Abstract
Progress in understanding the pathogenesis of hepatitis C virus (HCV) has been slowed by the absence of tractable small animal models. Whereas GB virus B (GBV-B, an unclassified flavivirus) shares a phylogenetic relationship and several biologic attributes with HCV, including hepatotropism, it is not known to cause persistent infection, a hallmark of HCV. Here, we document persistent GBV-B infection in one of two healthy tamarins (Saguinus oedipus) inoculated intrahepatically with infectious synthetic RNA. High-titer viremia (108 to 109 genome equivalents per ml) and transiently elevated serum alanine transaminase activities were present from weeks 4 to 12 postinoculation in both animals. However, whereas GBV-B was eliminated from one animal by 20 weeks, the second animal remained viremic (103 to 107 genome equivalents per ml) for >2 years, with alanine transaminase levels becoming elevated again before spontaneous resolution of the infection. A liver biopsy taken late in the course of infection demonstrated hepatitis with periportal mononuclear infiltrates, hepatocellular microvesicular changes, cytoplasmic lipid droplets, and disordered mitochondrial ultrastructure, findings remarkably similar to chronic hepatitis C. GBV-B-infected hepatocytes contained numerous small vesicular membranous structures resembling those associated with expression of HCV nonstructural proteins, and sequencing of GBV-B RNA demonstrated a rate of molecular evolution comparable to that of HCV. We conclude that GBV-B is capable of establishing persistent infections in healthy tamarins, a feature that substantially enhances its value as a model for HCV. Mitochondrial structural changes and altered lipid metabolism leading to steatosis are conserved features of the pathogenesis of chronic hepatitis caused by these genetically distinct flaviviruses.
Chronic hepatitis C is a major threat to the public health. Although the underlying molecular mechanisms remain uncertain, hepatitis C virus (HCV) usually escapes both innate and adaptive immune responses, resulting in long-term persistence of the virus, and placing chronically infected individuals at risk for cirrhosis and hepatocellular carcinoma (1). Available therapies include IFN-α and ribavirin, but their efficacy is limited, and even lengthy treatment frequently fails to achieve virus elimination (reviewed in ref. 2). Unfortunately, the search for better and more specific antiviral therapies is complicated by the lack of a robust, fully permissive cell culture system, as well as the absence of a readily available animal model of HCV infection.
Other than humans, the only animal species that has been shown to be reproducibly susceptible to virus challenge is the chimpanzee (Pan troglodytes) (3, 4). Chimpanzees may also be infected by inoculation of synthetic, genome-length viral RNA directly into the liver (5–9). Although there is typically little evidence of liver disease, the chimpanzee model has allowed characterization of HCV replication in vivo, the host response to the virus, and molecular evolution of the virus during persistent infection (10–13). However, chimpanzees are endangered, and are available on only a very limited basis, and their use is prohibitively expensive. There is thus a critical need for an alternative and more readily available animal model.
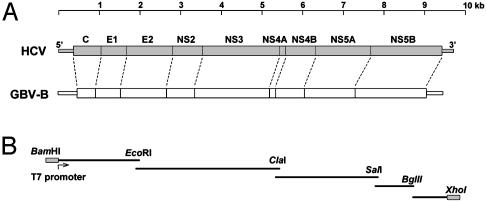
The GB hepatitis agent was initially recovered from a surgeon who presented with jaundice. Serial passage of the infectious material in tamarins was associated with an acute, inflammatory hepatitis (14, 15). However, the nature of the responsible agent remained an enigma until Simons et al. (16) molecularly cloned its genome and demonstrated that it was a previously unknown flavivirus-like virus, which they termed GB virus B (GBV-B). GBV-B is hepatotropic and causes acute hepatitis in several species of New World monkeys, including tamarins (Saguinus sp.) (14, 17), owl monkeys (Aotus trivirgatus) (18), and marmosets (Callithrix jacchus) (19). It is more closely related to HCV than any other known animal virus in terms of both its genome organization (Fig. 1A) and sequence relatedness (16, 20–22). For these reasons, it is a potentially useful surrogate for modeling host–pathogen interactions in hepatitis C. To date, however, GBV-B has not been shown to establish persistent infection, nor cause chronic hepatic inflammation, both of which are key features of hepatitis C.
Fig. 1.
(A) Alignments of the HCV and GBV-B genomes, showing putative protein assignments in the GBV-B polyprotein coding sequence and the similarities in GBV-B and HCV genome structure. (B) Overlapping segments of the GBV-B genome that were amplified by RT-PCR and were assembled into the full-length clone pGBV-B/2.
Here, we describe the long-term persistence of GBV-B in a tamarin infected by intrahepatic inoculation of synthetic viral RNA. We show that chronic GBV-B infection leads to many of the pathologic and virologic changes associated with chronic hepatitis C.
