Abstract
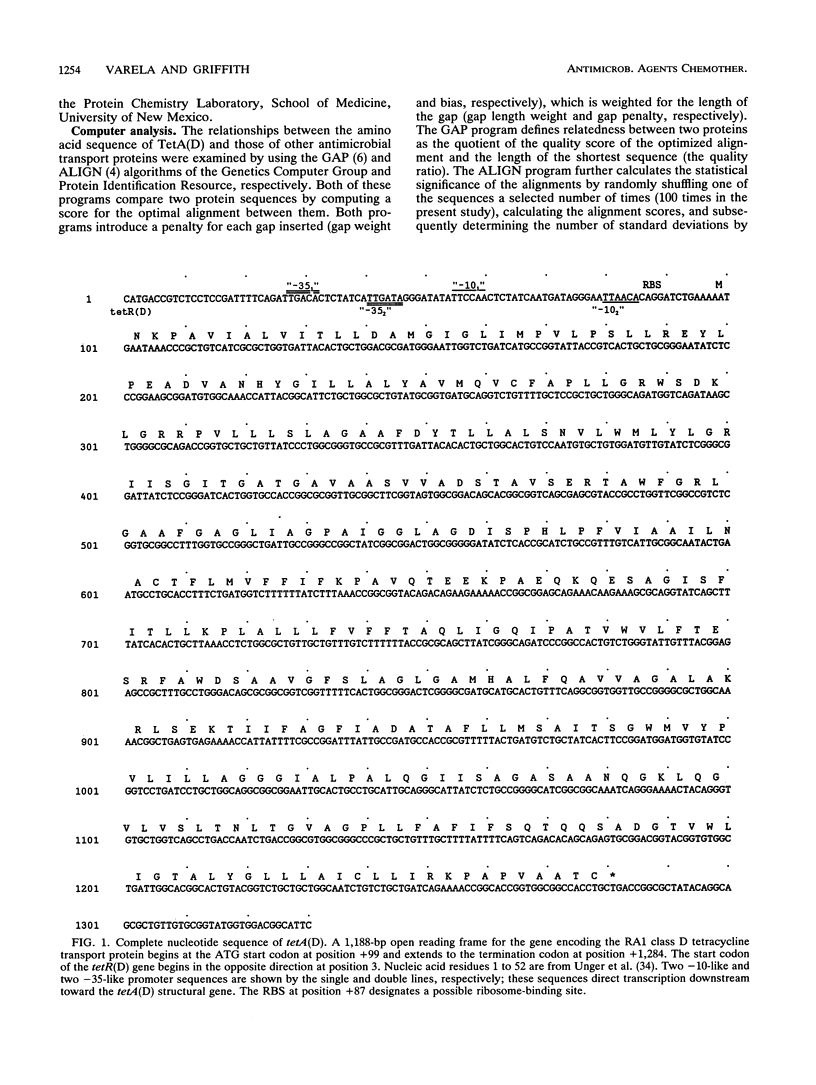
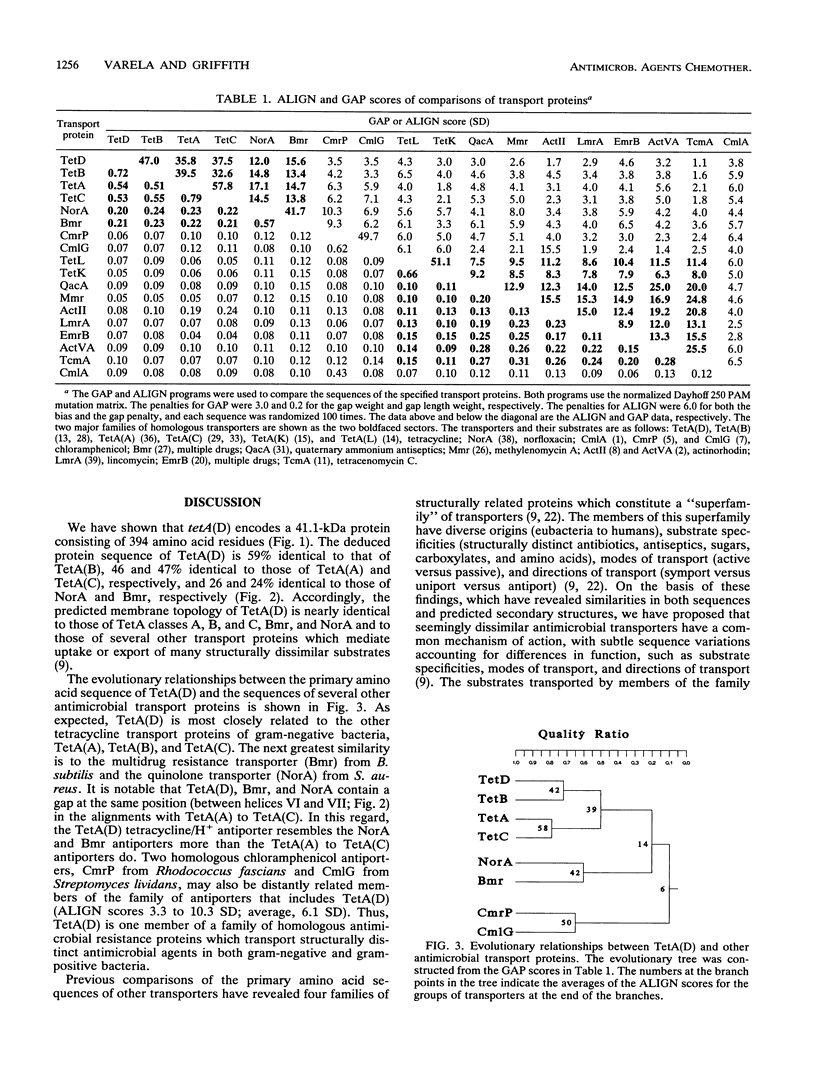
The nucleotide sequence of the plasmid pRA1 gene encoding the TetA(D) tetracycline/H+ antiporter was determined. The deduced amino acid sequence was compared with those of other antimicrobial and antiseptic transporters. The deduced product of tetA(D) is a 41.1-kDa protein consisting of 394 amino acids comprising 12 membrane-spanning domains. Three classes of amino acid motifs found in TetA(D) are highly conserved in other transporters, implying that they participate in structures necessary for substrate recognition, binding, or translocation. A common mechanism of transport is suggested, with subtle sequence variations accounting for varied substrate specificities, modes of transport, and directions of transport.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bissonnette L., Champetier S., Buisson J. P., Roy P. H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol. 1991 Jul;173(14):4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero J. L., Martinez E., Malpartida F., Hopwood D. A. Organisation and functions of the actVA region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor. Mol Gen Genet. 1991 Dec;230(3):401–412. doi: 10.1007/BF00280297. [DOI] [PubMed] [Google Scholar]
- Chopra I., Hawkey P. M., Hinton M. Tetracyclines, molecular and clinical aspects. J Antimicrob Chemother. 1992 Mar;29(3):245–277. doi: 10.1093/jac/29.3.245. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Desomer J., Vereecke D., Crespi M., Van Montagu M. The plasmid-encoded chloramphenicol-resistance protein of Rhodococcus fascians is homologous to the transmembrane tetracycline efflux proteins. Mol Microbiol. 1992 Aug;6(16):2377–2385. doi: 10.1111/j.1365-2958.1992.tb01412.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich W., Betzler M., Schrempf H. An amplifiable and deletable chloramphenicol-resistance determinant of Streptomyces lividans 1326 encodes a putative transmembrane protein. Mol Microbiol. 1991 Nov;5(11):2789–2797. doi: 10.1111/j.1365-2958.1991.tb01987.x. [DOI] [PubMed] [Google Scholar]
- Fernández-Moreno M. A., Caballero J. L., Hopwood D. A., Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991 Aug 23;66(4):769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- Griffith J. K., Baker M. E., Rouch D. A., Page M. G., Skurray R. A., Paulsen I. T., Chater K. F., Baldwin S. A., Henderson P. J. Membrane transport proteins: implications of sequence comparisons. Curr Opin Cell Biol. 1992 Aug;4(4):684–695. doi: 10.1016/0955-0674(92)90090-y. [DOI] [PubMed] [Google Scholar]
