Abstract
In Xenopus, estrogen induces the stabilization of vitellogenin mRNA and the destabilization of albumin mRNA. These processes correlate with increased polysomal activity of a sequence-selective mRNA endonuclease, PMR-1, and a hnRNP K homology-domain RNA-binding protein, vigilin. Vigilin binds to a region of the vitellogenin mRNA 3′-untranslated region (3′-UTR) implicated in estrogen-mediated stabilization. The vigilin-binding site in the vitellogenin B1 mRNA 3′-UTR contains two consensus PMR-1 cleavage sites. The availability of purified PMR-1 and recombinant vigilin made it possible to test the hypothesis that RNA-binding proteins interact with cis-acting elements to stabilize target mRNAs by blocking cleavage by site-specific mRNA endonucleases. Vigilin binds to the vitellogenin mRNA 3′-UTR site with at least 30-fold higher affinity than it exhibits for the albumin mRNA segment containing the mapped PMR-1 cleavage sites. This differential binding affinity correlates with differential in vitro susceptibility of the protein–RNA complexes to cleavage by PMR-1. Whereas recombinant vigilin has no detectable protective effect on PMR-1 cleavage of albumin mRNA, it retards in vitro cleavage of the vitellogenin mRNA 3′-UTR by purified PMR-1. The PMR-1 sites in the vitellogenin mRNA 3′-UTR are functional because they are readily cleaved in vitro by purified PMR-1. These results provide direct evidence for differential susceptibility to endonuclease-mediated mRNA decay resulting from the differential affinity of a RNA-binding protein for cis-acting stability determinants.
For many mRNAs, cytoplasmic stability is determined by endonuclease cleavage within the body of the message (1–6). Endonuclease-catalyzed decay is likely to be regulated by RNA-binding proteins interacting with cis-acting mRNA stability elements to block cleavage by site-selective mRNA endonucleases (3, 7–9). A recent example is the identification of an endonuclease cleavage site within the α-complex binding region of the α-globin 3′-untranslated region (3′-UTR) (10).
Progress toward understanding estrogen regulation of mRNA turnover in Xenopus liver made it possible for us to determine directly the relationship between cleavage by a mRNA endonuclease and protection from cleavage by a sequence- and structure-specific mRNA-binding protein. Estrogen stabilizes the mRNA encoding the egg yolk precursor protein, vitellogenin (11), and destabilizes albumin mRNA (12). Current evidence indicates that the first committed step in albumin mRNA decay is the activation of an endoribonuclease termed PMR-1 (polysomal ribonuclease 1) (13). PMR-1 is a Mr = 62,000 protein (14) that is a member of the peroxidase gene family (15). In vitro PMR-1 selectively degrades serum protein mRNAs, but not ferritin mRNA or mRNAs coding for other intracellular proteins (13). PMR-1 preferentially cuts (14) RNA within a single-stranded consensus pentamer APyrUGA (16), and at a number of nonconsensus sites. Intriguingly, two copies of the consensus APyrUGA pentamer were found in a region of the vitellogenin mRNA 3′-UTR, which has been implicated in the control of vitellogenin mRNA stability (17). This region of vitellogenin mRNA is bound by an estrogen-induced polysome-associated protein (18), which was identified as vigilin/scp160/DDP1, a 155-kDa RNA-binding protein bearing 15 hnRNP K homology domains (19). Although little is known about the function of vertebrate vigilin in mRNA metabolism, yeast Scp160p/vigilin is part of a large polysome-associated messenger RNA–protein complex that includes poly(A)-binding protein (20). With the availability of purified PMR-1 and recombinant vigilin, we sought to directly test the hypothesis that mRNAs can be protected from endonuclease cleavage by the differential affinity of a RNA-binding protein for a particular transcript. We show that PMR-1 cleaves the vitellogenin 3′-UTR in vitro at the expected sites. Vigilin binds the vitellogenin 3′-UTR with much higher affinity than the portion of albumin mRNA bearing the PMR-1 cleavage sites, and this differential affinity directly influences the susceptibility of these RNAs to in vitro cleavage by PMR-1.
