) Effect of XBP-1u on XBP-1s-dependent UPR element (UPRE) activation in PI-treated NIH 3T3 cells with 8-fold excess of XBP-1u plasmids. Transfected cells were treated with MG-132 for 16 h before harvesting for luciferase assays. Values represent fold induction of activity compared with the reporter alone after normalizing to Renilla. (E) Inhibition of XBP-1s-dependent activation of the UPRE reporter in NIH 3T3 cells by XBP-1u lysine to arginine mutants.
XBP-1s, but not XBP-1u, possesses a potent transactivation domain (14) and reconstitutes Ig secretion in B cells (22). Our results raised the question as to what, if any, the function of the unspliced protein might be in the setting of treatment with PIs. Because XBP-1u shares the leucine zipper motif at the N terminus, it was possible that it might partner with XBP-1s to regulate its activity. To test this, NIH 3T3 cells were cotransfected with an XBP-1s expression plasmid in the presence or absence of XBP-1u and UPRE-luciferase reporter plasmid. In the absence of treatment, XBP-1s, but not XBP-1u, greatly increased reporter activity (Fig. 3D). In the presence of MG-132, however, XBP-1u now significantly suppressed transactivation of the reporter by XBP-1s, suggesting that the accumulated, stabilized XBP-1u protein acted as a dominant negative to suppress the activity of the spliced species (Fig. 3D).
To more directly investigate whether XBP-1u protein acted as a dominant-negative inhibitor of XBP-1s, it was necessary to avoid other potentially complicating actions of the PIs. We produced more stable forms of XBP-1u by changing lysine residues in the C terminus, the site of potential XBP-1u ubiquitination, to arginine. XBP-1uKK and XBP-1uKKK, mutant proteins in which two and three C-terminal lysines, respectively, have been replaced with arginine, are expressed at a higher level than the original XBP-1u protein, consistent with a role for ubiquitination-dependent degradation (Fig. 3C). These more stable mutant forms of XBP-1u inhibited the transactivation of the reporter by XBP-1s, even in the absence of PIs (Fig. 3E). We conclude that the unspliced version of XBP-1u can act as a dominant-negative inhibitor of the spliced form when its expression is stabilized by interference with its degradation by ubiquitination, a situation that occurs in myeloma cells in the presence of PIs.
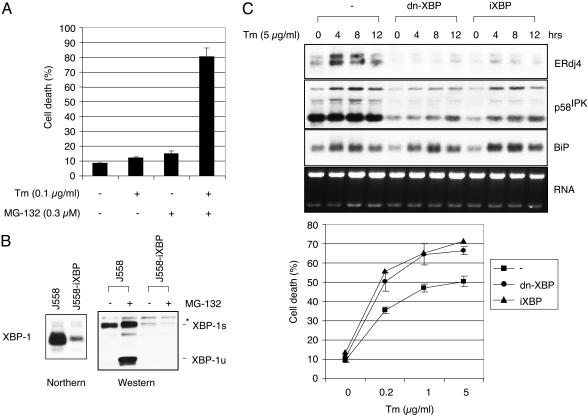
Absence of Functional XBP-1 Increases ER Stress-Induced Apoptosis of Myeloma Cells. Although PIs heighten ER stress in myeloma cells by preventing the degradation of ERAD substrates, they paradoxically inhibit UPR activation. Therefore, we hypothesized that PIs would induce apoptosis in part by inducing ER stress and subsequent apoptotic signaling pathways while simultaneously preventing an appropriate UPR. Consistent with this hypothesis, Tm and MG-132 synergistically induced apoptosis in J558 myeloma cells (Fig. 4A). These results indicate that Tm and MG-132 augmented ER stress by increasing the input of misfolded proteins and blocking ERAD degradation, respectively.
Fig. 4.
Cells with an impaired UPR are more sensitive to ER stress-induced apoptosis. (A) Synergistic effect of Tm and MG-132 on apoptosis. Annexin V-positive cells were counted after treating J558 cells for 18 h with suboptimal concentrations of Tm and MG-132 as indicated. (B) Generation of J558-iXBP cells by retroviral transduction of J558 cells with the U6 promoter-based XBP-1 RNAi vector. (C) XBP-1-dependent gene expression in J558 cells that express control GFP, dn-XBP, or iXBP-1 treated with Tm. Generation of dn-XBP-1 J558 cells by infection with a retrovirus containing dn-XBP cDNA inserted into the GFP-RV vector (22). ERdj4, p58IPK, and BiP gene expression was examined by Northern blot analysis. (D) Increased apoptosis in iXBP-1- and dn-XBP-1-expressing J558 cells. Cells were treated with the indicated amounts of Tm for 48 h, and dead cells were counted after annexin V staining.