Methods
Virus and Antibodies. Tamarin sera containing infectious GBV-B were obtained from Jens Bukh (Laboratory for Viral Diseases, National Institute of Allergy and Infectious Diseases, Bethesda) and from an animal infected with GBV-B reference material provided by the American Type Culture Collection. Rabbit antisera to recombinant GBV-B NS3 and GBV-B NS5B were obtained from Michael Murray (Schering–Plough Research Institute, Kenilworth, NJ).
cDNA Cloning, Plasmid Construction, and RNA Transcription. Viral RNA was extracted from the serum of a GBV-B-infected tamarin, converted to cDNA by reverse transcription, and amplified by PCR, using oligonucleotide primers based on the reported GBV-B sequence (ref. 16 and Fig. 1B). cDNA representing the then unknown 3′ terminal sequences of the virus was obtained by using the approach described by Kolykhalov et al. (23). Further details of these methods are described in Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org. A genome-length cDNA copy was assembled downstream of the T7 promoter in pACNR1180 (gift of Peter Bredenbeek, Leiden University, Leiden, The Netherlands), resulting in the plasmid pGBV-B/2. Two micrograms of XhoI-digested plasmid DNA were transcribed in vitro by using a T7 MEGAscript kit (Ambion, Austin, TX). The RNA product was examined by nondenaturing agarose gel electrophoresis and stored frozen at –70°C until the tamarins were inoculated.
Tamarin Inoculations. RNA (≈75 μg) diluted in PBS was inoculated at laparotomy into the livers of GBV-B-naïve tamarins (Saguinus oedipus). Serum samples were collected at regular intervals postinoculation and, where indicated, liver biopsies were obtained at laparoscopy. All animal protocols were approved by the Institutional Animal Care and Use Committee of the Southwest Foundation for Biomedical Research.
TaqMan Quantification of GBV-B RNA. GBV-B RNA was extracted from tamarin serum samples and quantified by TaqMan real-time RT-PCR assays with primers targeting the core (24) or NS5A regions of the genome. Details are provided in Supporting Methods.
GBV-B NS3 ELISA. Tamarin sera were tested for the presence of antibodies to GBV-B NS3 by ELISA as described (24), with the exception that purified NS3 (a gift of Michael Murray, Schering–Plough Research Institute) was used in lieu of GST-NS3 fusion protein.
Determination of GBV-B Sequence. GBV-B RNA was isolated from serum by using the QIAamp viral extraction kit (Qiagen, Valencia, CA) and heated to 65°C for 5 min before conversion to cDNA by using hexamer oligonucleotides, and either the Advantage RT-PCR kit (CLONTECH) or SuperScript III reverse transcriptase (Invitrogen). The resulting cDNAs programmed PCR amplification of 0.5- to 2.0-kb DNA fragments by using GBV-B-specific primers, and the Advantage 2 PCR enzyme system (CLONTECH) or Platinum Taq polymerase (Invitrogen). Purified amplimers were sequenced directly on an Applied Biosystems automatic sequencer.
Results
Construction of a Genome-Length, Infectious Molecular Clone of GBV-B. Viral RNA was extracted from the serum of experimentally infected tamarins and was reverse-transcribed into cDNA. The GBV-B genome was then PCR amplified in four segments (see Fig. 1B) and cloned into plasmid DNA. The 3′ terminal viral sequence, not reported by Simons et al. (16), was amplified by RT-PCR after ligation of a synthetic oligonucleotide to the 3′ end of the viral RNA (see Methods). The resulting cDNA fragments were assembled into a genome-length clone (pGBVB/2) downstream of the T7 RNA polymerase promoter. Excluding the poly(U) track within the 3′UTR (23 nt), the pGBVB/2 sequence (GenBank accession no. AY243572) shares 99.9% and 99.6% nucleotide identity with infectious molecular clones reported by Bukh et al. (ref. 25; GenBank accession no. AF179612) and Sbardellati et al. (ref. 26; EMBL accession no. AJ277947), and encodes a polyprotein differing from these at only three and seven residues, respectively.
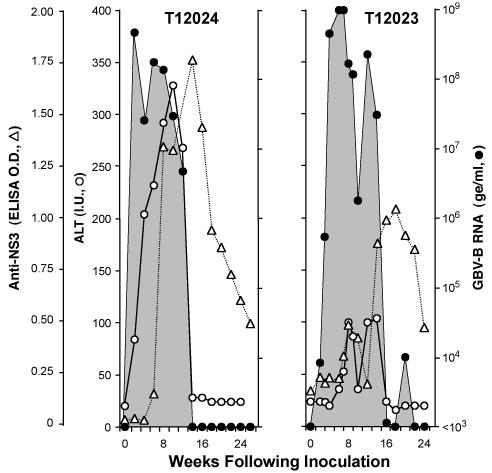
To assess the infectivity of RNA transcribed from pGBV-B/2, the products of two separate in vitro T7 transcription reactions were inoculated into the livers of two healthy tamarins. Infection was documented in each by the presence of viral RNA in subsequent serum samples (detected by quantitative RT-PCR), and the development of antibodies reactive with the viral NS3 protein. Fig. 2 compares the course of infection in one of these tamarins (T12023) with the typical course of infection in a tamarin inoculated intravenously with wild-type virus (T12024). Both of these tamarins developed an acute, self-limited infection characterized by a transient, high-titer viremia associated with significant elevation of the serum alanine transaminase (ALT) activity (Fig. 2).