Materials and Methods
Mapping PMR-1 Cleavage Sites in Vitellogenin mRNA 3′-UTR.
To map in vitro endonuclease cleavage sites, RNA for the vitellogenin 3′-UTR was 5′ end labeled by transcription in the presence of [γ-32P]GTP. The RNA was incubated with 30 units of PMR-1 in 10 mM Tris⋅HCl (pH 7.5)/1 mM DTT/75 mM KCl for 30 min at 23°C. One unit of PMR-1 is the amount needed to completely cleave 7 fmol of albumin substrate transcript in 30 min at 23°C. The reaction was terminated by addition of an equal volume of 98% formamide dye loading solution and heating for 3 min at 100°C. Cleavage fragments were separated on a 6% polyacrylamide-urea gel. The location of cleavage sites within vitellogenin mRNA was determined by coelectrophoresis of a dideoxy sequencing ladder prepared from pB1UTR-15 by using a primer (5′-GAATACAAGCTTATGAATAAG) corresponding to the 5′ 21 bp of the transcribed RNA. pB1UTR-15 contains the 3′ 97 nt of the vitellogenin B1 mRNA 3′-UTR, including the polyadenylation sequence, with short <7-nt polylinkers at each end.
Expression of Recombinant Vigilin.
For recombinant vigilin expression and purification, a NdeI site was introduced at the 5′ end of the cDNA of the human vigilin (HDL-BP)-coding region at nucleotide 154 (21) and an XbaI site was added downstream of the poly(A) signal (nucleotide 4,336) by PCR amplification between oligonucleotides containing the restriction sites at their 5′ ends. The product was digested with NdeI and XbaI and cloned into pTetCMV-Fo(AS) (22). To generate pVLFlagVig, an XbaI and partial BglII digest were used to clone the full-length FLAG-vigilin cDNA into pVL1392 (Invitrogen). Baculovirus expressing FLAG-vigilin was generated in Sf9 cells as described (23). Cells were lysed by sonication in 10 mM Tris⋅HCl (pH 7.9)/500 mM KCl/10% glycerol/0.1% NP-40 with protease inhibitors. Debris was removed by centrifugation and the supernatant was incubated with M2 agarose (Eastman Kodak). FLAG-vigilin was eluted with Flag peptide and stored at −80°C. Purified FLAG-vigilin was characterized by SDS/PAGE and Western blotting.
Purification of PMR-1 and Preparation of Polysome Extracts.
The purification of PMR-1 and the preparation of polysomal salt extracts used as a source of vigilin were done as described (14, 18). Male Xenopus obtained from Xenopus One were housed in Plexiglas aquaria and kept on a 12-h light-dark cycle.
Comparision of Vigilin-Binding Activity of Polysome Salt Extract for Albumin mRNA and Vitellogenin 3′-UTR.
32P-labeled vitellogenin mRNA 3′-UTR (Vit B1–15) (2 × 104 cpm) (23), or the portion of albumin mRNA bearing mapped PMR-1 cleavage sites (pXAlb160) (16), were incubated with increasing amounts of polysome salt extract for 20 min on ice. Incubations were carried out in a 10-μl volume containing 6 mM Tris⋅HCl (pH 7.6)/0.6 mM DTT/6% glycerol/0.96 mM EDTA/60 mM KCl/200 ng BSA/8 units of Rnasin/10 μg each of tRNA and heparin. The amount of RNA bound was determined by fractionation on a 4% native polyacrylamide gel and binding was quantified by PhosphorImager analysis.
Vigilin Endonuclease Protection Assay.
