Abstract
The oligopeptide transport system (Opp) of Lactococcus lactis has the unique capacity to mediate the transport of peptides from 4 up to at least 18 residues. The substrate specificity of this binding protein-dependent ATP-binding cassette transporter is determined mainly by the receptor protein OppA. To study the specificity and ligand-binding mechanism of OppA, the following strategy was used: (i) OppA was purified and anchored via the lipid moiety to the surface of liposomes; (ii) the proteoliposomes were used in a rapid filtration-based binding assay with radiolabeled nonameric bradykinin as a reporter peptide; and (iii) combinatorial peptide libraries were used to determine the specificity and selectivity of OppA. The studies show that (i) OppA is able to bind peptides up to at least 35 residues, but there is a clear optimum in affinity for nonameric peptides; (ii) the specificity for nonameric peptides is not equally distributed over the whole peptide, because positions 4, 5, and 6 in the binding site are more selective; and (iii) the differences in affinity for given side chains is relatively small, but overall hydrophobic residues are favored—whereas glycine, proline, and negatively charged residues lower the binding affinity. The data indicate that not only the first six residues (enclosed by the protein) but also the C-terminal three residues interact in a nonopportunistic manner with (the surface of) OppA. This binding mechanism is different from the one generally accepted for receptors of ATP-binding cassette-transporter systems.
The oligopeptide transport system (Opp) and the proteinase PrtP of Lactococcus lactis are the most critical components of the proteolytic system of this bacterium. In the degradation pathway for exogenous proteins, there is no alternative activity that can substitute these enzymes (1). In vivo studies on the uptake by Opp of peptides from complex β-casein mixtures, generated by PrtP activity, showed that peptides of five up to at least 10 residues are taken up via Opp (2). Subsequent kinetic studies showed that peptides up to at least 18 residues can be handled by the system (3). This unusual length selectivity sets Opp apart from other homologous systems involved in peptide uptake.
The Opp system of L. lactis belongs to the superfamily of ATP-binding cassette transporters (4) and consists of five proteins: the integral membrane proteins OppB and OppC, the ATP-binding proteins OppD and OppF, and the receptor protein OppA. The substrate specificity of these systems is generally determined by the receptor protein. The binding mechanism of these receptors is thought to be the “Venus-flytrap” mechanism, in which the binding site is formed by a cleft between two domains that are connected by a flexible hinge. On binding of the ligand, the cleft closes, and the ligand is completely enclosed (5). The conformational change on ligand binding to OppA results in a fluorescence blue shift in the emission spectrum, and this property has been used to determine the dissociation constants (Kd) for a limited set of peptides (6, 7). Importantly, these studies have also indicated that the first six residues of nonameric peptides are enclosed by the protein, whereas the remaining three interact with the surface of the protein. By selectively modifying cysteines in the peptides, it could be shown that the tolerance for large bulky groups at positions 4, 5, and 6 is smaller than that of positions 1 and 3, whereas positions 7 and 9 seemed most promiscuous (7).
An in-depth analysis of the peptide-binding properties of OppA, with complex peptide libraries, is reported in this paper. The libraries consist of randomized peptide mixtures that share one defined amino acid at a given position, whereas the other positions are randomized. For every position, there are thus 19 peptide mixtures (cysteine is not included in the library; refs. 8 and 9). These combinatorial peptide libraries have been successfully applied to decipher the binding motif of the transporter associated with antigen processing (10), and the approach allows one to determine the effects of individual residues at a given position in a peptide, independent of the sequence context. Because of the limited availability and the large number of different peptides, a rapid filtration assay was developed to study peptide binding to OppA.
Experimental Procedures
Bacterial Strain and Growth Conditions.
L. lactis strain AMP2/pAMP31 (11) is a MG1363-derivative that lacks the genes pepX, pepT, pepC, pepN, and oppA. Plasmid pAMP31 is a pGK13-derivative with an insert (1,802-bp NcoI–BamHI fragment) that codes for OppA with a C-terminal six-histidine tag. The AMP2/pAMP31 strain was grown in M17 broth (Difco) supplemented with 0.5% (wt/vol) glucose plus 5 μg/ml erythromycin. For large-scale purification of OppA, the AMP2/pAMP31 cells were grown in batch in a 10-liter fermentor with pH control (ADI 1065 fermentor; Applicon Dependable Instruments B.V., Schiedam, The Netherlands). The pH was kept constant at pH 6.5 by the addition of 1 M KOH. The cells were harvested at OD660 of 5.0, which corresponds to the late exponential phase of growth.