Fig. 2.
Profiles of acute GBV-B infections in tamarins: T12024, which was inoculated intravenously with GBV-B-positive tamarin serum, and T12023, which was infected by intrahepatic inoculation of synthetic pGBV-B/2 RNA. •, viremia measured by quantitative RT-PCR with a sensitivity of detection of ≈103 ge/ml (except at weeks 143 and 147, when testing of a larger volume of serum improved sensitivity to ≈50 ge/ml). ○, serum ALT (IU); (▵), anti-NS3 (ELISA OD at 405 nm).
There was a subtle difference in these infection profiles, however, in terms of the rapidity with which maximum viremia (≈109 genome equivalents (ge)/ml in both tamarins) was achieved. Peak viremia in T12023 did not occur until 4 weeks after the intrahepatic inoculation of synthetic RNA, with significantly less viral RNA present in the serum at 2 and 3 weeks, whereas viremia was maximal at the time of the first bleed at 2 weeks in T12024 and other tamarins inoculated with virus (data not shown). This difference in the kinetics of viremia was reproduced in the second tamarin inoculated with RNA (see below). Its basis is uncertain, but likely reflects a difference in the specific infectivity of the inocula. Viremia persisted at high levels for 14 weeks in T12023, then rapidly declined in magnitude coincident with resolution of the ALT abnormalities (Fig. 2). Multiple serum samples collected at late time points contained no detectable viral RNA and declining levels of antibodies to the GBV-B NS3 protein. These results confirm the infectivity of synthetic RNA transcribed from pGBV-B/2.
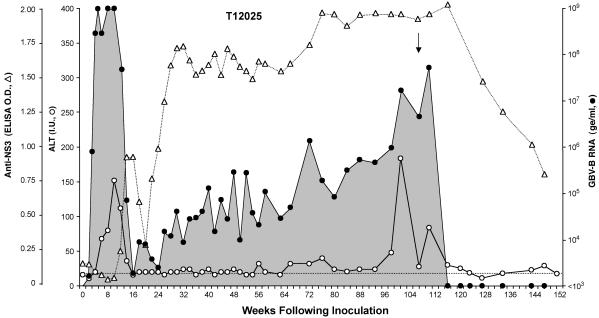
Persistent GBV-B Infection. Whereas the absence of viral persistence has been considered an important difference between GBV-B and HCV infection, a second tamarin (T12025) inoculated with synthetic GBV-B RNA developed persistent infection and chronic viral hepatitis (Fig. 3). As was the case with T12023 (Fig. 2), T12025 developed a high-titer viremia after a delay of 4 weeks. This viremia was accompanied by elevated ALT levels during the second and third months of the infection. A significant drop in the magnitude of the viremia between 12 and 16 weeks after infection was associated with a return of the ALT activity to baseline. However, T12025 failed to completely eliminate the infection, and viral RNA remained detectable in serum samples collected over the ensuing 23 months (Fig. 3). Antibodies to GBV-B NS3, which were present by 14 weeks after RNA inoculation, persisted for the ensuing 30 months.
Fig. 3.
Persistent GBV-B infection in tamarin T12025, which was infected by intrahepatic inoculation of synthetic RNA (see the Fig. 2 legend). The arrow marks the timing of the liver biopsy. The dashed line represents preinfection serum ALT activity.
After a nadir in the viremia of ≈103 ge/ml at 16 weeks postinoculation, the quantity of circulating virus increased slowly in T12025, reaching ≈5 × 107 ge/ml by 110 weeks (Fig. 3). This increase in viremia was matched by a gradual elevation of the serum ALT activity, which was over twice baseline by week 66, and demonstrated a sharp spike 101 weeks after RNA inoculation (Fig. 3). Approximately 10 weeks later, the viremia abruptly declined, with apparent clearance of the virus by week 120. Viral RNA was not detected in five subsequent serum samples collected over the next 28 weeks, even when larger volumes of serum were tested to improve the sensitivity of detection to ≈50 ge/ml. Decreasing levels of NS3 antibody activity after week 120 were also consistent with the elimination of GBV-B (Fig. 3). Thus, GBV-B infection persisted in T12025 for >2 years, with viremia and chemical evidence of hepatitis, but eventually spontaneously resolved after a second episode of acute hepatocellular injury and marked ALT elevation.