To study the effects of vigilin binding on cleavage by PMR-1, 500 fmol of the albumin or vitellogenin mRNA segments containing the PMR-1 sites was incubated with or without 40 units of PMR-1 and 500 ng of purified recombinant vigilin in 10 mM Tris⋅HCl (pH 7.5)/1 mM DTT/75 mM KCl for 45 min at 23°C. The reaction was terminated by addition of an equal volume of formamide dye loading solution and heating for 3 min at 100°C. Cleavage products were separated by electrophoresis on a 6% polyacrylamide-urea gel and the degradation of input RNA was visualized by PhosphorImager analysis.
Results
Vitellogenin mRNA 3′-UTR Is a Substrate for Cleavage by PMR-1.
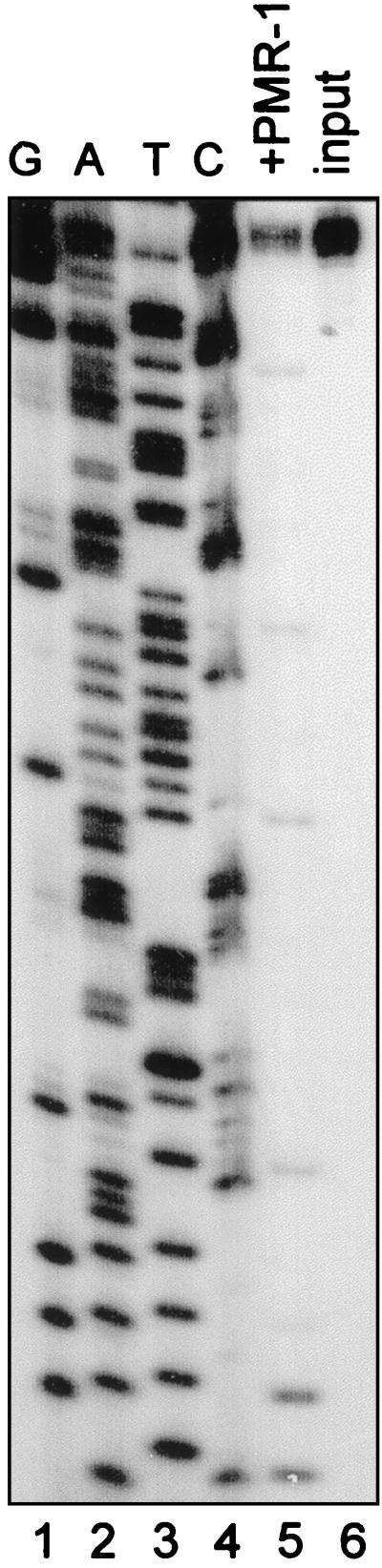
PMR-1 was purified based on its ability to generate a unique doublet product by cleaving within two copies of the overlapping sequence APyrUGA located 160 nt from the 5′ end of albumin mRNA (14). These two overlapping APyrUGA elements reside within a single-stranded loop of a highly structured portion of albumin mRNA (16). The structure of this portion of albumin mRNA is shown in Fig. 1 Upper, with the consensus PMR-1 cleavage sites boxed. Below this is shown a portion of the vitellogenin B1 mRNA 3′-UTR (B1–15) that is bound by the estrogen-induced polysomal protein vigilin (18, 19). This portion of vitellogenin mRNA contains two consensus PMR-1 cleavage sites (boxed). Earlier work suggested that this region is important in the estrogen-induced stabilization of vitellogenin mRNA (17), and the presence of PMR-1 cleavage sites raised the possibility that mRNA stabilization might result from inhibition of PMR-1 endonuclease cleavage by vigilin binding over and occluding these sites. The feasibility of this concept was tested in the experiment shown in Fig. 2. In this experiment, a 5′ end-labeled transcript for the vitellogenin mRNA 3′-UTR was subjected to partial digestion with a limiting amount of purified PMR-1 to generate a series of cleavage products (24). These were then separated on a high-resolution polyacrylamide-urea gel and compared with a DNA-sequencing ladder prepared from the same region to identify cleavage sites. The strongest cleavages occurred within the first APyrUGA element at the 5′ end, with weaker cleavage in the downstream consensus element and at several nonconsensus sites. Arrows in Fig. 1 (Lower) identify the location of these cleavages on the vitellogenin mRNA 3′-UTR sequence, with the size of the arrow indicating the relative strength of cleavage at a particular site. The ability of PMR-1 to cleave at both consensus and nonconsensus sites has been documented previously (15, 16).