Purification and Membrane Reconstitution of OppA-His6.
Inside-out membrane vesicles of L. lactis AMP2/pAMP31 were isolated by lysis of the cells in a French press (12). The membranes were solubilized in 20 mM KPi/100 mM KCl/10% (wt/vol) glycerol, pH 8.0 (buffer A) with 0.5% (wt/vol) dodecyl-β-d-maltoside (Sigma) for 20 min on ice. The insoluble material was removed by centrifugation (15 min; 280,000 × g), and 10 mM imidazole was added to the supernatant. This solution was incubated with Ni-nitrilo-triacetic acid resin (Qiagen, Chatsworth, CA) for 2 h at 4°C under continuous shaking. The column was washed subsequently with buffer A containing 10 mM imidazole plus 0.05% (wt/vol) Triton X-100 and buffer A with 25 mM imidazole plus 0.05% (wt/vol) Triton X-100. The protein was eluted with 20 mM KPi/100 mM KCl/10% (wt/vol) glycerol/300 mM imidazole, pH 7.0, plus 0.05% (wt/vol) Triton X-100. The purified OppA was incorporated into preformed liposomes (4 mg of lipid per ml) via its lipid anchor (three palmitoyl phosphatidyl ethanolamine moieties) with a lipid to protein ratio of 20:1 (wt/wt). The liposomes consisted of Escherichia coli lipids and egg yolk l-α-phosphatidyl choline in a ratio of 3:1 (wt/wt). After 20 min of incubation at room temperature under continuous shaking, Biobeads SM2 (Bio-Rad) were added (wet weight 80 mg/ml) to remove the detergent. After 1 h incubation at 4°C, fresh beads were added, and the proteoliposomes were incubated for another 1 h at 4°C. The proteoliposomes were washed and resuspended in assay buffer [25 mM KPi/100 mM KCl/1 mM K-EDTA plus 10% (wt/vol) glycerol, pH 6.0] and stored in small aliquots in liquid nitrogen.
Peptide-Binding Assay.
Peptide binding to OppA was measured by making use of the high-affinity reporter ligand [3,4(n)-3H]bradykinin (Amersham Pharmacia), which is a nonameric peptide with the sequence RPPGFSPFR. [3H]Bradykinin (0.1 μM, unless indicated otherwise) with or without given peptide library (see below) was incubated with the proteoliposomes containing OppA at 25°C for 4 min in assay buffer with a total volume of 100 μl; the final OppA concentration was 20 μg/ml. This incubation was followed by a 1-min incubation with antibodies raised against OppA by using a titer of 1:10. Subsequently, the assay mix was diluted with 2 ml of ice-cold 8% (wt/vol) polyethylene glycol (PEG) 6000 and filtered over 0.2-μm pore-size cellulose acetate (OE66) filters (Schleicher & Schüll, Dassel, Germany), after which the filters were washed again with 2 ml of ice-cold 8% (wt/vol) PEG 6000. The radioactivity on the filter was determined by liquid scintillation counting (13).
Most of the randomized peptide libraries that are used in this work have been described (8). In brief, 19 amino acids (cysteine was excluded) were coupled in randomized sequence positions by using solid-phase peptide synthesis with fluorenylmethoxycarbonyl/tertiair-butyl chemistry. The amino acid composition of the peptide libraries was determined by pool sequencing (14), electrospray mass spectrometry (15), and amino acid analysis. Deviations from the equimolar representation of the amino acid in randomized sequence positions were found to be within the error limits of the analytical methods (16). The inhibition of bradykinin binding to OppA by the randomized peptide libraries was tested in 100 μl of assay mixture at fixed reporter and inhibitor concentration (0.1 μM [3H]bradykinin and 1.0 or 5.0 μM inhibitor peptide). Every peptide was tested at least three times, and the arithmetic average and the standard deviation (Xσn − 1) are presented. The amount of bound, labeled peptide was corrected for unspecific binding, which was determined in the presence of a 10,000-fold molar excess of unlabeled bradykinin.
Miscellaneous.
Peptide concentrations were determined by using the bicinchoninic acid assay (Pierce), and protein concentrations were determined by the method of Lowry et al. (17) with BSA as the standard.