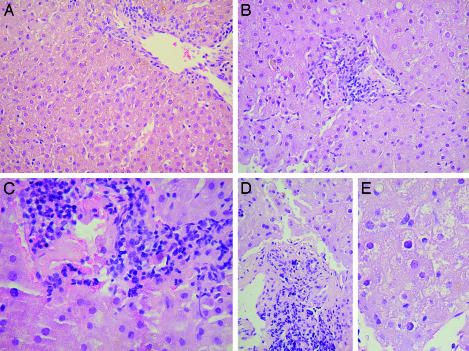
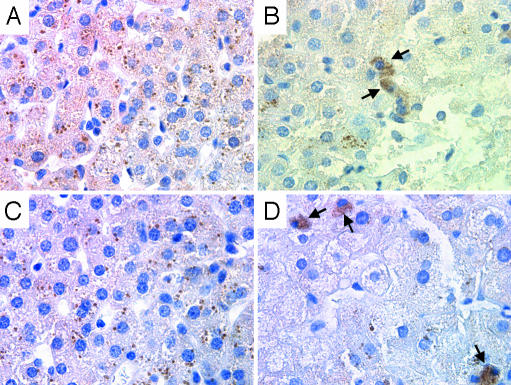
Pathologic Changes in Persistent GBV-B Infection. A biopsy of the liver at week 107 confirmed ongoing hepatitis in T12025 after almost 2 years of persistent viremia (Fig. 3). Although the serum ALT was within the normal range at the time of biopsy [28 international units (IU)], as were serum aspartate aminotransferase, γ-glutamyl transpeptidase, albumin, bilirubin, and total serum protein levels (data not shown), the portal tracts were expanded with lymphocytic infiltrates (Fig. 4 B–D), findings that were not present in uninfected liver (Fig. 4A). Focal disruption of the portal tracts was associated with spillover of lymphocytes into the parenchyma (Fig. 4C). There was relatively little inflammatory change within the liver parenchyma, but microvesicular changes were widespread within the hepatocytes of the persistently infected tamarin (Fig. 4E). Lipofuchsin deposits, which were present in both infected and uninfected tamarins, were probably unrelated to GBV-B. Immunohistochemical staining demonstrated the presence of GBV-B NS3 and GBV-B NS5B proteins within the cytoplasm of <1% of hepatocytes in the infected liver (Fig. 5 B and D). Antigen-laden cells were located outside of the periportal regions where inflammation was most prominent, and were not identified in uninfected control tissue (Fig. 5 A and C).
Fig. 4.
Hematoxylin/eosin-stained sections of normal tamarin liver (A) and liver from tamarin T12025 (B–E), 107 weeks after intrahepatic inoculation of RNA. See text for findings. Original magnification: A, B, and D, ×50; C, ×100; E, ×200.
Fig. 5.
Immunohistochemical staining for GBV-B NS3 (A and B) and GBV-B NS5B (C and D) antigens in normal tamarin liver (A and C) and liver taken from T12025 (B and D), 107 weeks after RNA inoculation. Arrows indicate cells containing abundant cytoplasmic viral antigen.
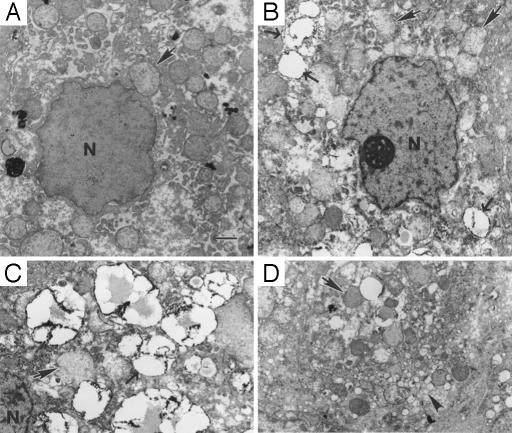
Examination of ultrathin sections by transmission electron microscopy demonstrated abnormalities in mitochondrial ultrastructure in the persistently infected liver tissue, with loss of both density and definition of cristae (Fig. 6; compare structures marked by solid arrows in A versus B–D). Most hepatocytes in the infected liver contained multiple small (≈2-μm diameter) lipid-containing vesicles (Fig. 6 B and C, open arrows), which were not present in uninfected tamarin liver (Fig. 6A). Interestingly, in some hepatocytes, these lipid droplets appeared to be within the nucleus (not shown). Some hepatocytes also contained clusters of smooth-surfaced cytoplasmic vesicles of various sizes (Fig. 6D, arrowhead). These membranous structures resemble those described in cells expressing HCV nonstructural proteins, and that have been proposed to contribute to the viral replication complex (27). These vesicles may thus be indicative of ongoing GBV-B replication in hepatocytes.
Fig. 6.