Figure 1.
RNA sequence elements identified as in vitro cleavage substrates for PMR-1. (Upper) The solved secondary structure of a region in the 5′ end of albumin mRNA bearing mapped PMR-1 cleavage sites (16). APyrUGA consensus cleavage sites are boxed. (Lower) The sequence of the vitellogenin B1 3′-UTR bound by vigilin containing the only two APyrUGA elements present in the 3′-UTR of vitellogenin mRNA (18). The sites for cleavage of albumin mRNA within the APyrUGA elements are indicated (Upper) with open arrows, and the PMR-1 cleavage sites in the vitellogenin mRNA 3′-UTR determined in Fig. 2 are indicated with filled arrows, with the relative strength of each site represented by the size of each arrow.
Figure 2.
Identification of PMR-1 cleavage sites in the vitellogenin mRNA 3′-UTR. Vitellogenin mRNA 3′-UTR labeled on the 5′ end with 32P was digested with 30 units of PMR-1 and the resulting products were separated on a 6% polyacrylamide-urea gel. The sites of RNA cleavage were determined by comparison to a DNA sequencing ladder prepared from the cloned vitellogenin cDNA by using a primer beginning at the 5′ end of the encoded transcript. Cleavage sites identified in this manner are indicated on the vitellogenin sequence in Fig. 1.
Vigilin Binds Selectively to the Vitellogenin mRNA 3′-UTR.
Binding of vigilin, a large, estrogen-inducible (18) multi-hnRNP K homology domain protein is proposed to protect vitellogenin mRNA against degradation during estrogen exposure. Because the degradation of albumin mRNA is increased in the presence of estrogen, vigilin would be expected to bind poorly, or not at all to the PMR-1-sensitive region(s) of albumin mRNA. The binding of vigilin to albumin and vitellogenin mRNAs was determined by electrophoretic mobility-shift assays (EMSA) by using either salt extracts from liver polysomes of control and estrogen-treated frogs, or recombinant human vigilin. Both recombinant vigilin and vigilin in polysome extract from estrogen-stimulated frogs exhibited significant binding activity for the vitellogenin mRNA 3′-UTR segment (Fig. 3A Right; Vigilin, + E extract). In this experiment, polysome extract from control frogs showed no binding activity (Fig. 3; − E extract), although in earlier work, a low level of vigilin-binding activity was detected in extracts from control frogs (18). In contrast to its strong binding to the vitellogenin mRNA 3′-UTR, vigilin binding to the segment of albumin mRNA containing the PMR-1 cleavage sites was barely detectable (Fig. 3A Left). UV crosslinking confirmed that the complexes seen in Fig. 3A resulted from binding of vigilin to the RNAs. The autoradiogram in Fig. 3B shows crosslinking of both mRNAs to recombinant vigilin yielded the expected Mr = 155,000 product (Vigilin). A protein of the identical size was also detected after UV crosslinking of both mRNAs to polysome extract from estrogen-treated frogs (Fig. 3B; + E extract). Because the same UV-crosslinking conditions were used to identify vigilin–RNA complexes by using the complex mixture of proteins in the polysome extract and by using purified vigilin, the intensities of the cross-linked bands may not quantitatively reflect the abundance of the initial RNA–protein complexes. No band was observed in reactions in which UV irradiation was performed without added protein (No protein) or with polysome extract from control frogs (Fig. 3B; − E extract). The Mr = 70,000 band observed here has been seen previously (18) and is thought to represent a vigilin breakdown product (unpublished observations). The Western blot in Fig. 3C confirmed that the difference in the amount of vigilin RNA-binding activity in polysome extracts from control and estrogen-treated frogs resulted from estrogen induction of this RNA-binding protein.
