Abstract
DNA repair mechanisms are essential for the maintenance of genomic integrity. Disruption of gene products responsible for DNA repair can result in chromosomal damage. Improperly repaired chromosomal damage can result in the loss of chromosomes or the generation of chromosomal deletions or translocations, which can lead to tumorigenesis. The MYC protooncogene is a transcription factor whose overexpression is frequently associated with human neoplasia. MYC has not been previously implicated in a role in DNA repair. Here we report that the overexpression of MYC disrupts the repair of double-strand DNA breaks, resulting in a several-magnitude increase in chromosomal breaks and translocations. We found that MYC inhibited the repair of γ irradiation DNA breaks in normal human cells and blocked the repair of a single double-strand break engineered to occur in an immortal cell line. By spectral karyotypic analysis, we found that MYC even within one cell division cycle resulted in a several-magnitude increase in the frequency of chromosomal breaks and translocations in normal human cells. Hence, MYC overexpression may be a previously undescribed example of a dominant mutator that may fuel tumorigenesis by inducing chromosomal damage.
Many genes have been identified that are responsible for repairing DNA breaks and preserving chromosomal integrity (1–3). The loss of function of some of these genes can result in widespread chromosomal damage and contribute to tumorigenesis (4–6). In mammalian cells, double-strand DNA breaks (DSBs) are repaired by homology-directed recombination (HDR), nonhomologous end joining (NHEJ), and singlestrand annealing (SSA) (7). Defects in any of these repair mechanisms can result in chromosomal breaks, fusions, and translocations (8). Many human cancers exhibit characteristic chromosomal translocations thought to be responsible for tumorigenesis (9). The mechanisms by which these chromosomal translocations occur are not clear.
MYC is a protooncogene that normally regulates cellular growth and proliferation and in some contexts induces apoptosis (10–12). MYC overexpression is thought to cause tumorigenesis by promoting unrestrained cellular proliferation and blocking differentiation. MYC overexpression may also contribute to tumorigenesis by inducing genomic destabilization (13–15). The genomic damage induced by MYC can be broadly grouped into two classes of abnormalities. First, overexpression of MYC induces loss of chromosomal integrity associated with chromosomal aberrations such as gene amplifications, double minutes, and fusions. Second, MYC overexpression can cause inappropriate DNA replication, resulting in endoreduplication.
We speculated that the former type of genomic abnormalities might be caused by defects in the repair of DSBs. Here we demonstrate that MYC overexpression disrupts the repair of DSBs. Moreover, we show that MYC overexpression in normal human cells results in a several-magnitude increase in chromosomal breaks and translocations. Hence, MYC may be a previously undescribed example of a gain-of-function mutation that can function as a dominant mutator. Our results have implications for how MYC induces and is restrained from causing tumorigenesis.
Materials and Methods
Immunocytochemistry and Staining with γ-H2AX Antibody. NHF-MYCER (NHF, normal human foreskin fibroblasts) were grown in DMEM without phenol red supplemented with 10% FBS and penstrep in four-well culture slides (Becton Dickinson). γ-H2AX staining was performed as described (16). MYC was activated by adding estradiol (E2) (1 μM) to the medium. For irradiation, cells were irradiated with 2 Gy on a 100-mm dish roundtable by using a 137 cesium source.