Transmission electron microscopy of normal tamarin liver (A) and
persistently infected tamarin liver from T12025 (B–D).  ,
mitochondria, with ill-defined cristae in the infected tissue. →, lipid
droplets with electron-dense surface deposits. ▸, clusters of small
smooth-surface vesicles. N, nucleus. (Bar, 1 μm.)
,
mitochondria, with ill-defined cristae in the infected tissue. →, lipid
droplets with electron-dense surface deposits. ▸, clusters of small
smooth-surface vesicles. N, nucleus. (Bar, 1 μm.)
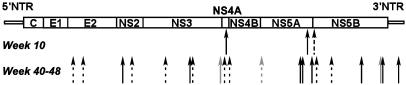
Molecular Evolution of GBV-B During Persistent Infection. To determine the rate of molecular evolution of the GBV-B genome during persistent infection, we amplified the viral sequence by RT-PCR from serum collected from T12025 40–48 weeks after initial inoculation and compared the dominant nucleotide sequence with that of the synthetic RNA inoculum. As summarized in Fig. 7, a total of 23 single-nucleotide substitutions were identified over the entire genome, 11 of which resulted in amino acid changes within the polyprotein (see Table 1, which is published as supporting information on the PNAS web site). Three single-nucleotide substitutions were present by week 10, two of which eliminated amino acid differences that distinguish our infectious GBV-B molecular clone from that reported by Bukh et al. (25): Ala-1594 → Thr in NS4A and Val-2236 → Ala in NS5A. Similar mutations (Ala-1594 → Val and Val-2236 → Ala) occurred during the self-limited infection of T12023 (Fig. 2). Mutations at these residues are likely to represent correction of suboptimal quasispecies polymorphisms present in pGBV-B/2. Three other nucleotide changes were present in the dominant sequence at weeks 40–42, but they either completely or partially reverted to the pGBV-B/2 sequence by weeks 44–48. Such results are consistent with ongoing quasispecies selection during GBV-B persistence. Excluding these six mutations, the GBV-B genome accumulated mutations at a rate of ≈2.4 × 10–3 nucleotide substitutions per base position per year during persistent infection, a value remarkably close to that reported for HCV in persistently infected chimpanzees (1.5–2.2 × 10–3 nucleotide substitutions per base position per year; refs. 10 and 11). Interestingly, none of the 11 mutated amino acid residues in GBV-B were within the envelope glycoproteins (Fig. 7).
Fig. 7.
Mutations present in virus circulating in T12025 between weeks 40 and 48 postinoculation of RNA. Those occurring by week 10 were also observed in acutely infected tamrins. Solid arrows indicate nonsilent mutations in the ORF or nucleotide substitutions in the noncoding regions of the viral genome. Lightly shaded arrows indicate mutations that were present at weeks 40–42 but reverted to wild-type sequence by weeks 44–48.
Passage of GBV-B Virus from a Persistently Infected Tamarin. To determine whether the unusually lengthy persistence of virus in T12025 was related to mutation in the GBV-B genome, we inoculated a naïve tamarin, T16441, intravenously with week-52 serum from T12025 containing ≈3 × 104 ge of virus. This tamarin sustained a typical acute, self-limited infection, which was marked by the presence of high-titer viremia from weeks 2–16 postinoculation, and with a prominent but transient increase in serum ALT activity associated with histopathologic evidence of acute hepatitis (data not shown). These results indicate that the long-term persistence of GBV-B in T12025 reflects an individual host response to the infection, rather than a unique genetic composition of the virus.
Discussion
Because animal modeling of HCV infections is restricted to chimpanzees, it is extraordinarily expensive and difficult to accomplish. GBV-B offers an excellent surrogate, although its natural host is uncertain. Attempts to demonstrate GBV-B in either the original human GB serum specimen or in other human or nonhuman primate materials failed (17, 28). Early cross-protection studies suggested that GBV-B may be of human origin (29), but the virus does not infect chimpanzees (18). Thus, GBV-B is likely to be native to one or more nonhuman primate species, probably of New World origin (14, 18, 19, 30). Whereas GBV-B and HCV are distinctly different agents, sharing only 27–33% amino acid identity within the polyprotein (20, 21), it would seem reasonable to assign them to the same genus, Hepacivirus, in the family Flaviviridae (31). The genomes have a similar overall organization. The putative structures of the internal ribosome entry sites of these viruses are remarkably similar, with the exception of two additional internal stem loops in GBV-B (32, 33). The NS3 serine proteinases of these viruses have closely related substrate specificities (34), requirements for analogous cognate NS4A cofactor domains, and susceptibility to candidate HCV NS3/4A protease inhibitors (35). Finally, like HCV, the 3′NTR of GBV-B contains a homopolymeric poly(U) track (25, 36).