Figure 3.
Characterization of vigilin binding to vitellogenin mRNA 3′-UTR and albumin mRNAs. (A) EMSA analysis was performed on 500 fmol of a 160-nt uniformly labeled albumin RNA containing the mapped PMR-1 cleavage sites (16) (Left) and vitellogenin 3′-UTR (Right). Lanes marked “No protein” contain input RNA. RNAs were incubated with either 5 μg of protein extract from liver polysomes of control animals (− E extract), 5 μg of extract from polysomes of estrogen-treated animals (+ E extract), or 750 ng of recombinant vigilin (Vigilin). The vigilin–RNA complex is indicated by the open arrow. The probes used had equal specific activity, and the albumin RNA EMSA gel was exposed for 20 vs. 4 h for vitellogenin RNA gel. (B) Complexes assembled on ice as in A were crosslinked for 5 min at 0.12 joules/cm2 at a distance of 5 cm. After digestion with RNase A to remove unbound probe, the protein samples were separated by SDS/PAGE on a 10% polyacrylamide gel and cross-linked peptides were visualized by PhosphorImager. The mobility of a 158-kDa maltose-binding protein-β-galactosidase fusion protein size marker is indicated. (C) Samples (10 μg) of extract from liver polysomes of control (− E) and estrogen-treated (+ E) frogs were analyzed by Western blot by using a polyclonal antibody to epitope-tagged hnRNP K homology domains 3–5 of human vigilin, which was expressed in Escherichia coli and then purified.
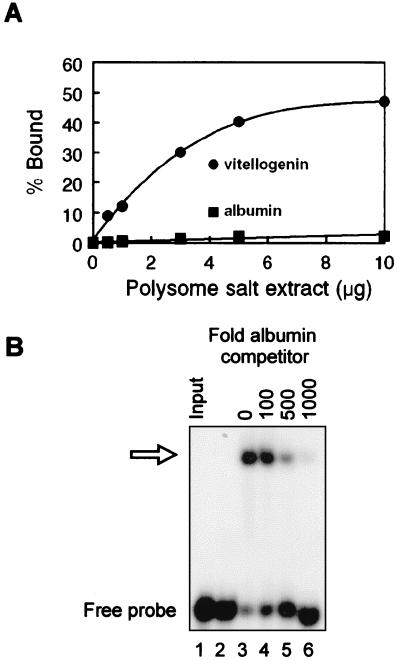
Protein excess binding curve EMSA was used to determine the relative binding affinity for vigilin present in polysome salt extract to the albumin and vitellogenin transcripts. In the experiment in Fig. 4A, a constant amount of each transcript was incubated with increasing amounts of polysome extract and the resulting slower-migrating RNA–protein complexes were quantified by PhosphorImager analysis. These data indicate that the affinity of vigilin for the vitellogenin mRNA 3′-UTR sequence bearing the PMR-1 cleavage sites is at least 30-fold higher than its affinity for the portion of albumin mRNA bearing these cleavage sites. This difference in binding affinity was confirmed by the experiment in Fig. 4B, where a greater than 100-fold molar excess of unlabeled albumin mRNA was required to compete 50% of vigilin binding to [32P]vitellogenin 3′-UTR. The same degree of competition was achieved with a 1.7-fold molar excess of unlabeled vitellogenin mRNA 3′-UTR (data not shown). These data demonstrate that vigilin binding to the albumin mRNA segment containing the PMR-1 cleavage sites is negligible under conditions in which vigilin efficiently binds to the vitellogenin mRNA 3′-UTR containing the PMR-1 sites.
Figure 4.