Results
Bradykinin Binding to OppA.
To study peptide binding to liposome-attached OppA with radiolabeled bradykinin as reporter peptide, it was necessary to determine first the efficacy and reproducibility of the assay and show that it reports quantitatively on the binding of bradykinin to OppA. Some aspects of the assay require explanation. Firstly, cellulose acetate filters were used to minimize aspecific binding of bradykinin to the filters. Secondly, antibodies raised against OppA together with PEG 6000 were used to collect the proteoliposomes on the filters more effectively. Without these treatments, more than 60% of the material passed through the filters. Similar binding kinetics was observed in the absence of the antibodies, but the amount of binding was lower because of the loss of OppA; small proteoliposomes are less well retained by the filters.
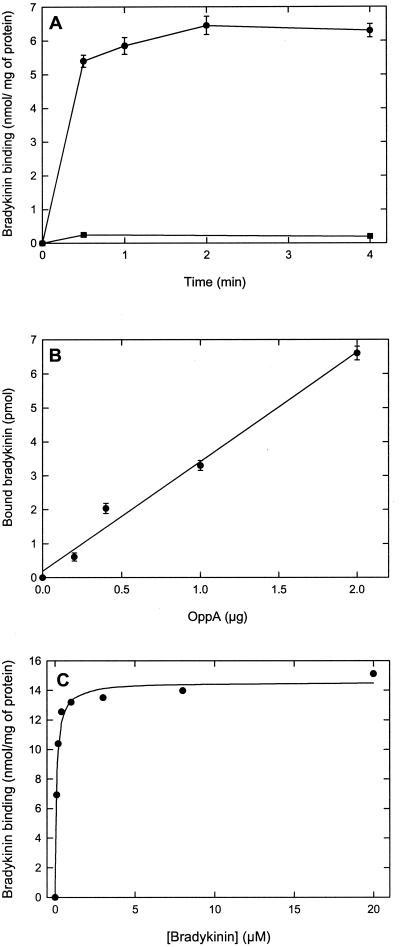
The binding of bradykinin, and that of other peptides to OppA, is a rapid process (6), and after 30 s of incubation, more than 90% of the maximal binding level was reached. After 2 min, the binding was stable and did not change significantly on prolonged incubation (Fig. 1A). Importantly, the observed binding of bradykinin was proportional to the OppA concentration over a broad range (Fig. 1B). The Kd value for bradykinin binding to OppA was 0.09 ± 0.01 μM, which is comparable to the Kd determined for the soluble protein (without lipid anchor) in the fluorimetric assay (6). The maximal level of bradykinin binding was 14.6 ± 0.3 nmol/mg protein (Fig. 1C), which corresponds to a binding stoichiometry of 1:1 mol of bradykinin per mol of protein. These results clearly indicate that the binding assay described herein is suitable for the analysis of the specificity of OppA.
Figure 1.
Characteristics of bradykinin binding to OppA. (A) Time-dependent binding of bradykinin to proteoliposomes containing OppA (●) and liposomes without OppA (■). The final OppA concentration was 20 μg/ml, which corresponds to a lipid concentration of the liposomes of 4 mg/ml; the [3H]bradykinin concentration was 0.1 μM. (B) Bradykinin binding as a function of the OppA concentration. (C) Determination of the dissociation constant (Kd) for bradykinin binding to OppA. The experimental data were fitted to a hyperbole from which the Kd (0.09 ± 0.01 μM) and the Bmax (14.6 ± 0.3 nmol/mg protein) were estimated.
Length Specificity of OppA.
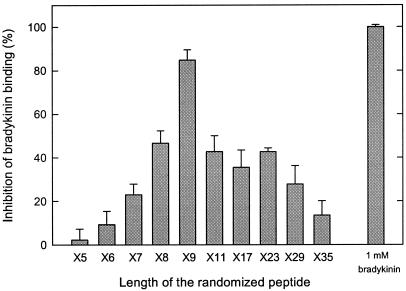
To determine the optimal length for peptides binding to OppA, fully randomized peptide libraries were used with different lengths in the range of 5–35 amino acids. All of the peptides were tested at a total concentration of 1 μM and in the presence of 0.1 μM labeled bradykinin. There was a sharp increase in affinity from 5 to 9 amino acids, whereas peptides of 11 up to 29 amino acids had an affinity comparable to the octameric library. Strikingly, the library with a length of 35 amino acids was still able to inhibit the bradykinin binding significantly (Fig. 2). Overall, the data indicate a clear optimum for peptide binding, and consistent with previous observations, pentameric peptides bind very poorly (6) even though they are well transported (3).