To further test the effect of an impaired UPR on the handling of ER stress, we generated myeloma cell lines functionally deficient in XBP-1 either by transducing J558 cells with a potent dominant-negative XBP-1 retrovirus (dn-XBP-1) (unpublished observations) or a small interfering RNA retrovirus (iXBP). Because dn-XBP-1 does not possess the C-terminal destabilization motif present in XBP-1u, it is expressed at high levels (Fig. 3C, lane 4) and inhibits XBP-1s-induced transactivation very efficiently (unpublished observations). Suppression of XBP-1 expression in the iXBP-transduced cells was confirmed by both Northern and Western blot analysis (Fig. 4B). The functional impairment of XBP-1 activity was demonstrated by the greatly reduced induction of XBP-1-dependent UPR target genes, ERdj4 and p58IPK (unpublished observations) (33, 34), by Tm in both the dn-XBP-1- and iXBP-1-transduced cells (Fig. 4C). In contrast, the induction of BiP was minimally affected by dn-XBP or XBP-1 RNAi. No effect on cell proliferation or viability was observed at baseline. However, on Tm treatment, dn-XBP-1 and iXBP-1 myeloma cells both displayed significantly increased apoptosis when compared with control GFP-transduced J558 cells (Fig. 4D), demonstrating that the IRE1α/XBP-1 pathway contributes to the survival of myeloma cells under ER stress conditions.
Discussion
We and others have previously suggested that activation of XBP-1–/– B cells fails to yield plasma cells because a functional UPR is necessary to protect them from stress-induced death (22, 35). Similarly, PIs may cause apoptosis of myeloma cells, the malignant counterpart of the plasma cell, by interfering with the UPR. Here we have provided evidence that PIs induce ER stress but disrupt the UPR. PIs caused a profound shift in the ratio of XBP-1u and -1s proteins both by inhibiting the activity of the endoribonuclease IRE1α, and hence XBP-1 mRNA splicing, and stabilizing the resulting XBP-1u protein. The unspliced form of XBP-1 acts as a dominant-negative inhibitor of the spliced, active XBP-1 species, thereby preventing an effective UPR. Thus, treatment with PIs, which heighten ER stress in myeloma cells by preventing the degradation of ERAD substrates, simultaneously cripples the UPR, resulting in increased apoptosis. Our data demonstrate that the UPR, and more particularly XBP-1, is an important target of PIs, although PIs certainly affect other cellular pathways that impinge on apoptosis (36).
Two additional UPR signaling pathways involve the activation of transcription factor ATF6 or translational repression mediated by PERK/eIF2α. ATF6, like XBP-1 a basic region/leucine zipper transcription factor, is a second ER transmembrane component that is constitutively expressed in an inactive form until ER stress results in proteolytic cleavage of its N-terminal cytoplasmic domain by the S2P serine protease to produce a potent transcriptional activator of chaperone genes (14, 26, 37–40). Although we could not assess whether PIs inhibited the processing of the inactive ATF6 precursor by S2P because the processed form of ATF6 is itself stabilized by PIs, we know that dn-XBP-1 potently inhibits the function of ATF6 through heterodimerization (unpublished observations). Cell death in dn-XBP-1-transduced myeloma cells did not, however, exceed that observed in the iXBP-1 J558 cell line (Fig. 4D), as would have been expected if both factors were significant targets for PIs.
PEK/PERK, like IRE1α, is a type 1 transmembrane serine/threonine protein kinase that undergoes ER stress-induced dimerization of its luminal domain, autophosphorylates, and then acts in the cytoplasm to phosphorylate eIF2α. Phosphorylation of eIF2α leads to translation attenuation in response to ER stress (19, 41). The induction of the stress response gene CHOP, shown to be PERK-dependent (42), is prevented by PIs (Fig. 1 A), suggesting that PIs might also target PERK. Further, because the ER luminal domain of IRE1 and PERK are interchangeable and conserved throughout evolution (43), it is tempting to speculate that the mechanism by which PIs inhibit IRE1α and PERK activation will be similar. Indeed, preliminary experiments demonstrate that MG-132 resulted in a very marked decrease in Tm-induced autophosphorylation of PERK, similar to what we had observed for IRE1α (unpublished observations). Little is known about the factors that control activation of IRE1α. To date, BiP and TRAF2 are the only proteins reported to interact with IRE1α (24, 44), and the mechanism by which PIs alter the activity of the endoribonuclease function of IRE1α is therefore unclear. One potential explanation is that PIs stabilize the expression of an unknown protein(s) that acts, in the resting state, as an inhibitor of IRE1α activity, possibly by stabilizing its association with BiP, or by preventing ER stress-induced dimerization of the ER luminal domain.