Vigilin-binding affinity for vitellogenin mRNA 3′-UTR and albumin mRNA. (A) A protein excess-binding curve EMSA was performed by using 32P-labeled albumin mRNA or vitellogenin 3′-UTR and increasing amounts of polysome extract from estrogen-treated frogs. Binding activity was quantified by using the PhosphorImager to compare the relative amount of radiolabeled RNA shifted into slower-migrating complexes versus unbound RNA at the bottom of the gel. (B) The ability of the albumin transcript bearing the PMR-1 cleavage sites to displace vigilin-bound vitellogenin mRNA 3′-UTR was determined by a competition EMSA. Polysome extract (5 μg) from estrogen-treated frogs was incubated with 500 fmol of 32P-labeled vitellogenin mRNA 3′-UTR plus the indicated amounts of unlabeled albumin mRNA competitor. The position of the retarded complex is indicated by the open arrow.
Vigilin Selectively Protects the Vitellogenin mRNA 3′-UTR from Cleavage by PMR-1.
The difference in binding affinity of vigilin for vitellogenin mRNA 3′-UTR as compared with albumin mRNA could modulate cleavage by PMR-1 and thus account for the vastly different fates of these mRNAs within the same cell after estrogen stimulation. To test cleavage by PMR-1 in the presence and absence of vigilin, a uniformly 32P-labeled albumin transcript bearing the mapped PMR-1 sites was incubated with 40 units of purified PMR-1 in the presence or absence of recombinant vigilin. Without added vigilin, 65% of the input albumin RNA transcript was cleaved by PMR-1 within the consensus APyrUGA elements to generate the doublet product characteristic of this endonuclease-substrate combination (16) (Fig. 5; Alb and PMR-1). Albumin mRNA was unaffected by incubation with recombinant vigilin (Fig. 5; Alb and Vigilin), indicating that vigilin lacks detectable RNase activity. In the presence of both PMR-1 and vigilin, the albumin transcript was cleaved in a manner similar to that seen with no added vigilin (Fig. 5; Alb, PMR-1, and Vigilin). Thus, consistent with its very low affinity for albumin mRNA, vigilin was unable to prevent degradation by PMR-1.
Figure 5.
Vigilin binding selectively inhibits PMR-1 cleavage of vitellogenin mRNA 3′-UTR; 500 fmol of uniformly labeled albumin mRNA (Alb) or vitellogenin mRNA 3′-UTR (Vit) were incubated with buffer alone (No protein), 40 units of PMR-1 (PMR-1), 500 ng of recombinant vigilin (Vigilin), or PMR-1 plus vigilin (PMR-1 and Vigilin). Samples recovered from each reaction were electrophoresed on a 6% polyacrylamide-urea gel and visualized by PhosphorImager. Lane 1 contains a size marker of HinfI-cut φX174 DNA.
When the segment of the vitellogenin mRNA 3′-UTR containing the PMR-1 cleavage sites was incubated with 40 units of PMR-1, 72% of input vitellogenin mRNA was degraded (Fig. 5; Vit and PMR-1). The greater number of degradation intermediates seen in this experiment compared with the experiment in Fig. 2 results from using uniformly labeled RNA. As with albumin mRNA, specific degradation products were not detected when vitellogenin mRNA 3′-UTR was incubated alone with recombinant vigilin (Fig. 5; Vit and Vigilin). However, vigilin binding significantly protected vitellogenin mRNA 3′-UTR from cleavage by PMR-1 (Fig. 5; Vit, PMR-1, and Vigilin).