DSB Assays. DSB repair was measured by using a GFP-based assay for homologous recombination (17). The hamster cell line, DRAA8, and the pCAGGS construct containing I-SceI were kindly provided by M. Jasin. DRAA8 cells were infected with retrovirus derived from LYNX-MYCER, and double-resistant clones (neomycin and puromycin) were isolated. The DRAA8-MYCER cells were electroporated (0.260 kV) with 20 μg of the expression vector pCAGGS and then immediately plated in medium for 24 or 48 h. To activate MYC, E2 (1 μM) was added to the medium. Cells were rinsed twice with PBS, recovered by trypsin/EDTA, concentrated by centrifugation, washed twice with PBS, resuspended in tissue culture medium, and then analyzed by flow cytometry as described (17). Southern blot hybridization was performed by using conventional techniques. To confirm that GFP+ cells had undergone short-track gene conversion, genomic DNA was isolated from FACS-sorted GFP+ cells, then digested with SalI, HindIII, and I-SceIor Bcg1, and then probed for the I-SceI-GFP as described (17). Global DSB repair was measured by a PCR-based assay as described in detail (18). Briefly, genomic DNA was isolated from DRAA8-MYCER cells that had been either not transfected or transfected with I-SceI and untreated or treated with E2. Then PCR amplification of the SceGFP region was performed by using AGGGCGGGGTTCTGG and CCTTCGGGCATGGCGGACTTGA as primers.
Flow Cytometry Analysis for DNA Content and Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling (TUNEL). To measure the DNA content, cells were fixed in ethanol, stained with propidium iodide, and then analyzed by flow cytometry, exactly as described (14). To measure TUNEL-positive cells, the Apo-Direct kit was used as described by the manufacturer (PharMingen, San Diego), and cells were subsequently measured by flow cytometry.
Global Genomic Repair Assay. Global genomic DNA repair was performed as published (19).
Spectral Karyotyping (SKY). Metaphase cells were prepared as described above and used for SKY. SKY was performed by using the Applied Spectral Imaging (ASI, Carlsbad, CA) skypaint kit for human chromosomes according to the manufacturer's protocol. SKY images were acquired with a Spectracube-2 (ASI) mounted on a Leica DMR microscope. Acquisition and analysis were performed by using the ASI image capturing (si 2.2) and analysis (skyview 1.6.1) software. At least 15 metaphase spreads of good spectral hybridization quality and banding morphology were analyzed for each preparation
Results
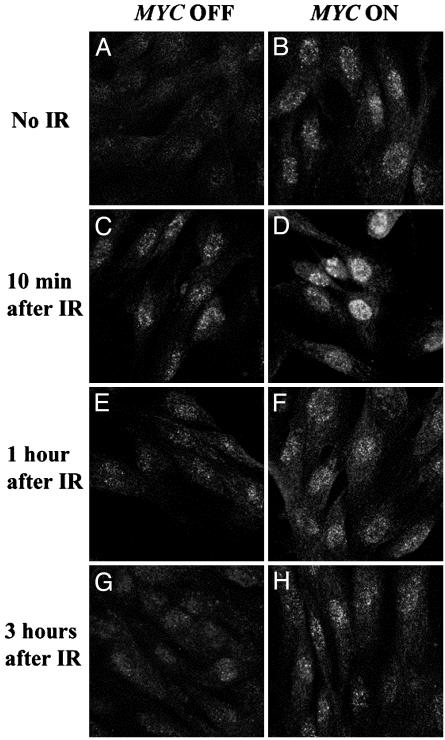
MYC Overexpression Causes DSBs in Normal Cells. To investigate whether MYC activation influenced the repair of DSBs caused by γ irradiation, we conditionally regulated MYC activation in NHF of early passage by introducing, through a retroviral vector, the MYCER chimeric gene product (NHF-MYCER), which exhibits MYC activation only in the presence of E2 (14). As a negative control, we generated NHF that contained the empty retroviral vector (NHF-BABE). Then, we exposed NHF to γ irradiation in the absence and presence of E2. To detect DNA breaks and repair, we measured the formation of γ-H2AX by fluorescent confocal microscopy at different times after irradiation (Fig. 1). MYC activation alone induced an increase in γ-H2AX (Fig. 1 A vs. B). As a negative control, we found that NHF-BABE treated with E2 did not exhibit increased γ-H2AX. As expected, we found that irradiation induced a marked increase in γ-H2AX compared with nonirradiated cells (Fig. 1 C, E, and G). The induction of MYC concurrently with irradiation resulted in an increase of γ-H2AX (Fig. 1 D, F, and H). One hour after irradiation, DNA repair had occurred in the irradiated NHF without MYC activation (Fig. 1E), showing almost identical γ-H2AX staining as the nonirradiated controls, consistent with previous reports that the repair of DNA damage caused by irradiation occurs rapidly (16). In contrast, when we irradiated cells in which we had also activated MYC, γ-H2AX staining persisted for up to 3 h after irradiation (Fig. 1H). We conclude that MYC overexpression interferes with the ability of normal human cells to repair DNA damage caused by γ irradiation.
