Abstract
Coactivators are believed to mediate estrogen-induced gene responses via interaction with estrogen receptors (ER). Currently, a major challenge is to determine the importance of each coactivator in a specific cell type and promoter context in response to a particular ligand. The potential of ER to interact with a growing list of coactivators has been shown in a variety of in vitro and gene transfer assays, yet very few data have demonstrated the interaction of endogenous coactivators with ER in intact cells. We report here a ligand-specific interaction of endogenous human ER (hER) and the AIB1 coactivator in MCF-7 human breast cancer cells by using immunoprecipitation analyses. Complexes between endogenously expressed hER and AIB1 were detected in estradiol-treated cells and to a much lesser extent in cells treated with the partial agonist, monohydroxytamoxifen. We were unable to detect an hER–SRC-1 complex in our immunoprecipitations from MCF-7 cells. The in vitro-binding affinity for mouse ER interaction with AIB1 was estimated to be 40–120 nM. We conclude that AIB1 is a major coactivator for hER in MCF-7 human breast cancer cells.
It is well established that estrogen action in the cell is mediated by estrogen receptors (ERs) α and β (1). Detailed regulation of gene expression is believed to be mediated by coregulator proteins that bind ER in a ligand-dependent manner (2, 3). Several structurally distinct subclasses of nuclear receptor coregulators have been identified, including the SRC family of coactivators. This family includes SRC-1 (also termed p160/NcoA-1/ERAP-160), TIF-2 (also termed SRC-2/GRIP-1), and AIB1 (also termed SRC-3/ACTR/RAC-3/TRAM-1) (4). These proteins have been shown to physically bind several agonist-occupied nuclear receptors in vitro and to enhance hormone-dependent transcription in transient transfection assays. It is probable that coactivators preferentially interact with receptors dependent on cell type, ligand, and promotor context, which could contribute to the specificity of the physiological response. However, very few data are available on the existence and importance of endogenous receptor–coactivator complexes actually formed in response to a specific ligand in the whole cell.
Several studies suggest that the newest member of the SRC family, AIB1, has a special role in breast tissue. AIB1 is expressed in a wide variety of tissues, but the highest expression is in breast and ovary. Mice that lack the ability to express AIB1 show greatly reduced sensitivity of breast tissue to estrogen and progesterone administration (5). In addition, recent findings demonstrating that SRC-1 does not colocalize with ERα in rat mammary epithelial cells suggest that other SRC family members likely play a more important role in ERα-mediated hormone actions in breast tissue (6). In addition, AIB1 was found to be overexpressed in 64% of primary breast tumors and in four of five ER+ breast and ovarian cancer cell lines (7). In a study of 1,157 human breast tumors, overexpression of AIB1 was shown to correlate with estrogen and progesterone receptor positivity (8). This study showed also that AIB1 amplification correlated directly with tumor size. Taken together, these data suggest that overexpression of AIB1 in some breast cancer cells may contribute to estrogen-induced promotion of tumorigenesis.
A number of studies have shown the potential for ER to interact with various proteins and to enhance estrogen-induced transcription in either an in vitro assay such as GST pulldown or an engineered expression system such as cell transfections or the yeast two-hybrid system (3, 9). These studies do not address the question of whether AIB1 is important for estrogen-induced gene responses in specific cells with endogenous concentrations of receptors and coactivators. The goal of this study was to determine whether AIB1 directly interacts with ER within a breast cancer cell. In this study, we use coimmunoprecipitation to show that human ER (hER) and AIB1 form a complex in a ligand-specific manner within the nucleus of MCF-7 cells. GST-AIB1 fusion protein and baculovirus expressed mouse ER (mER) were used to estimate the binding affinity of mER for AIB1 in vitro.
Materials and Methods
Cell Culture.
All cell lines were obtained from American Type Culture Collection and were routinely cultured in phenol red-free DMEM with 10% (vol/vol) FBS. MCF-7 cells were switched to bovine calf serum for several passages before experiments to reduce estrogen.
Extractions.