Discussion
The changes in mRNA metabolism that occur after estrogen stimulation in Xenopus liver provide a useful model for investigating biochemical events involved in mRNA stabilization and destabilization. The availability of PMR-1 and vigilin enabled us to test the idea that mRNA stability can be modulated by inhibition of endonuclease cleavage by a sequence- and structure-selective RNA-binding protein. Whereas there is compelling evidence that PMR-1 plays a central role in the destabilization of serum protein mRNAs (13–15, 25), several observations indicate it would also likely degrade vitellogenin mRNA in the absence of a protecting factor. We have found that PMR-1 is present in an inactive form in albumin mRNA-containing messenger RNA–protein complexes and with albumin-synthesizing polysomes before estrogen stimulation (K.S.C., C. Tang, and D.R.S., unpublished work). Estrogen stimulation activates polysome-associated PMR-1, resulting in rapid degradation of albumin mRNA; however, after the disappearance of cytoplasmic albumin mRNA, PMR-1 colocalizes with the peak of vitellogenin-synthesizing polysomes on sucrose density gradients (25). This suggests a pattern where PMR-1 forms a complex with its target mRNA before initiating mRNA decay. This complex is stable until such time as PMR-1 becomes activated, or until a protective RNA-binding protein dissociates from a critical region of a target mRNA. Because PMR-1 cleavage sites are present in the vitellogenin mRNA 3′-UTR and PMR-1 cleaves naked vitellogenin mRNA 3′-UTR preferably within consensus APyrUGA elements as efficiently as it cleaves within these elements in albumin mRNA (Figs. 2 and 5), it seems reasonable that these sites are a target for PMR-1 cleavage in vivo. This view is supported by recent experiments with ligation-mediated PCR to identify in vivo degradation intermediates of albumin and vitellogenin mRNA (M. N. Hanson and D.R.S., unpublished work). Whereas the complete 6.2-kb sequence of the B1 vitellogenin mRNA is not available, only two APyrUGA are elements present in the related A2 vitellogenin mRNA, and they are positioned similarly to the elements in the 3′-UTR of B1 vitellogenin mRNA. This segment of vitellogenin mRNA is implicated in estrogen regulation of vitellogenin mRNA stability (17). In contrast to vitellogenin mRNA, APyrUGA PMR-1 cleavage sites are scattered throughout the sequences of the serum protein mRNAs.
The fact that albumin mRNA is degraded by PMR-1 in the presence of vigilin confirms that protection of vitellogenin mRNA 3′-UTR comes from the greater affinity of vigilin for this RNA and not by inhibition of PMR-1 activity. We have looked without success for similar proteins whose binding to albumin mRNA might be altered by estrogen treatment, supporting the idea that the limiting factor in albumin mRNA destabilization is the activation of PMR-1. As noted above, PMR-1 only cleaves APyrUGA when this sequence is single stranded (16). The APyrUGA elements in the portion of albumin mRNA examined here reside in either a loop or a bulged stem of a highly structured RNA molecule (16). Computer modeling predicts that the vigilin-binding region within the vitellogenin mRNA 3′-UTR is largely unstructured, and by using in vitro selection, we have shown vigilin binds with highest affinity to unstructured RNA largely free of G residues (26). Thus, the protection of the vitellogenin mRNA 3′-UTR from cleavage by PMR-1 is determined by the high-affinity binding of vigilin to this relatively unstructured sequence, whereas vigilin binds albumin mRNA poorly and is thus unable to protect it from cleavage by PMR-1. We expect that protection from cleavage by the differential affinity of RNA-binding proteins for regions containing mRNA endonuclease cleavage sites will be a common theme in mRNA decay.
Acknowledgments
This work was supported by National Institutes of Health Grants GM38277 to D.R.S. and DK50080 to D.J.S. All procedures followed the guidelines of the Animal Welfare Act and the Department of Health, Education, and Welfare (National Institutes of Health) for use of experimental animals.