Figure 2.
Length specificity of OppA. Fully randomized peptide libraries that differ in length from 5 to 35 amino acid residues were tested at 1.0 μM in the presence of 0.1 μM [3H]bradykinin. The OppA concentration was 20 μg/ml; error bars indicate the standard deviation (Xσn − 1), n = 3.
Influence of d-Amino Acids on Peptide Binding.
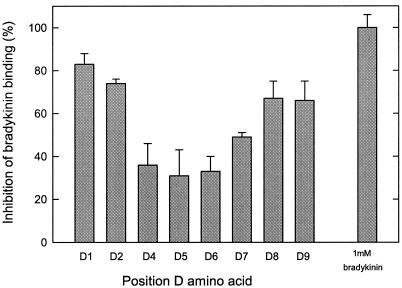
To investigate whether peptides with d-amino acids in the sequence have a destabilizing effect on peptide binding, randomized nonameric peptide libraries were tested with d-amino acids on a defined position. These peptides were tested at a 50-fold excess over bradykinin, because the affinity of OppA for the libraries with d-amino acids at position 1, 2, 8, or 9 was ±10 times lower than for all l-isomer nonameric libraries. The effect of the d-amino acids was not equally distributed over the nonameric peptide (Fig. 3). The reduced inhibitory effect on bradykinin binding of the d-isomers was most pronounced at positions 4, 5, and 6 of the nonameric peptides, whereas peptides with d-amino acids at the N- and C-terminal positions had much less effect on the affinity. This inhibition pattern might indicate that the peptide backbone at positions 4, 5, and 6 contributes more to the overall binding affinity than the peptide backbones at the other positions do or that the binding regions of OppA for the N- and C-terminal residues are more flexible/adaptive.
Figure 3.
Effects of D-amino acids on peptide binding. Nonameric fully randomized peptide libraries containing one D-amino acid residue at a defined position were tested at 5.0 μM in the presence of 0.1 μM [3H]bradykinin. The OppA concentration was 20 μg/ml; error bars indicate the standard deviation (Xσn − 1), n = 3.
Nonameric Randomized Peptide Libraries.
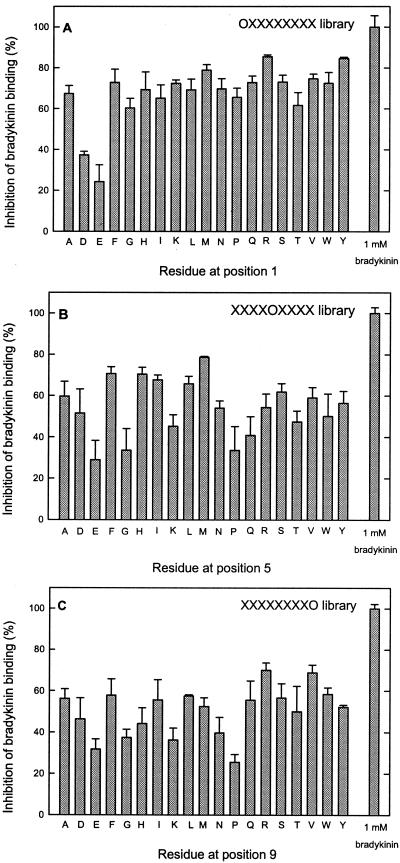
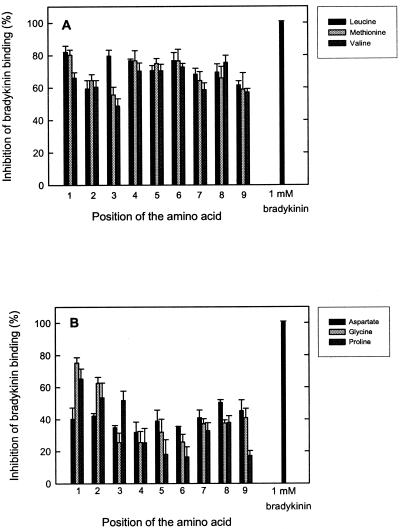
The synthetic nonameric peptides are randomized at all positions except one, which has a defined amino acid. For every defined position, there are 19 different libraries (cysteine is not included in the libraries); thus, in total, the complete set of nonameric peptides consists of 9 × 19 = 171 libraries. All these peptide libraries were tested for binding to OppA. Fig. 4A shows that OppA tolerates all amino acids at the first position of a nonameric peptide (OX8 library). Only the negatively charged amino acids glutamate and aspartate inhibited the bradykinin binding significantly less than the other residues. OppA was more selective at position 5 of the nonameric peptide library (X4OX4), which is most manifest from the lower affinity for peptides with glutamate, glycine, lysine, proline, or glutamine at this position (Fig. 4B). A similar pattern of selectivity was observed for the X8O library (Fig. 4C), although the absolute differences were smaller than for position 5 (Fig. 4B). Overall, the differences in inhibition for the peptides of the different libraries were not extreme; the largest difference was 5.3-fold (16.5% inhibition for proline at position 6 vs. 86.7% for isoleucine at position 6). However, it is clear that OppA prefers certain amino acids, particularly at the positions 4, 5, and 6. Hydrophobic