We have shown that PIs modestly induce UPR target genes, consistent with previous reports (29, 30). However, PIs inhibit the stress-induced UPR, as evidenced by suppression of IRE1-mediated XBP-1 mRNA splicing and stabilization of XBP-1u protein as well as PERK autophosphorylation. The mechanism by which PIs induce UPR target gene induction is unclear, but is unlikely to be one of the known UPR signal pathways. Thus, we could not find any evidence that PIs induced IRE1α or PERK phosphorylation or XBP-1 mRNA splicing. Similarly, MG-132, but not Tm, treatment of XBP-1-deficient MEFs resulted in the normal induction of ERdj4, an XBP-1-dependent UPR target gene, suggesting that MG-132 and Tm induce UPR target genes through distinct mechanisms (unpublished observations). One possibility is that PIs induce distinct transcription factors [e.g., heat shock factors (30)] that may in turn induce both cytosolic Hsps and ER-resident chaperones.
Most data suggest that proteasome inhibition induces cell death in proliferating cells while it inhibits apoptosis in differentiated cells such as thymocytes and sympathetic neurons. Thus, PIs induced apoptosis in human glioma cells, human T-cell leukemia cells, and PC-12 cells while etoposide-induced apoptosis in thymocytes was suppressed with peptide aldehyde PIs (45–47). Apoptosis in glioma cells is morphologically characterized by dilated rough ER, cytoplasmic vacuoles, and dense mitochondrial deposits. Interestingly, this histologic picture was not affected by the broad caspase inhibitor zVADfmk, although apoptosis was inhibited (45). Another study demonstrated that PI-induced glioma cell death was associated with mitochondria-independent caspase-3 activation (46). Here we have shown that PIs induce apoptosis of myeloma cells by disrupting the UPR. We demonstrate that blockade of the IRE1α/XBP-1 pathway by PIs contributes to the death of myeloma cells under ER stress conditions. It may be that the mechanisms by which PIs induce apoptosis will depend not only on the status of differentiation, proliferation, or activation of a given cell, but also on its function. Thus, secretory cells that require an active UPR and ERAD to ensure proper processing of client proteins in the ER may be particularly susceptible to apoptosis by agents that evoke ER stress but disrupt the UPR. Compounds that inhibit the UPR by targeting the activity of IRE1/XBP-1, in combination with PIs (48), may prove to be potent therapeutic agents for the treatment of multiple myeloma and other tumors, such as adenocarcinomas of the prostate, breast, and ovary, that originate from secretory cells.
Acknowledgments
We thank Drs. H. Ovaa and H. Ploegh for providing proteasome inhibitors; Dr. J. Yuan for anti-caspase 12 antibody; Drs. K. Mowen, M. Oukka, and H. Ploegh for thoughtful comments on the manuscript; and C. McCall for help with manuscript preparation. This work was supported by National Institutes of Health Grant AI32412 (to L.H.G.), an award from the Multiple Myeloma Research Foundation (to L.H.G.), and an Irvington Institute postdoctoral fellowship award (to N.N.I.).
Abbreviations: ER, endoplasmic reticulum; ERAD, ER-associated degradation; MM, multiple myeloma; Tm, tunicamycin; UPR, unfolded protein response.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 30, 2002.