However, two putative attributes of GBV-B have distinguished it from HCV. First, long-term persistence of virus has not been observed in tarmarins infected with GBV-B. Second, there has been little genetic variation among reported GBV-B sequences (16, 25, 26). This latter observation is likely to reflect the common origin of the GBV-B sequences reported to date, because we observed a rate of molecular evolution of GBV-B similar to that reported for HCV during virus persistence (10, 11). Interestingly, no mutations occurred within the structural proteins during the persistence of virus in T12025 (Fig. 7). The analogous proteins of HCV also undergo little molecular evolution in persistently infected chimpanzees, even within the hypervariable regions (10, 11, 13). This finding suggests that persistent GBV-B infection, like persistent HCV infection, is not caused by escape from a neutralizing antibody response, at least in these nonhuman primate models. As with HCV, the evolution of acute GBV-B infection to long-term persistence is likely to depend on a complex interplay with the host's immune response, particularly in the early stages of the infection (37), and may be indicative of specific virus disruption of one or more immune defense mechanisms.
Of ≈30 tamarins inoculated intravenously with GBV-B and reported in the literature, all cleared the virus between 9 and 26 weeks postinoculation and none developed chronic infection (17–19, 25, 26). Whereas this result suggests that persistent GBV-B infection may be relatively uncommon, the course of infection in T12025 demonstrates that at least some normal tamarins fail to clear the acute infection and go on to develop a chronic hepatitis associated with persistent viremia. Because there are too few reports (six tamarins) of animals challenged by direct intrahepatic inoculation of synthetic RNA (this article and refs. 25 and 26), it is unclear as to whether this route of infection enhances the likelihood of persistent infection. We cannot rule out the possibility that the pGBV-B/2 sequence might uniquely predispose to persistence, although T12023 developed a typical acute infection after inoculation with this RNA (Fig. 2). We have observed the consistent selection of alternative sequence polymorphisms at amino acid residues in both NS4A and NS5A after infection of tamarins with synthetic RNA (Fig. 7), and such polymorphisms could be important to the generation of persistence. Further studies will be needed to determine the frequency and mechanisms of GBV-B persistence, as well as the immunologic responses that led to eventual elimination of the infection in T12025. Such spontaneous resolution of HCV infection is rare once chronicity is established (38).
A biopsy of the liver 107 weeks after the inoculation of T12025 with synthetic RNA, while viremia was still at high titer (Fig. 3), revealed pathologic features characteristic of the liver injury in chronic hepatitis C. In addition to lymphocytic infiltration of portal tracts and occasional focal disruption of the limiting plate (Fig. 4 B–D), there were widespread microvesicular changes in hepatocytes (Fig. 4E), with numerous small lipid droplets visualized by transmission electron microscopy (Fig. 6B). Steatosis is an important and relatively unique hallmark of chronic hepatitis C in humans (39). The core protein of HCV has been closely associated with lipid accumulation in both cellular and transgenic animal models (40–42), and it localizes to the surface of such droplets (40, 43). Both HCV and GBV-B core proteins have been suggested to contain a proline motif resembling the lipid-interaction domain of plant oleosins (44). The lipid droplets in the hepatocytes of T12025 represent a previously unrecognized feature of GBV-B-associated liver injury. They support the contention that the GBV-B and HCV core proteins share a common capacity for interacting with pathways involved in lipid metabolism, and suggest that lipid droplet formation may be important in the viral life cycle.
The changes in mitochondrial ultrastucture noted in T12025 (Fig. 6) also parallel observations in HCV-infected livers (45, 46), as well as in transgenic animal and cellular models of HCV protein expression (47). Such common histopathology suggests common pathogenetic mechanisms, and indicates that more could be learned by further studies of chronic GBV-B hepatitis in New World monkeys.
Supplementary Material
Acknowledgments
We thank Deborah Chavez, Bradley Pfeiffer, Hui-Qun Wang, and Teri Chapa for expert technical assistance; Shu-Yuan Xiao for interpretation of liver histology; Jens Bukh for providing GBV-B inoculum; Peter Bredenbeek for pACNR1180; and Michael Murray for GBV-B antibodies and antigen. This work was supported in part by National Institutes of Health Grants R24-RR15081, U19-AI40035, RO1-AI49574, and P51-RR13986, the Schering-Plough Research Institute, and the French Ministère de la Recherche (“Emergence, Jeune Equipe” and “Réseau Fondamental Hépatite C”).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GBV-B, GB virus B; ge, genome equivalents; HCV, hepatitis C virus; ALT, alanine transaminase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY243572).