Abbreviations
- PMR-1
polysomal ribonuclease 1
- 3′-UTR
3′-untranslated region
- EMSA
electrophoretic mobility-shift assay
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220425497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220425497
References
- 1.Schoenberg D R, Chernokalskaya E. In: mRNA Metabolism and Posttranscriptional Gene Regulation. Harford J, Morris D R, editors. New York: Wiley; 1997. pp. 217–240. [Google Scholar]
- 2.Binder R, Hwang S P, Ratnasabapathy R, Williams D L. J Biol Chem. 1989;264:16910–16918. [PubMed] [Google Scholar]
- 3.Binder R, Horowitz J A, Basilion J P, Koeller D M, Klausner R D, Harford J B. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Dijk E L, Sussenbach J S, Holthuizen P E. RNA. 1998;4:1632–1635. doi: 10.1017/s1355838298981316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C H, Leeds P, Ross J. J Biol Chem. 1998;273:25261–25271. doi: 10.1074/jbc.273.39.25261. [DOI] [PubMed] [Google Scholar]
- 6.Gallouzi I, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, Capony J P, Tocque B, Tazi J. Mol Cell Biol. 1998;18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demaria C T, Brewer G. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 8.Doyle G A R, Betz N A, Leeds P F, Fleisig A J, Prokipcak R D, Ross J. Nucleic Acids Res. 1998;26:5036–5044. doi: 10.1093/nar/26.22.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prokipcak R D, Herrick D J, Ross J. J Biol Chem. 1994;269:9261–9269. [PubMed] [Google Scholar]
- 10.Wang Z, Kiledjian M. EMBO J. 2000;19:295–305. doi: 10.1093/emboj/19.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brock M L, Shapiro D J. Cell. 1983;34:207–214. doi: 10.1016/0092-8674(83)90151-4. [DOI] [PubMed] [Google Scholar]
- 12.Pastori R L, Moskaitis J E, Buzek S W, Schoenberg D R. Mol Endocrinol. 1991;5:461–468. doi: 10.1210/mend-5-4-461. [DOI] [PubMed] [Google Scholar]
- 13.Pastori R L, Moskaitis J E, Schoenberg D R. Biochemistry. 1991;30:10490–10498. doi: 10.1021/bi00107a018. [DOI] [PubMed] [Google Scholar]
- 14.Dompenciel R E, Garnepudi V R, Schoenberg D R. J Biol Chem. 1995;270:6108–6118. doi: 10.1074/jbc.270.11.6108. [DOI] [PubMed] [Google Scholar]
- 15.Chernokalskaya E, DuBell A N, Cunningham K S, Hanson M N, Dompenciel R E, Schoenberg D R. RNA. 1998;4:1537–1548. doi: 10.1017/s1355838298980451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chernokalskaya E, Dompenciel R E, Schoenberg D R. Nucleic Acids Res. 1997;25:735–742. doi: 10.1093/nar/25.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen D A, Shapiro D J. Mol Cell Biol. 1990;10:371–376. doi: 10.1128/mcb.10.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodson R E, Shapiro D J. Mol Cell Biol. 1994;14:3130–3138. doi: 10.1128/mcb.14.5.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodson R E, Shapiro D J. J Biol Chem. 1997;272:12249–12252. doi: 10.1074/jbc.272.19.12249. [DOI] [PubMed] [Google Scholar]
- 20.Lang B D, Fridovich-Keil J L. Nucleic Acids Res. 2000;28:1576–1584. doi: 10.1093/nar/28.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKnight G L, Reasoner J, Gilbert T, Sundquist K O, Hokland B, McKernan P A, Champagne J, Johnson C J, Oram J F. J Biol Chem. 1992;267:12131–12141. [PubMed] [Google Scholar]
- 22.Wu S-Y, Chiang C-M. BioTechniques. 1996;21:718–722. doi: 10.2144/96214rr05. [DOI] [PubMed] [Google Scholar]
- 23.Chiang C-M, Roeder R G. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 24.Schoenberg D R, Cunningham K S. Methods Companion Methods Enzymol. 1999;17:60–73. doi: 10.1006/meth.1998.0708. [DOI] [PubMed] [Google Scholar]
- 25.Pastori R L, Schoenberg D R. Arch Biochem Biophys. 1993;305:313–319. doi: 10.1006/abbi.1993.1428. [DOI] [PubMed] [Google Scholar]
- 26.Kanamori H, Dodson R E, Shapiro D J. Mol Cell Biol. 1998;18:3991–4003. doi: 10.1128/mcb.18.7.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]