- Griffith J. K., Kogoma T., Corvo D. L., Anderson W. L., Kazim A. L. An N-terminal domain of the tetracycline resistance protein increases susceptibility to aminoglycosides and complements potassium uptake defects in Escherichia coli. J Bacteriol. 1988 Feb;170(2):598–604. doi: 10.1128/jb.170.2.598-604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoile P. G., Hutchinson C. R. Sequence and transcriptional analysis of the Streptomyces glaucescens tcmAR tetracenomycin C resistance and repressor gene loci. J Bacteriol. 1992 Jun;174(11):3651–3658. doi: 10.1128/jb.174.11.3651-3658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson P. J., Maiden M. C. Homologous sugar transport proteins in Escherichia coli and their relatives in both prokaryotes and eukaryotes. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1236):391–410. doi: 10.1098/rstb.1990.0020. [DOI] [PubMed] [Google Scholar]
- Hillen W., Schollmeier K. Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Res. 1983 Jan 25;11(2):525–539. doi: 10.1093/nar/11.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T., Ikeda T., Tomizuka N., Furukawa K. Nucleotide sequence of the tetracycline resistance gene of pTHT15, a thermophilic Bacillus plasmid: comparison with staphylococcal TcR controls. Gene. 1985;37(1-3):131–138. doi: 10.1016/0378-1119(85)90265-3. [DOI] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983 Nov;10(3):251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Levy S. B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992 Apr;36(4):695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O., Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marger M. D., Saier M. H., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993 Jan;18(1):13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- Marshall B., Tachibana C., Levy S. B. Frequency of tetracycline resistance determinant classes among lactose-fermenting coliforms. Antimicrob Agents Chemother. 1983 Dec;24(6):835–840. doi: 10.1128/aac.24.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicholas P., Chopra I., Rothstein D. M. Genetic analysis of the tetA(C) gene on plasmid pBR322. J Bacteriol. 1992 Dec;174(24):7926–7933. doi: 10.1128/jb.174.24.7926-7933.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal R. J., Chater K. F. Nucleotide sequence analysis reveals similarities between proteins determining methylenomycin A resistance in Streptomyces and tetracycline resistance in eubacteria. Gene. 1987;58(2-3):229–241. doi: 10.1016/0378-1119(87)90378-7. [DOI] [PubMed] [Google Scholar]