References
- 1.Orlowski, R. Z., Stinchcombe, T. E., Mitchell, B. S., Shea, T. C., Baldwin, A. S., Stahl, S., Adams, J., Esseltine, D. L., Elliott, P. J., Pien, C. S., et al. (2002) J. Clin. Oncol. 20, 4420–4427. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, K. C. (2002) Semin. Oncol. 29, 17–20. [DOI] [PubMed] [Google Scholar]
- 3.Barlogie, B., Shaughnessy, J., Zangari, M. & Tricot, G. (2002) Semin. Oncol. 29, 26–33. [DOI] [PubMed] [Google Scholar]
- 4.Adams, J. (2002) Curr. Opin. Oncol. 14, 628–634. [DOI] [PubMed] [Google Scholar]
- 5.Kisselev, A. F. & Goldberg, A. L. (2001) Chem. Biol. 8, 739–758. [DOI] [PubMed] [Google Scholar]
- 6.Mitsiades, N., Mitsiades, C. S., Richardson, P. G., Poulaki, V., Fanourakis, G., Tai, Y. T., Chauhan, D., Gu, X., Bailey, C., Joseph, M., et al. (2003) Blood 101, 2377–2380. [DOI] [PubMed] [Google Scholar]
- 7.Mitsiades, N., Mitsiades, C. S., Poulaki, V., Chauhan, D., Fanourakis, G., Gu, X., Bailey, C., Joseph, M., Libermann, T. A., Treon, S. P., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 14374–14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hideshima, T. & Anderson, K. C. (2002) Nat. Rev. Cancer 2, 927–937. [DOI] [PubMed] [Google Scholar]
- 9.Hideshima, T., Mitsiades, C., Akiyama, M., Hayashi, T., Chauhan, D., Richardson, P., Schlossman, R., Podar, K., Munshi, N. C., Mitsiades, N. & Anderson, K. C. (2003) Blood 101, 1530–1534. [DOI] [PubMed] [Google Scholar]
- 10.LeBlanc, R., Catley, L. P., Hideshima, T., Lentzsch, S., Mitsiades, C. S., Mitsiades, N., Neuberg, D., Goloubeva, O., Pien, C. S., Adams, J., et al. (2002) Cancer Res. 62, 4996–5000. [PubMed] [Google Scholar]
- 11.Kozutsumi, Y., Segal, M., Normington, K., Gething, M. J. & Sambrook, J. (1988) Nature 332, 462–464. [DOI] [PubMed] [Google Scholar]
- 12.Ruegsegger, U., Leber, J. H. & Walter, P. (2001) Cell 107, 103–114. [DOI] [PubMed] [Google Scholar]
- 13.Shen, X., Ellis, R. E., Lee, K., Liu, C. Y., Yang, K., Solomon, A., Yoshida, H., Morimoto, R., Kurnit, D. M., Mori, K. & Kaufman, R. J. (2001) Cell 107, 893–903. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida, H., Matsui, T., Yamamoto, A., Okada, T. & Mori, K. (2001) Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- 15.Calfon, M., Zeng, H., Urano, F., Till, J. H., Hubbard, S. R., Harding, H. P., Clark, S. G. & Ron, D. (2002) Nature 415, 92–96. [DOI] [PubMed] [Google Scholar]
- 16.Cox, J. S., Shamu, C. E. & Walter, P. (1993) Cell 73, 1197–1206. [DOI] [PubMed] [Google Scholar]
- 17.Cox, J. S. & Walter, P. (1996) Cell 87, 391–404. [DOI] [PubMed] [Google Scholar]
- 18.Tirasophon, W., Welihinda, A. A. & Kaufman, R. J. (1998) Genes Dev. 12, 1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding, H. P., Zhang, Y. & Ron, D. (1999) Nature 397, 271–274. [DOI] [PubMed] [Google Scholar]
- 20.Lee, K., Tirasophon, W., Shen, X., Michalak, M., Prywes, R., Okada, T., Yoshida, H., Mori, K. & Kaufman, R. J. (2002) Genes Dev. 16, 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reimold, A. M., Iwakoshi, N. N., Manis, J., Vallabhajosyula, P., Szomolanyi-Tsuda, E., Gravallese, E. M., Friend, D., Grusby, M. J., Alt, F. & Glimcher, L. H. (2001) Nature 412, 300–307. [DOI] [PubMed] [Google Scholar]
- 22.Iwakoshi, N. N., Lee, A. H., Vallabhajosyula, P., Otipoby, K. L., Rajewsky, K. & Glimcher, L. H. (2003) Nat. Immunol. 4, 321–329. [DOI] [PubMed] [Google Scholar]
- 23.Reimold, A. M., Ponath, P. D., Li, Y. S., Hardy, R. R., David, C. S., Strominger, J. L. & Glimcher, L. H. (1996) J. Exp. Med. 183, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertolotti, A., Zhang, Y., Hendershot, L. M., Harding, H. P. & Ron, D. (2000) Nat. Cell Biol. 2, 326–332. [DOI] [PubMed] [Google Scholar]