MCF-7 cells were grown to 70–80% confluency and exposed to 20 nM 17β-estradiol (E2), 500 nM monohydroxytamoxifen (MHT), or 20 nM ICI 182,780 (ICI). Nuclear extracts were prepared as previously described (10). Briefly, cells were rinsed in calcium- and magnesium-free Hanks' balanced salt solution and lifted with 3 mM EDTA. Detached cells were rinsed and then lysed in four packed cell volumes (pcv) of hypotonic buffer containing 10 mM Tris⋅Cl, pH 7.5, 1 mM EDTA, 1 mM DTT, 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 10 μM leupeptin, 2 μg/ml aprotinin, and 4 μM pepstatin by using a Dounce homogenizer. Homogenates were centrifuged and the cytosolic extract was recovered. The nuclear pellets were resuspended in 4× pcv of hypotonic buffer containing either 2 mM vanadate, decavanadate form (DV, NEv), or 0.6 M NaCl (NEs) for 30 min on ice followed by centrifugation. The supernatants represented nuclear extracts (NEv or NEs), which were used for immunoprecipitation and Western blot analysis. Vanadate solutions were prepared as previously described (10). Briefly, a 500 mM solution of orthovanadate was titrated to pH 7.5 with HCl to induce polymer formation to the decavanadate form, indicated by the bright orange color. Concentration is given in terms of free vanadate, but the solutions were always in the decavanadate form. DV solutions were used within 24 h.
Immunoprecipitations.
Immunoprecipitation reactions contained nuclear extract, Ig at a final concentration of 0.5 μg/100 μl, and 285 mM NaCl. Either hypotonic buffer or buffer containing 0.6 M NaCl was used to adjust the DV or the salt nuclear extract to the same final salt concentration in all immunoprecipitation reactions. Protein A- or A/G-Sepharose beads were added, and samples were incubated at 4°C with rocking for at least 30 min. Beads were washed with 10–20 volumes of ice-cold TBS (24 mM Tris⋅Cl, pH 7.4/2 mM KCl/163 mM NaCl), extracted with SDS sample buffer at 85°C for 10 min, and the soluble sample recovered after centrifugation. Ig was either rabbit antibody against hER (HC-20, Santa Cruz Biotechnology) or normal rabbit Ig (Santa Cruz Biotechnology).
Western Blotting.
Cytosolic and nuclear extracts were diluted in SDS sample buffer and heated at 95°C for 3–5 min just before loading onto the gel. SDS/PAGE was performed by standard methods (11). Samples were run along with prestained molecular weight markers (Amersham). Proteins separated by SDS/PAGE were transferred onto nitrocellulose membranes (Hybond ECL, Amersham). Membranes were cut in two pieces, horizontally, along the 97,000 Mr size marker allowing primary detection of hER (Mr, 66,000) and AIB1 (Mr, 160,000) from the same blot. Membranes were blocked for 1 h at room temperature in 5% dried milk in TBST (TBS + 0.2% (vol/vol) Tween-20, Bio-Rad). Primary antibody was diluted in blocking solution and membranes incubated for 1 h at room temperature with shaking. A horseradish peroxidase (HRP)-linked secondary antibody was diluted in blocking solution and membranes incubated for 1 h at room temperature with shaking. After each incubation, blots were washed five times in TBST at room temperature. ER was detected with the ECL chemiluminesence substrate from Amersham. SRC-1 and AIB1 immunocomplexes were less abundant, either because of the amount of coactivator or the strength of the primary antibody, and detection was with Renaissance from NEN. Bands were visualized by exposure to autoradiography film (Hyperfilm ECL, Amersham) and quantitated by using nih image public domain software (http://rsb.info.nih.gov/nih-image/index.html). Only bands on the same blot, detected with the same primary antibody, were compared quantitatively. Primary antibodies and their dilutions were as follows: 1:1,000 for rabbit anti-hER (HC-20, Santa Cruz Biotechnology), 1:1,000 for rabbit anti-mER (affinity purified ER1438; ref. 10), 1:500 for mouse anti-hER (SRA-1000, StressGen Biotechnologies, Victoria, BC, Canada) used for blotting samples immunoprecipitated with a rabbit anti-ER antibody, 1:10 for mouse anti-AIB1 from hybridoma cell-conditioned media, or 1:1,000 for ascites [AIB1 antibodies were AC3 for full length protein and AX15.1 for GST-AIB1 fusion protein (12)], and 1:200 for mouse anti-SRC-1 (clone 1135/H4, Affinity BioReagents, Golden, CO). HRP-linked secondary antibodies (Amersham) and their dilutions were as follows: anti-mouse IgG was 1:2,000 for ER, 1:1,000 for SRC-1, and 1:10,000 for AIB1 detection; anti-rabbit IgG was 1:2000 for ER detection.