residues are clearly favored at all positions (Fig. 5A), whereas aspartate, glycine, and proline are accommodated in the binding pocket with a much lower affinity (Fig. 5B). To summarize all data obtained with the screening of the complete set of peptides in the 171 libraries, the average inhibition values over the nine positions for peptides with a defined amino acid and the standard deviations of these average numbers were calculated (Table 1). For tyrosine-, alanine-, leucine-, and isoleucine-containing peptides, high inhibition values with low standard deviation values were observed. This result implies that the overall affinity for these residues is high and that there is not much variation in affinity over the nine positions. High standard deviation values are mainly the result of peptides with certain residues at the middle positions (positions 4, 5, and 6) and the C terminus of the nonameric peptides, which results in lower affinities. The standard deviations are particularly high for lysine, glycine, and proline. There are also amino acids, like glutamate and aspartate, that show low inhibition and low standard deviation values, indicating an overall low affinity for peptides with these amino acids.
Figure 4.
Inhibition of bradykinin binding by randomized peptide libraries with defined residues at given positions. The effect of defined residues at position 1 (A), position 5 (B), and position 9 (C) of nonameric peptide libraries are shown. All peptides were tested at 1.0 μM in the presence of 0.1 μM [3H]bradykinin. Amino acids are indicated in single-letter code. The OppA concentration was 20 μg/ml; error bars indicate the standard deviation (Xσn − 1), n = 3.
Figure 5.
Effects of defined amino acid residues at different positions in a nonameric randomized peptide. (A) Effect of leucine, methionine, and valine residues at different positions in the nonameric peptide library on bradykinin binding. (B) Effects of aspartate, glycine, and proline at different positions. All competitor peptides were tested at 1.0 μM in the presence of 0.1 μM [3H]bradykinin. The OppA concentration was 20 μg/ml; error bars indicate the standard deviation (Xσn − 1), n = 3.
Table 1.
Average inhibition of bradykinin binding (and standard deviation) by randomized nonameric peptides with a defined amino acid residue at every position
| Amino acid | Average inhibition over nine positions, % | Standard deviation Xσn − 1, % | Amino acid | Average inhibition over nine positions, % | Standard deviation Xσn − 1, % |
|---|---|---|---|---|---|
| Tyr | 75.6 | 4.7 | Trp | 63.2 | 10.9 |
| Ala | 72.8 | 5.0 | Arg | 63.2 | 13.3 |
| Leu | 71.6 | 7.9 | His | 60.9 | 13.2 |
| Ile | 71.3 | 8.2 | Lys | 59.3 | 18.1 |
| Met | 68.7 | 8.7 | Thr | 56.0 | 10.5 |
| Asn | 65.8 | 7.4 | Asp | 41.1 | 6.8 |
| Ser | 65.1 | 8.5 | Gly | 40.3 | 17.5 |
| Gln | 64.9 | 8.3 | Glu | 35.2 | 9.9 |
| Phe | 64.8 | 4.1 | Pro | 34.3 | 16.2 |
| Val | 64.5 | 8.8 |
All peptides were tested at 1.0 μM in the presence of 0.1 μM [3H]bradykinin; the OppA concentration was 20 μg/ml, n = 3 for every peptide. The data on the inhibition values of the individual amino acids at every position are available on request to the authors.