Fig. 1.
Overexpression of MYC induces formation of γ-H2AX foci in normal human fibroblasts. NHF-MYCER cells were stained for γ-H2AX (green) after 2 h without (A) and with (B) MYC activation and analyzed by confocal microscopy. NHF-MYCER cells without (C, E, and G) and with (D, F, and H) MYC activation were γ-irradiated and stained for formation of γ-H2AX-foci 10 min (C and D), 1h(E and F),and3h(G and H) after irradiation (IR). Representative data from one of three experiments are shown.
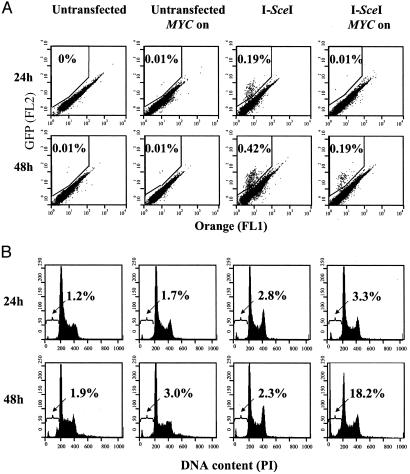
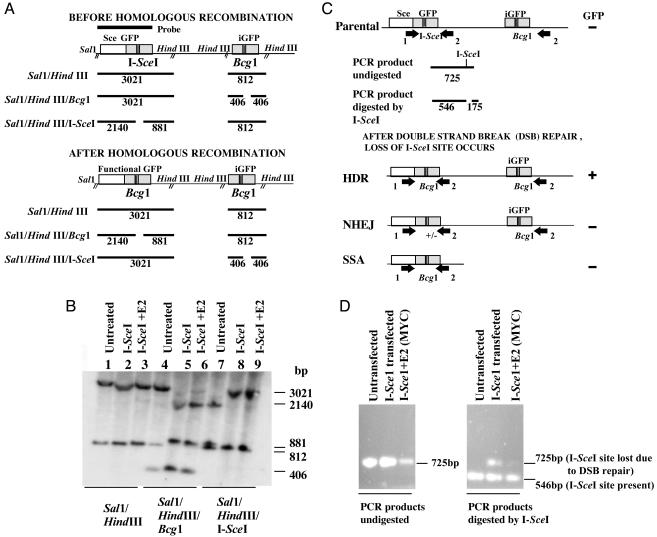
MYC Overexpression Disrupts DNA Break Repair. To determine whether MYC overexpression was directly influencing DSB repair, we examined whether MYC blocked the repair of a single DSB. For this purpose, we used a cell line, DRAA8, which had previously been engineered to contain the recombination substrate, DR-GFP (17). Transfection of I-SceI into these cells results in a single DSB. The repair of this DNA break occurs by homologous recombination resulting in the formation of a functional GFP. We introduced MYCER into the DRAA8 cell line through a retroviral vector and examined the influence of MYC activation on the repair of a DSB (Fig. 2). As expected, the DRAA8 or DRAA8 MYCER cells exhibited a low background level of GFP expression. MYC activation alone did not influence the background frequency of cells expressing GFP. The introduction of I-SceI resulted in an anticipated increase in the number of cells expressing GFP. However, we found that MYC activation for 24 h after I-SceI transfection reduced the frequency of GFP-positive cells to almost background levels (Fig. 2 A). Therefore, it seems that MYC activation inhibits the repair of DSBs by interfering with HDR. We confirmed by Southern analysis that cells that were GFP positive had indeed undergone the predicted recombination through short-track gene conversion (Fig. 3 A and B).