- Neyfakh A. A., Bidnenko V. E., Chen L. B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. T., Postle K., Bertrand K. P. Sequence homology between the tetracycline-resistance determinants of Tn10 and pBR322. Gene. 1983 Nov;25(1):83–92. doi: 10.1016/0378-1119(83)90170-1. [DOI] [PubMed] [Google Scholar]
- Peden K. W. Revised sequence of the tetracycline-resistance gene of pBR322. Gene. 1983 May-Jun;22(2-3):277–280. doi: 10.1016/0378-1119(83)90112-9. [DOI] [PubMed] [Google Scholar]
- Reeck G. R., de Haën C., Teller D. C., Doolittle R. F., Fitch W. M., Dickerson R. E., Chambon P., McLachlan A. D., Margoliash E., Jukes T. H. "Homology" in proteins and nucleic acids: a terminology muddle and a way out of it. Cell. 1987 Aug 28;50(5):667–667. doi: 10.1016/0092-8674(87)90322-9. [DOI] [PubMed] [Google Scholar]
- Rouch D. A., Cram D. S., DiBerardino D., Littlejohn T. G., Skurray R. A. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol. 1990 Dec;4(12):2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Unger B., Klock G., Hillen W. Nucleotide sequence of the repressor gene of the RA1 tetracycline resistance determinant: structural and functional comparison with three related Tet repressor genes. Nucleic Acids Res. 1984 Oct 25;12(20):7693–7703. doi: 10.1093/nar/12.20.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Waters S. H., Rogowsky P., Grinsted J., Altenbuchner J., Schmitt R. The tetracycline resistance determinants of RP1 and Tn1721: nucleotide sequence analysis. Nucleic Acids Res. 1983 Sep 10;11(17):6089–6105. doi: 10.1093/nar/11.17.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Someya Y., Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. The role of a conserved sequence motif, GXXXXRXGRR, in a putative cytoplasmic loop between helices 2 and 3. J Biol Chem. 1992 Sep 25;267(27):19155–19162. [PubMed] [Google Scholar]
- Yoshida H., Bogaki M., Nakamura S., Ubukata K., Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990 Dec;172(12):6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Z., Schmidt H., Piepersberg W. Molecular cloning and characterization of two lincomycin-resistance genes, lmrA and lmrB, from Streptomyces lincolnensis 78-11. Mol Microbiol. 1992 Aug;6(15):2147–2157. doi: 10.1111/j.1365-2958.1992.tb01388.x. [DOI] [PubMed] [Google Scholar]