Discussion
In this paper, we define the substrate binding activity of OppA of L. lactis by making use of combinatorial peptide libraries. These libraries have been used to determine the length and isomer specificity of OppA as well as the influence of a specific residue at a given position in the (nonameric) peptide. Because the peptide libraries contain aromatic residues, it was not possible to assess the binding activity via changes in intrinsic protein fluorescence (6, 7). With the purified OppA attached to the liposomes via its lipid anchor, it was possible to test all of the peptides of the library in a rapid filtration assay by using only small quantities of the synthetic peptides. To our knowledge, this work is the first time that the properties of a membrane-associated binding protein are studied in this manner. There are a number of aspects of the developed binding assay that require clarification. First, the reporter peptide needs to be bound with high affinity and needs to have a low off rate, conditions that are met for bradykinin (6). Second, aspecific binding of bradykinin to commonly used filters such as cellulose nitrate and poly(vinylidene difluoride) was high, and only filters of the cellulose acetate type gave significantly low backgrounds (less than 2% of the binding of bradykinin to OppA). Third, to prevent passage of the proteoliposomes through the cellulose acetate filters, OppA antibodies and PEG 6000 were included in the wash buffer. These inclusions resulted in minimal loss of OppA as was confirmed by immunoblotting of the filtrate and the protein on the filter (data not shown).
Analysis of the inhibition of bradykinin binding to OppA by randomized peptide libraries shows that (i) the affinity of OppA for nonameric peptides is the highest, but peptides up to 35 amino acid residues are still bound; (ii) the effect of d-amino acid residues in a nonameric peptide is most pronounced at positions 4, 5, and 6; d-amino acid residues at the N- and C-terminal positions are better tolerated but not as good as all l-isomer peptides; and (iii) all nonameric peptides are bound by OppA, but the selectivity is determined mainly by the amino acids in the middle positions and the C terminus of the peptide.
The large upper-size limit for peptide binding to OppA of L. lactis is striking, because other oligopeptide-binding proteins, e.g., those of Salmonella typhimurium and E. coli, seem to bind peptides in the range of 2–5 residues with an optimum in binding affinity for tripeptides (18). The di/tripeptide-binding protein DppA of L. lactis also shows strict size-exclusion limits and exclusively binds dipeptides and tripeptides (19), indicating that the broad size specificity is not a general aspect of peptide-binding proteins in L. lactis. It would be interesting to investigate whether peptides of 35 amino acids can still be transported by the Opp system. It is possible that the transport proteins OppB and C determine the upper-size limit of peptide transport, but it should be emphasized that a peptide with a length of 18 residues has been found to be transported (3).
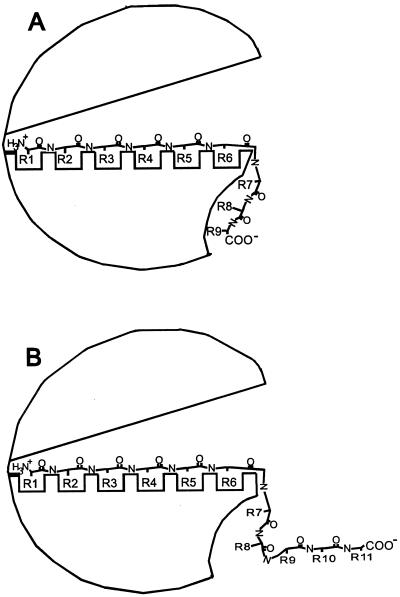
The general mechanism of substrate association to binding proteins is described by the Venus flytrap mechanism, that is, the ligand binds in the cleft between two globular domains, and on binding of the ligand, the protein closes (5). For the OppAs of Gram-negative bacteria, it is known that the peptide is enclosed completely by the binding protein (18, 20). In view of the different properties of OppA of L. lactis, it is very unlikely that the very long peptides are bound in a comparable manner. And indeed, studies with single cysteine containing nonameric peptides, selectively labeled with large bulky fluorescent and nonfluorescent groups, indicate that only the first six residues are enclosed by OppA of L. lactis (7). There is, however, not a direct relationship between the distribution in affinities of the peptides with a particular residue at a given position and the parts of the peptide that are enclosed (residues 1–6) and those that stick out (residues 7–9). Looking at the effect of d-amino acids in the nonameric randomized peptides, it is clear that the d-isomers contribute most to a decrease in affinity at positions 4, 5, and 6 (and 7). These results thus indicate that the first three binding pockets of OppA are more promiscuous than the pockets 4, 5, and 6. If residues at