Fig. 2.
MYC overexpression prevents the repair of a DSB through homologous recombination and induces apotosis. DRAA8-MYCER cells were untreated or electroporated with I-SceI in the presence or absence of E2 and analyzed 24 or 48 h later. (A) GFP expression was measured by flow cytometry to determine the frequency of cells that successfully underwent homologous recombination. (B) Propodium iodide staining of cells was analyzed by flow cytometry to measure apoptosis. The percentage of cells that had undergone homologous recombination or were undergoing apoptosis is shown. FL1, green fluorescence; FL2, orange fluorescence. Representative data from one of three experiments are shown.
Fig. 3.
MYC overexpression globally suppresses DNA repair of DSBs. (A) Map of the DR-GFP substrate before and after homologous recombination. Sizes of expected fragments are indicated in base pairs. (B) Southern analysis confirmed that the GFP-positive cells have undergone repair of their DSBs through short-track gene conversion or homologous recombination. Genomic DNA from DRAA8-MYCER cells that were untransfected and GFP-positive cells that were I-SceI-transfected in the presence or absence of MYC activation with E2 was digested with the indicated restriction enzymes and analyzed by Southern analysis. The 812-bp band in lane 4 is due to incomplete digestion by Bcg1. (C) DSB repair mediated through HDR, NHEJ, and SSA results in the loss of the I-SceI site. The PCR-based assay using genomic DNA and primers 1 and 2 (arrow) results in a 725-bp fragment, which can then be cleaved by I-SceI to yield 546- and 175-bp fragments. After I-SceI transfection and DSB repair, the PCR product can no longer be cleaved by I-SceI due to loss of the I-SceI site. (D) Agarose gel analysis of the PCR products from the untransfected, I-SceI-transfected, and I-SceI-transfected and MYC activated cells were either digested by I-SceI (Right) or not (Left). In all three cell lines, the PCR product was a 725-bp fragment (Left). When the PCR products were digested by I-SceI (Right), the untransfected cell line yielded a 546-bp fragment and a 175-bp fragment that is not shown; I-SceI-transfected cells yielded 546- and 725-bp fragments caused by DSB repair, whereas I-SceI-transfected and MYC activated cells were found to yield only a 546-bp fragment due to impairment of DSB repair.
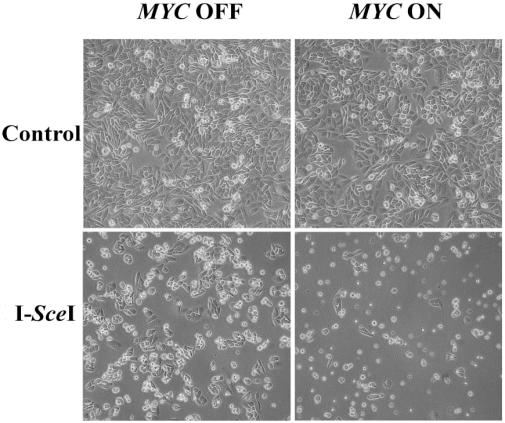
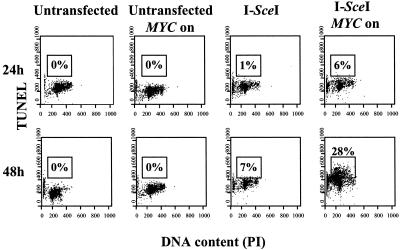
DSB repair can occur through many different pathways. To evaluate whether MYC was globally interfering with DSB repair, we used a PCR-based assay that would detect repair mediated by HDR, NHEJ, or SSA (Fig. 3 C and D; ref. 18). Indeed, we found that MYC suppressed DSB repair through HDR, NHEJ, and SSA. As an apparent consequence, MYC activation caused >90% of the I-SceI-transfected DRAA8 cells to die by 48 h, as observed by phase microscopy (Fig. 4). Among the rare living cells, some had succeeded in undergoing homologous recombination and were GFP positive. The cells appeared to have died from apoptosis, which we confirmed by finding a 40-fold relative increase in number of cells with a subdiploid DNA content (Fig. 2B). Many of these cells were TUNEL positive (Fig. 4). Hence, MYC may induce cells to undergo apoptosis by impairing the repair of DSBs.