References
- 1.Seeff, L. B. & Hoofnagle, J. H. (2002) Hepatology 36, S1–S2. [DOI] [PubMed] [Google Scholar]
- 2.Zein, C. O. & Zein, N. N. (2002) Microbes Infect. 4, 1237–1246. [DOI] [PubMed] [Google Scholar]
- 3.Abe, K., Inchauspe, G., Shikata, T. & Prince, A. M. (1992) Hepatology 15, 690–695. [DOI] [PubMed] [Google Scholar]
- 4.Farci, P., London, W. T., Wong, D. C., Dawson, G. J., Vallari, D. S., Engle, R. & Purcell, R. H. (1992) J. Infect. Dis. 165, 1006–1011. [DOI] [PubMed] [Google Scholar]
- 5.Kolykhalov, A. A., Agapov, E. V., Blight, K. J., Mihalik, K., Feinstone, S. M. & Rice, C. M. (1997) Science 277, 570–574. [DOI] [PubMed] [Google Scholar]
- 6.Beard, M. R., Abell, G., Honda, M., Carroll, A., Gartland, M., Clarke, B., Suzuki, K., Lanford, R., Sangar, D. V. & Lemon, S. M. (1999) Hepatology 30, 316–324. [DOI] [PubMed] [Google Scholar]
- 7.Yanagi, M., Purcell, R. H., Emerson, S. U. & Bukh, J. (1997) Proc. Natl. Acad. Sci. USA 94, 8738–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanagi, M., St. Claire, M., Shapiro, M., Emerson, S. U., Purcell, R. H. & Bukh, J. (1998) Virology 244, 161–172. [DOI] [PubMed] [Google Scholar]
- 9.Yanagi, M., Purcell, R. H., Emerson, S. U. & Bukh, J. (1999) Virology 262, 250–263. [DOI] [PubMed] [Google Scholar]
- 10.Thomson, M., Nascimbeni, M., Gonzales, S., Murthy, K. K., Rehermann, B. & Liang, T. J. (2001) Gastroenterology 121, 1226–1233. [DOI] [PubMed] [Google Scholar]
- 11.Major, M. E., Mihalik, K., Fernandez, J., Seidman, J., Kleiner, D., Kolykhalov, A. A., Rice, C. M. & Feinstone, S. M. (1999) J. Virol. 73, 3317–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomson, M., Nascimbeni, M., Havert, M. B., Major, M., Gonzales, S., Alter, H., Feinstone, S. M., Murthy, K. K., Rehermann, B. & Liang, T. J. (2003) J. Virol. 77, 862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassett, S. E., Thomas, D. L., Brasky, K. M. & Lanford, R. E. (1999) J. Virol. 73, 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karayiannis, P., Petrovic, L. M., Fry, M., Moore, D., Enticott, M., McGarvey, M. J., Scheuer, P. J. & Thomas, H. C. (1989) Hepatology 9, 186–192. [DOI] [PubMed] [Google Scholar]
- 15.Deinhardt, F., Holmes, A. W., Capps, R. B. & Popper, H. (1967) J. Exp. Med. 125, 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simons, J. N., Pilot-Matias, T. J., Leary, T. P., Dawson, G. J., Desai, S. M., Schlauder, G. G., Muerhoff, A. S., Erker, J. C., Buijk, S. L., Chalmers, M. L., et al. (1995) Proc. Natl. Acad. Sci. USA 92, 3401–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlauder, G. G., Dawson, G. J., Simons, J. N., Pilot-Matias, T. J., Gutierrez, R. A., Heynen, C. A., Knigge, M. F., Kurpiewski, G. S., Buijk, S. L., Leary, T. P., et al. (1995) J. Med. Virol. 46, 81–90. [DOI] [PubMed] [Google Scholar]
- 18.Bukh, J., Apgar, C. L., Govindarajan, S. & Purcell, R. H. (2001) J. Med. Virol. 65, 694–697. [DOI] [PubMed] [Google Scholar]
- 19.Lanford, R. E., Chavez, D., Notvall, L. & Brasky, K. M. (2003) Virology 311, 72–80. [DOI] [PubMed] [Google Scholar]
- 20.Ohba, K., Mizokami, M., Lau, J. Y., Orito, E., Ikeo, K. & Gojobori, T. (1996) FEBS Lett. 378, 232–234. [DOI] [PubMed] [Google Scholar]
- 21.Muerhoff, A. S., Leary, T. P., Simons, J. N., Pilot-Matias, T. J., Dawson, G. J., Erker, J. C., Chalmers, M. L., Schlauder, G. G., Desai, S. M. & Mushahwar, I. K. (1995) J. Virol. 69, 5621–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karayiannis, P. & McGarvey, M. J. (1995) J. Viral Hepat. 2, 221–226. [DOI] [PubMed] [Google Scholar]
- 23.Kolykhalov, A. A., Feinstone, S. M. & Rice, C. M. (1996) J. Virol. 70, 3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beames, B., Chavez, D., Guerra, B., Notvall, L., Brasky, K. M. & Lanford, R. E. (2000) J. Virol. 74, 11764–11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bukh, J., Apgar, C. L. & Yanagi, M. (1999) Virology 262, 470–478. [DOI] [PubMed] [Google Scholar]
- 26.Sbardellati, A., Scarselli, E., Verschoor, E., De Tomassi, A., Lazzaro, D. & Traboni, C. (2001) J. Gen. Virol. 82, 2437–2448. [DOI] [PubMed] [Google Scholar]
- 27.Egger, D., Wolk, B., Gosert, R., Bianchi, L., Blum, H. E., Moradpour, D. & Bienz, K. (2002) J. Virol. 76, 5974–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlauder, G. G., Pilot-Matias, T. J., Gabriel, G. S., Simons, J. N., Muerhoff, A. S., Dawson, G. J. & Mushahwar, I. K. (1995) Lancet 346, 447–448. [DOI] [PubMed] [Google Scholar]
- 29.Holmes, A. W., Deinhardt, F., Wolfe, L., Froesner, G., Paterson, D., Casto, B. & Conrad, M. E. (1973) Nature 243, 419–420. [DOI] [PubMed] [Google Scholar]
- 30.Bukh, J., Apgar, C. L. & Purcell, R. H. (1997) in Viral Hepatitis and Liver Disease, eds. Rizzetto, M., Purcell, R. H., Gerin, J. L. & Verme, G. (Minerva Medica, Turin, Italy), pp. 392–395.