positions 7, 8, and 9 associate with the surface of OppA in an unspecific manner without the peptide backbone and side chains making specific contacts with the protein (“opportunistic binding”), one would expect an inhibition of bradykinin binding by the D7, D8, and D9 libraries comparable to that of all l-isomer peptides. This inhibition is clearly not observed. In fact, the affinity of OppA for peptides with d-amino acids at positions 8 or 9 is comparable to the affinities of OppA for those peptides with d-amino acids at position 1, 2, or 3, and this affinity is much lower than that of all l-isomer peptide libraries. These observations suggest that the interaction of the residues 8 and 9 with the surface of OppA is not (entirely) opportunistic and that specific contacts with the protein are made. The observation that the binding affinity sharply drops with peptides longer than nine residues—in fact the binding affinities of octameric and undecameric (and longer) peptides are similar (Fig. 2)—suggests that steric hindrance of the additional residues (e.g., going from the 9-mer to the 11-mer) prevents critical contacts from being made between the ninth residue and OppA (Fig. 6). Consistent with this interpretation is the observation that amidation of the C terminus of the nonameric peptide SLSQSLSQS results in a 7-fold increase in the Kd (7).
Figure 6.
Schematic representation of the peptide-binding mechanism of OppA of L. lactis. Possible binding to OppA of nonameric (A) and undecameric (B) peptides. The open-liganded conformation is depicted.
The absolute difference in affinity of OppA for the nonameric peptides from the library is small. This result implies that the side chains of the amino acid residues of the peptide contribute relatively little to the overall affinity. The affinity would thus be determined mainly by the electrostatic interactions and hydrogen bonding of the peptide backbone and the binding protein. Overall, peptides with hydrophobic and aromatic residues bind with higher affinity, and those with proline, glycine, and negatively charged amino acids exhibit the lowest affinity. Strikingly, the observed differences in affinity for the peptides resemble those of di/tripeptide-binding protein DppA of L. lactis. Glycine and glutamate containing dipeptides and tripeptides bind with low affinity; triglycine did not seem to be a substrate; and dipeptides and tripeptides with hydrophobic residues were all high-affinity substrates of DppA (19). Also, for the human peptide transporter associated with antigen processing, it has been found that hydrophobic and aromatic residues are favored, whereas negatively charged residues are disfavored at the C-terminal position of nonameric peptides. Proline at the second position of a nonameric peptide resulted in a dramatic drop of the affinity (8). Finally, small peptides in water have been subdivided into nine different classes on the basis of conformational analysis. It has been suggested that the relative binding (and transport) of a particular peptide relates to its conformation in water (21). It is possible that the conformation of the peptides tested here also affects the binding to OppA, but at present there are too few data on the conformations of nonameric peptides in solution to test this hypothesis.
In conclusion, the results clearly add to our understanding of the peptide-binding mechanism of OppA of L. lactis, and the observed features set this protein apart from the homologs in E. coli and S. typhimurium of which the three-dimensional structures have been determined. It should be stressed, however, that (detailed) binding studies with peptides larger than five residues have not been performed with these proteins. It is worth noting that some studies indicate a chaperone-like activity for OppA of E. coli (22), implying that larger peptides interact with the surface of this protein. At present, it is unclear how this proposed chaperone-like activity relates to the peptide-binding mechanism of the protein.
Acknowledgments
This work was supported financially by the Ministry of Economic Affairs, the Ministry of Education, Culture, and Science, and the Ministry of Agriculture, Nature Management, and Fishery in the framework of an industrially relevant research program of the Netherlands Association of Biotechnology Centres in The Netherlands, by European Union Grant Bio-4-CT-960016, and by the Deutsche Forschungsgemeinschaft (to R.T.).