Fig. 4.
Cell death caused by MYC overexpression in the presence of a DSB. DRAA8-MYCER cells with or without MYC activation were electroporated with I-SceI, cultured for 48 h, then examined by phase microscopy. Over 90% of the I-SceI-transfected cells overexpressing MYC have died, as shown by the low cell density and large number of floating cells. I-SceI-transfected cells not overexpressing MYC grow at a higher cell density and do not show a large number of floating cells. Untransfected cells were included as a control. Representative data from one of three experiments are shown.
MYC Overexpression Has No Effect on Nucleotide Excision Repair. To determine whether MYC overexpression disrupted other types of DNA repair, we analyzed the effect of MYC to influence nucleotide excision DNA repair of UV irradiation-induced DNA photoproducts. Normal human fibroblasts were UV irradiated in the presence or absence of MYC activation, and global genomic DNA repair activity was measured (Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). We noted that MYC overexpression did not appear to influence the ability of cells to efficiently repair UV-induced cyclobutane pyrimidine dimers or 6-4 pyrimidine–pyrimidone photoproducts. Therefore, MYC overexpression does not perturb all types of DNA repair. Furthermore, because global genomic DNA repair of UV-induced pyrimidine dimers in primary human fibroblasts requires normal p53 function (19, 20), MYC activation is not inhibiting this aspect of p53 activity.
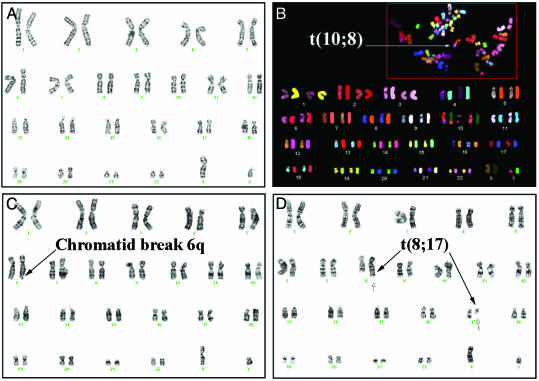
MYC Causes Chromosomal Breaks and Translocations in Normal Human Cells. We reasoned that if MYC overexpression interferes with the repair of spontaneously occurring DSBs, then MYC overexpression might induce DSBs in normal cells. However, previously we failed to see evidence that MYC caused chromosomal aberrations in normal cells (14). At that time, we had examined the consequences of 48 h of MYC activation in asynchronously growing cells. Because normal cells are very sensitive to DNA damage, we thought it would be likely that even if MYC caused DSBs, these cells would be rapidly eliminated (1, 2). To increase the chances of detecting DSBs, we examined the consequences of MYC activation during the one cell division cycle in NHF previously synchronized in G0 (Table 1; Fig. 5). We found, by both karyotypic analysis and SKY, that MYC activation in synchronized NHF-MYCER induced a frequency of 12% chromatid breaks, 3% deletions, and 3% chromosomal translocations (Table 1). In asynchronous NHF-MYCER, the frequency of damage caused by MYC was reduced to 2% breaks, 1% deletions, and 0.8% translocations. Both the breaks and the translocations appeared to be random and were found to involve multiple different chromosomes (Fig. 6 B–D). As a negative control, we examined metaphases prepared from NHF-BABE, NHF-BABE treated with E2 for 48 h, and NHF-MYCER not treated with E2 and detected rare chromatid breaks and no deletions or chromosomal translocations (Fig. 6A), consistent with reports that the frequency of chromosomal rearrangements in normal cells is <10–7 (21). Hence, MYC activation within one cell division cycle increased the frequency of chromosomal breaks, deletions, and translocations by up to several magnitudes.