GST-AIB1 Pull Down of mER.
mER prepared from baculovirus-infected Sf9 insect cells was incubated with ethanol vehicle, 1 μM E2, 1 μM MHT, or 1 μM ICI for at least 1 h on ice (13). GST or GST-AIB1 (AIB1 amino acids nos. 605-1294) fusion protein was bacterially expressed and bound to glutathione–agarose (7). The presence of protein on the beads was confirmed by extracting beads with SDS sample buffer followed by SDS/PAGE. GST was detected by Coomassie blue staining of the gel, whereas GST-AIB1 was confirmed by anti-AIB1 Western blot. Agarose-bound GST or GST-AIB1 was incubated with liganded mER on ice for at least 1 h in binding buffer (BB) (10% glycerol/50 mM Tris⋅Cl, pH 8.0/100 mM NaCl/1 mM DTT/0.1% IGEPAL/0.5 mg/ml BSA). Beads were washed with BB, protein extracted with SDS sample buffer at 85°C for 10 min, and soluble fraction recovered by centrifugation. Proteins were separated by SDS/PAGE and transferred to nitrocellulose membranes. Blots were cut horizontally between the 68,000 and 97,000 Mr size markers and GST-AIB1 (Mr, 100,000) and mER (Mr, 66,000) detected and quantitated by Western blot, as described above.
Results
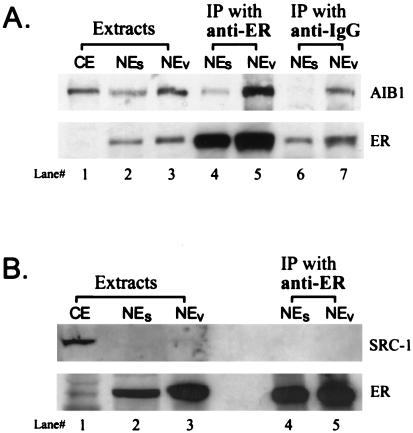
We examined the effect of estradiol administration on AIB1 and hER interactions in whole cells by looking at the endogenous complexes recovered from MCF-7 cell nuclear extracts. We treated MCF-7 cells with E2, lysed cells in low-salt buffer, and prepared nuclei. In the absence of ligand, very little ER is associated with the nuclei, whereas the liganded ER is tightly bound. Traditionally, high-salt buffers are used to extract the liganded ER from nuclei, and this probably disrupts protein–protein associations. We extracted the hER with associated proteins from nuclei by using the polyanion decavanadate. Previous work from our laboratory demonstrated that this gentle nuclear extraction method releases hER in a large 5–8S complex as compared with the 4–5S peak of salt extracted hER, as assessed by sucrose density gradients, suggesting that a protein complex might be maintained (10). The traditional method of making nuclear extracts with 0.6 M NaCl was also used for comparison. The Western blot of the samples is shown in Fig. 1A. The top half of the blot was immunostained for AIB1 (Mr, 160,000) and the bottom half for hER (Mr, 66,000). After E2 treatment of whole cells, the hER was absent from the cytosolic fraction (lane 1), exhibiting the ligand-induced tight nuclear binding that has been routinely observed (14). The E2-occupied nuclear hER was extracted from nuclei with buffer containing either 0.6 M NaCl (lane 2) or DV (lane 3). AIB1 is seen in the cytosolic fraction (lane 1) as well as in both nuclear extracts (lanes 2, 3). We have seen no effect of estradiol on the biochemical fractionation of endogenous AIB1 in MCF-7 cells (D.J.C., M.K.T. & F.E.M., unpublished data). Anti-hER antibody efficiently immunoprecipitated hER from either the salt (NEs) or DV (NEV) nuclear extracts, as shown in lanes 4 and 5. However, the quantity of AIB1 coimmunoprecipitating with the hER was much greater from the NEv extract (lane 5). This is consistent with our previous data that DV extraction preserves protein–protein interactions better than traditional NaCl extraction. Nonspecific antibody (anti-IgG) immunoprecipitations from NEs (lane 6) and NEv (lane 7) show the background. These results provide evidence for endogenous hER–AIB1 complex formation in MCF-7 human breast cancer cells after exposure to E2.