Abbreviations
- Opp
oligopeptide transport system
- PEG
polyethylene glycol
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220308797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220308797
References
- 1.Kunji E R S, Hagting A, de Vries C J, Juillard V, Haandrikman A J, Poolman B, Konings W N. J Biol Chem. 1995;270:1569–1574. doi: 10.1074/jbc.270.4.1569. [DOI] [PubMed] [Google Scholar]
- 2.Kunji E R S, Fang G, Jeronimus-Stratingh C M, Bruins A P, Poolman B, Konings W N. Mol Microbiol. 1998;27:1107–1118. doi: 10.1046/j.1365-2958.1998.00769.x. [DOI] [PubMed] [Google Scholar]
- 3.Detmers F J M, Kunji E R S, Lanfermeijer F C, Poolman B, Konings W N. Biochemistry. 1998;37:16671–16679. doi: 10.1021/bi981712t. [DOI] [PubMed] [Google Scholar]
- 4.Tynkkynen S, Buist G, Kunji E R S, Kok J, Poolman B, Venema G, Haandrikman A J. J Bacteriol. 1993;175:7523–7532. doi: 10.1128/jb.175.23.7523-7532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quiocho F A, Ledvina P S. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 6.Lanfermeijer F C, Picon A, Konings W N, Poolman B. Biochemistry. 1999;38:14440–14450. doi: 10.1021/bi9914715. [DOI] [PubMed] [Google Scholar]
- 7.Lanfermeijer F C, Detmers F J M, Konings W N, Poolman B. EMBO J. 2000;19:3649–3656. doi: 10.1093/emboj/19.14.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uebel S, Kraas W, Kienle S, Wiezmüller K-H, Jung G, Tampé R. Proc Natl Acad Sci USA. 1997;94:8976–8981. doi: 10.1073/pnas.94.17.8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uebel S, Wiesmüller K-H, Jung G, Tampé R. In: Current Topics in Microbiology and Immunology. Winnacker E L, Wong P, Famulok M, editors. Heidelberg: Springer; 1999. pp. 1–27. [DOI] [PubMed] [Google Scholar]
- 10.Uebel S, Tampé R. Curr Opin Immunol. 1999;11:203–208. doi: 10.1016/s0952-7915(99)80034-x. [DOI] [PubMed] [Google Scholar]
- 11.Picon A, Kunji E R S, Lanfermeijer F C, Konings W N, Poolman B. J Bacteriol. 2000;182:1600–1608. doi: 10.1128/jb.182.6.1600-1608.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Putman M, van Veen H W, Poolman B, Konings W N. Biochemistry. 1999;38:1002–1008. doi: 10.1021/bi981863w. [DOI] [PubMed] [Google Scholar]
- 13.Biessen E A L, Horn A S, Robillard G T. Biochemistry. 1990;29:3349–3354. doi: 10.1021/bi00465a029. [DOI] [PubMed] [Google Scholar]
- 14.Stevanovic S, Jung G. Anal Biochem. 1993;212:212–230. doi: 10.1006/abio.1993.1314. [DOI] [PubMed] [Google Scholar]
- 15.Metzger J W, Kempter C, Wiesmüller K-H, Jung G. Anal Biochem. 1994;219:261–277. doi: 10.1006/abio.1994.1266. [DOI] [PubMed] [Google Scholar]
- 16.Wiesmüller K-H, Feiertag S, Fleckenstein B, Kienle S, Stoll D, Hermann M, Jung G. In: Peptide and Cyclopeptide Libraries. Jung G, editor. Weinheim, Germany: Verlag Chemie; 1996. pp. 203–246. [Google Scholar]
- 17.Lowry O H, Rosenbrough N J, Farr A J, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 18.Sleigh S H, Seavers P R, Wilkinson A J, Ladbury J E, Tame J R H. J Mol Biol. 1999;291:393–415. doi: 10.1006/jmbi.1999.2929. [DOI] [PubMed] [Google Scholar]
- 19.Sanz Y, Lanfermeijer F C, Konings W N, Poolman B. Biochemistry. 2000;39:4855–4862. doi: 10.1021/bi992720s. [DOI] [PubMed] [Google Scholar]
- 20.Davies T G, Hubbard R E, Tame J R H. Protein Sci. 1999;8:1432–1444. doi: 10.1110/ps.8.7.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Payne G W, Grail B M, Marshall N J. Biochem Biophys Res Commun. 2000;267:283–289. doi: 10.1006/bbrc.1999.1967. [DOI] [PubMed] [Google Scholar]
- 22.Richarme G, Dantas Caldas T. J Biol Chem. 1997;272:15607–15612. doi: 10.1074/jbc.272.25.15607. [DOI] [PubMed] [Google Scholar]