Table 1. MYC overexpression induces chromosomal breaks, deletions, and translocations.
|
Karyotypic abnormality, %
|
||||
|---|---|---|---|---|
| Cell line | Treatment, h | Chromatid breaks | Deletions | Translocations |
| NHF-BABE | ||||
| Synchronized | None | 0 | 0 | 0 |
| Synchronized | E2, 48 | 1 | 0 | 0 |
| Asynchronized | None | 0 | 0 | 0 |
| Asynchronized | E2, 48 | 1 | 0 | 0 |
| NHF-MYCER | ||||
| Synchronized | None | 0 | 1 | 0 |
| Synchronized | E2, 48 | 12 | 3 | 3 |
| Asynchronized | None | 0 | 1 | 0 |
| Asynchronized | E2, 48 | 2 | 1 | 0.8 |
At least 100 metaphases were analyzed per treatment.
Fig. 5.
MYC overexpression impairs DSB repair and induces apoptosis. DRAA8-MYCER cells with or without MYC activation were electroporated with I-SceI, cultured for the indicated times, then analyzed by flow cytometry for DNA content by propidium iodide staining and DSB by TUNEL. The percentage of cells that are TUNEL-positive is indicated. Representative data from one of at least three experiments are shown.
Fig. 6.
Overexpression of MYC induces chromosomal breaks and translocations. (A) Normal karyotype of NHF-MYCER in the absence of E2. Shown is nonreciprocal translocation (B), t(10, 8) by SKY and chromatid break (C), and reciprocal chromosomal translocation, t(8, 17)(q24.3;q21) (D) by routine karyotypic analysis in NHF-MYCER after 48 h of MYC activation. Representative metaphases were chosen.
Discussion
We conclude that MYC overexpression interferes with the repair of DSBs. Because DSBs are thought to occur spontaneously at a high frequency in normal cells, the impairment of DNA repair alone is sufficient to cause DNA damage (6, 8). The inability to efficiently repair these breaks is likely responsible for the inappropriate chromosomal rearrangements resulting in chromosomal translocations described here. Our results may explain our previous observations that MYC overexpression results in the increased sensitivity to DNA-damaging agents, the induction of p53 protein levels, and cell cycle arrest (14, 22). Our results are consistent with the possibility that MYC may cause apoptosis by interfering with the repair of DSBs.
MYC has not been previously shown to have a role in DNA repair. Numerous studies illustrate that MYC can induce genomic destabilization and many mechanisms are possible (10, 11, 13–15, 23–26). In particular, it has been suggested that MYC may contribute to DNA damage through the increased formation of reactive oxygenated species (ROS) (26). Those experiments were performed with cells growing in low serum, which in itself is genotoxic and known to greatly increase the ability of MYC to induce apoptosis. It is unclear whether these results are relevant to our observations derived from cells that were grown under more ideal growth conditions. Regardless, the production of ROS could not account for our observation that MYC interferes with the ability of cells to repair a single DSB (Fig. 2), nor could ROS account for our observations that MYC activation alone induces chromosomal translocations in normal cells (Table 1; Fig. 5). In addition, the treatment of the NHF with either NAC or vitamin E together with MYC activation did not affect the results of the karyotypic analysis (data not shown). We think a more likely explanation is that MYC overexpression interrupts the function of some cell cycle checkpoints that are required for appropriate DNA repair. In yeast, the loss of S phase checkpoints alone can result in chromosomal damage, including chromosomal translocations (4, 6). Hence, MYC overexpression in human cells may similarly accentuate the consequences of endogenous DNA damage by compromising S phase checkpoints.