- 31.Robertson, B., Myers, G., Howard, C., Brettin, T., Bukh, J., Gaschen, B., Gojobori, T., Maertens, G., Mizokami, M., Nainan, O., et al. (1998) Arch. Virol. 143, 2493–2503. [DOI] [PubMed] [Google Scholar]
- 32.Rijnbrand, R., Abell, G. & Lemon, S. M. (2000) J. Virol. 74, 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grace, K., Gartland, M., Karayiannis, P., McGarvey, M. J. & Clarke, B. (1999) J. Gen. Virol. 80, 2337–2341. [DOI] [PubMed] [Google Scholar]
- 34.Scarselli, E., Urbani, A., Sbardellati, A., Tomei, L., De Francesco, R. & Traboni, C. (1997) J. Virol. 71, 4985–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butkiewicz, N., Yao, N., Zhong, W., Wright-Minogue, J., Ingravallo, P., Zhang, R., Durkin, J., Standring, D. N., Baroudy, B. M., Sangar, D. V., et al. (2000) J. Virol. 74, 4291–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sbardellati, A., Scarselli, E., Tomei, L., Kekule, A. S. & Traboni, C. (1999) J. Virol. 73, 10546–10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thimme, R., Oldach, D., Chang, K. M., Steiger, C., Ray, S. C. & Chisari, F. V. (2001) J. Exp. Med. 194, 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokosuka, O., Kojima, H., Imazeki, F., Tagawa, M., Saisho, H., Tamatsukuri, S. & Omata, M. (1999) J. Hepatol. 31, 394–399. [DOI] [PubMed] [Google Scholar]
- 39.Goodman, Z. D. & Ishak, K. G. (1995) Semin. Liver Dis. 15, 70–81. [DOI] [PubMed] [Google Scholar]
- 40.Barba, G., Harper, F., Harada, T., Kohara, M., Goulinet, S., Matsuura, Y., Eder, G., Schaff, Z., Chapman, M. J., Miyamura, T., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriya, K., Yotsuyanagi, H., Shintani, Y., Fujie, H., Ishibashi, K., Matsuura, Y., Miyamura, T. & Koike, K. (1997) J. Gen. Virol. 78, 1527–1531. [DOI] [PubMed] [Google Scholar]
- 42.Lerat, H., Honda, M., Beard, M. R., Loesch, K., Sun, J., Yang, Y., Okuda, M., Gosert, R., Xiao, S. Y., Weinman, S. A., et al. (2002) Gastroenterology 122, 352–365. [DOI] [PubMed] [Google Scholar]
- 43.Hope, R. G. & McLauchlan, J. (2000) J. Gen. Virol. 81, 1913–1925. [DOI] [PubMed] [Google Scholar]
- 44.Hope, R. G., Murphy, D. J. & McLauchlan, J. (2002) J. Biol. Chem. 277, 4261–4270. [DOI] [PubMed] [Google Scholar]
- 45.Barbaro, G., Di Lorenzo, G., Asti, A., Ribersani, M., Belloni, G., Grisorio, B., Filice, G. & Barbarini, G. (1999) Am. J. Gastroenterol. 94, 2198–2205. [DOI] [PubMed] [Google Scholar]
- 46.Anderson, M., Murray-Lyon, I. M., Coleman, J. C., McCaul, T. F., Bird, R. G., Tovey, G. & Zuckerman, A. J. (1982) J. Med. Virol. 9, 217–229. [DOI] [PubMed] [Google Scholar]
- 47.Okuda, M., Li, K., Beard, M. R., Showalter, L. A., Scholle, F., Lemon, S. M. & Weinman, S. A. (2002) Gastroenterology 122, 366–375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.