- 25.Campanero, M. R. & Flemington, E. K. (1997) Proc. Natl. Acad. Sci. USA 94, 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, Y., Shen, J., Arenzana, N., Tirasophon, W., Kaufman, R. J. & Prywes, R. (2000) J. Biol. Chem. 275, 27013–27020. [DOI] [PubMed] [Google Scholar]
- 27.Sui, G., Soohoo, C., Affar el, B., Gay, F., Shi, Y. & Forrester, W. C. (2002) Proc. Natl. Acad. Sci. USA 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindemann, D., Patriquin, E., Feng, S. & Mulligan, R. C. (1997) Mol. Med. 3, 466–476. [PMC free article] [PubMed] [Google Scholar]
- 29.Bush, K. T., Goldberg, A. L. & Nigam, S. K. (1997) J. Biol. Chem. 272, 9086–9092. [DOI] [PubMed] [Google Scholar]
- 30.Kawazoe, Y., Nakai, A., Tanabe, M. & Nagata, K. (1998) Eur. J. Biochem. 255, 356–362. [DOI] [PubMed] [Google Scholar]
- 31.Nakagawa, T., Zhu, H., Morishima, N., Li, E., Xu, J., Yankner, B. A. & Yuan, J. (2000) Nature 403, 98–103. [DOI] [PubMed] [Google Scholar]
- 32.Kessler, B. M., Tortorella, D., Altun, M., Kisselev, A. F., Fiebiger, E., Hekking, B. G., Ploegh, H. L. & Overkleeft, H. S. (2001) Chem. Biol. 8, 913–929. [DOI] [PubMed] [Google Scholar]
- 33.Kurisu, J., Honma, A., Miyajima, H., Kondo, S., Okumura, M. & Imaizumi, K. (2003) Genes Cells 8, 189–202. [DOI] [PubMed] [Google Scholar]
- 34.Shen, Y., Meunier, L. & Hendershot, L. M. (2003) J. Biol. Chem. 277, 15947–15956. [DOI] [PubMed] [Google Scholar]
- 35.van Anken, E., Romijn, E. P., Maggioni, C., Maezghrani, A., Sitia, R., Braakman, I. & Heck, A. J. R. (2003) Immunity 18, 243–253. [DOI] [PubMed] [Google Scholar]
- 36.Yang, Y., Fang, S., Jensen, J. P., Weissman, A. M. & Ashwell, J. D. (2000) Science 288, 874–877. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida, H., Haze, K., Yanagi, H., Yura, T. & Mori, K. (1998) J. Biol. Chem. 273, 33741–33749. [DOI] [PubMed] [Google Scholar]
- 38.Shen, J., Chen, X., Hendershot, L. & Prywes, R. (2002) Dev. Cell 3, 99–111. [DOI] [PubMed] [Google Scholar]
- 39.Ye, J., Rawson, R. B., Komuro, R., Chen, X., Dave, U. P., Prywes, R., Brown, M. S. & Goldstein, J. L. (2000) Mol. Cell 6, 1355–1364. [DOI] [PubMed] [Google Scholar]
- 40.Li, M., Baumeister, P., Roy, B., Phan, T., Foti, D., Luo, S. & Lee, A. S. (2000) Mol. Cell. Biol. 20, 5096–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi, Y., Vattem, K. M., Sood, R., An, J., Liang, J., Stramm, L. & Wek, R. C. (1998) Mol. Cell. Biol. 18, 7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urano, F., Bertolotti, A. & Ron, D. (2000) J. Cell Sci. 113, 3697–3702. [DOI] [PubMed] [Google Scholar]
- 43.Liu, C. Y., Schroder, M. & Kaufman, R. J. (2000) J. Biol. Chem. 275, 24881–24885. [DOI] [PubMed] [Google Scholar]
- 44.Urano, F., Wang, X., Bertolotti, A., Zhang, Y., Chung, P., Harding, H. P. & Ron, D. (2000) Science 287, 664–666. [DOI] [PubMed] [Google Scholar]
- 45.Wagenknecht, B., Hermisson, M., Groscurth, P., Liston, P., Krammer, P. H. & Weller, M. (2000) J. Neurochem. 75, 2288–2297. [DOI] [PubMed] [Google Scholar]
- 46.Kitagawa, H., Tani, E., Ikemoto, H., Ozaki, I., Nakano, A. & Omura, S. (1999) FEBS Lett. 443, 181–186. [DOI] [PubMed] [Google Scholar]
- 47.Stefanelli, C., Bonavita, F., Stanic, I., Pignatti, C., Farruggia, G., Masotti, L., Guarnieri, C. & Caldarera, C. M. (1998) Biochem. J. 332, 661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldberg, A. L. & Rock, K. (2002) Nat. Med. 8. [DOI] [PubMed]