Because we found that MYC does not prevent appropriate γ-H2AX formation, we infer that at least the initial recognition of DSBs is intact. Previously, we and others have shown that MYC activation results in the induction of p53 expression but appears to prevent the arrest in DNA replication that is normally associated with DNA damage (22, 26). Hence, rather than directly interrupting the DNA repair response, MYC most likely indirectly interfere with gene products that mediate HDR, NHEJ, and/or SSA. As a consequence, the DSBs that occur through exogenous mutagens or spontaneously during replication would most likely not be efficiently repaired. Instead, the cells may undergo apoptosis or repair the DSBs through defective NHEJ, resulting in chromosomal translocations.
Whether the mechanism is direct or indirect, MYC overexpression can function as a potent dominant mutator inducing chromosomal damage that can fuel tumorigenesis. Our results are consistent with our previous observations that even transient MYC activation can contribute to tumorigenesis in immortal rodent cell lines (14). The ability to induce genomic destabilization has led MYC to be castigated as the gene from hell (27). Fortunately, MYC's nefarious characteristics are frequently readily subdued on MYC inactivation, on which most tumors differentiate and/or undergo apoptosis (12, 28, 29). Our present results suggest a possible mechanism. The inactivation of MYC may cause tumor cells to lose their neoplastic properties, because the consequent restoration of some critical aspect of cell cycle checkpoint function may permit cells to recognize that they are genomically damaged and as a consequence enforces their differentiation and/or apoptosis.
The ability of MYC to impair DNA repair and induce genomic instability may play many roles in the ability of MYC to induce and maintain tumorigenesis. MYC may be restrained from causing tumorigenesis, because when it is overexpressed, it induces DNA damage that causes cell cycle arrest and/or apoptosis (14, 22). When these initial cell responses have been overcome, the ability of MYC to induce DNA damage may lead to the acquisition of additional genomic lesions that could fuel tumorigenesis. Finally, although MYC-induced tumorigenesis is reversible, tumors can relapse (28, 29). Recently, we have found that the tumors that relapsed despite the loss of MYC expression exhibit novel chromosomal translocations (30). We infer that the ability of MYC to induce chromosomal translocations may be responsible for the observed emergence of tumor cells that have escaped the requirement for persistent MYC overexpression.
Supplementary Material
Acknowledgments
We thank the members of the Felsher laboratory for suggestions; M. Jasin A. Pierce, and J. Stark (all from the Memorial Sloan–Kettering Cancer Center, New York), for generously providing guidance and reagents required for the DSB repair assays; D. Tree for help with confocal microscopy; D. Menke for help with γ irradiation; and K. Cimprich and T. Wang for a critical reading of the manuscript. This work was supported by National Cancer Institute Grants K08-CA75967-01 and R01-CA89305-01; the Elsa Pardee Foundation; the Esther Ehrman Lazard Faculty Scholar Award (to D.W.F.); and National Cancer Institute Grant T32 CA09151 (to D.D.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DSB, double-strand DNA break; HDR, homology-directed recombination; NHEJ, nonhomologous end joining; SSA, single-strand annealing; E2, estradiol; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling; SKY, spectral karyotyping; NHF, normal human foreskin fibroblast.
References
- 1.Zhou, B. B. & Elledge, S. J. (2000) Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 2.Khanna, K. K. & Jackson, S. P. (2001) Nat. Genet. 27, 247–254. [DOI] [PubMed] [Google Scholar]
- 3.Pierce, A. J., Stark, J. M., Araujo, F. D., Moynahan, M. E., Berwick, M. & Jasin, M. (2001) Trends Cell. Biol. 11, S52–S59. [DOI] [PubMed] [Google Scholar]
- 4.Myung, K., Chen, C. & Kolodner, R. D. (2001) Nature 411, 1073–1076. [DOI] [PubMed] [Google Scholar]
- 5.Weiss, R. S., Matsuoka, S., Elledge, S. J. & Leder, P. (2002) Curr. Biol. 12, 73–77. [DOI] [PubMed] [Google Scholar]
- 6.Myung, K., Datta, A. & Kolodner, R. D. (2001) Cell 104, 397–408. [DOI] [PubMed] [Google Scholar]
- 7.Richardson, C. & Jasin, M. (2000) Mol. Cell. Biol. 20, 9068–9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson, C. & Jasin, M. (2000) Nature 405, 697–700. [DOI] [PubMed] [Google Scholar]
- 9.Mitelman, F., Mertens, F. & Johansson, B. (1997) Nat. Genet. 15, Spec. No. 417–474. [DOI] [PubMed]
- 10.Oster, S. K., Ho, C. S., Soucie, E. L. & Penn, L. Z. (2002) Adv. Cancer Res. 84, 81–154. [DOI] [PubMed] [Google Scholar]
- 11.Grandori, C., Cowley, S. M., James, L. P. & Eisenman, R. N. (2000) Annu. Rev. Cell. Dev. Biol. 16, 653–699. [DOI] [PubMed] [Google Scholar]
- 12.Evan, G. I. & Vousden, K. H. (2001) Nature 411, 342–348. [DOI] [PubMed] [Google Scholar]
- 13.Mai, S., Hanley-Hyde, J. & Fluri M. (1996) Oncogene 12, 277–288. [PubMed] [Google Scholar]
- 14.Felsher, D. W. & Bishop, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3940–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Q. & Dang, C. V. (1999) Mol. Cell. Biol. 19, 5339–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. (1999) J. Cell. Biol. 146, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. (1999) Genes Dev. 13, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark, J. M., Hu, P., Pierce, A. J., Moynaham, M. E., Ellis, N. & Jasin, M. (2002) J. Biol. Chem. 277, 20185–20194. [DOI] [PubMed] [Google Scholar]
- 19.Ford, J. M., Baron, E. L. & Hanawalt, P. C. (1998) Cancer Res. 58, 599–603. [PubMed] [Google Scholar]
- 20.Ford, J. M. & Hanawalt, P. C. (1995) Proc. Natl. Acad. Sci. USA 92, 8876–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schar, P. (2001) Cell 104, 329–332. [DOI] [PubMed] [Google Scholar]
- 22.Felsher, D. W., Zetterberg, A., Zhu, J., Tlsty, T. & Bishop, J. M. (2000) Proc. Natl. Acad. Sci. USA 97, 10544–10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermeking, H., Funk, J. O., Reichert, M., Ellwart, J. W. & Eick, D. (1995) Oncogene 11, 1409–1415. [PubMed] [Google Scholar]
- 24.Amundson, S. A., Zhan, Q., Penn, L. Z. & Fornace, A. J., Jr. (1998) Oncogene 17, 2149–2154. [DOI] [PubMed] [Google Scholar]
- 25.Menssen, A. & Hermeking, H. (2002) Proc. Natl. Acad. Sci. USA. 99, 6274–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vafa, O., Wade, M., Kern, S., Beeche, M., Pandita, T. K., Hampton, G. M. & Wahl, G. M. (2002) Mol. Cell 9, 1031–1044. [DOI] [PubMed] [Google Scholar]
- 27.Soucek, L. & Evan, G. (2002) Cancer Cell 1, 406–408. [DOI] [PubMed] [Google Scholar]
- 28.Felsher, D. W. & Bishop, J. M. (1999) Mol. Cell 4, 199–207. [DOI] [PubMed] [Google Scholar]
- 29.Jain. M., Arvanitis, C., Chu, K., Dewey, W., Leonhardt, E., Trinh, M., Sundberg, C., Bishop, J. M. & Felsher, D. W. (2002) Science 297, 102–104. [DOI] [PubMed] [Google Scholar]
- 30.Karlsson, A., Giuriato, S., Tang, F., Fung-Weier, J., Levan, G. & Felsher, D. W. (2003) Blood 7, 2797–2803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.