Figure 1.
Interaction of endogenous hER with AIB1 is demonstrated by coimmunoprecipitation. MCF-7 cells were treated with E2 for 0.5 h and extracts prepared as described in Materials and Methods (CE, lane 1, NEs, lane 2, or NEv, lane 3). Equal volumes of extract per packed cell volume were loaded to allow direct visual comparison of lanes. Immunoprecipitation of NEs (lane 4) or NEv (lane 5) extracts by using anti-ER antibody was performed and analyzed by Western blot. (A) Blots were cut in half horizontally along the 97,000 marker. The top half was immunostained for AIB1(Mr, 160,000), and the bottom half for hER (Mr, 66,000). Nonspecific binding of NEs (lane 6) and NEv (lane 7) extracts is shown by using anti-IgG immunoprecipitation. The original extracts (lanes 2 and 3) represent 15% of the input into immunoprecipitation reactions. (B) Experiment conducted as in A, except the top half was immunostained for SRC-1 (Mr, 160,000). The original extracts (lanes 2 and 3) represent 13% of the input into immunoprecipitation reactions.
We were unable to detect SRC-1 coimmunoprecipitating with hER as shown in Fig. 1B. Cells were treated with 20 nM E2, protein extracted, and immunoprecipitation with anti-hER antibody conducted as in Fig. 1A. hER is localized in the nuclear extracts (lanes 2 and 3) and efficiently immunoprecipitated with anti-hER antibody (lanes 4 and 5) the same as seen in Fig. 1A. SRC-1 is clearly visible in the cytosolic extract (lane 1) but very low in the nuclear extracts (lanes 2 and 3). We were unable to detect SRC-1 coimmunoprecipitating with hER (lanes 4 and 5). We also were unable to detect hER–AIB1 complexes in extracts from T-47D human breast cancer cells (data not shown). We believe these results may be because of the lower endogenous concentrations of the SRC-1 in MCF-7 cells and AIB1 in T-47D cells.
We had previously observed that the decavanadate-extracted hER sedimented more densely on sucrose gradients than the salt-extracted form (10). We investigated whether AIB1 cofractionated with hER on sucrose density gradients by Western blot analysis of the fractions (data not shown). We observed a portion of hER cofractionating with the peak of AIB1 protein in nuclear extracts from E2-treated cells. Cofractionation alone is not direct evidence of a hER–AIB1 complex. However, these data are consistent with the existence of a complex, as shown in Fig. 1.
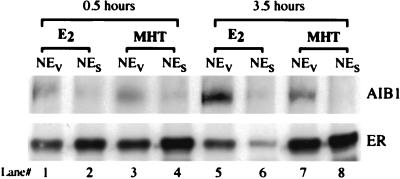
We wanted to determine whether any hER–AIB1 complex is formed in vivo in MCF-7 cells after treatment with the partial agonist/antagonist MHT. After an 18-hr incubation in stripped serum to deplete estrogen, cells were treated with E2 or MHT for 0.5 or 3.5 h. NEs and NEv were prepared and immunoprecipitated with anti-hER antibody as in Fig. 1. Fig. 2 shows the AIB1 and hER Western blots of the immunoprecipitated complexes. Both ligands showed pharmacological activity in MCF-7 cells by inducing the expected tight nuclear binding of the occupied hER (data not shown). As in Fig. 1, after 0.5 h of treatment with E2, more AIB1 coimmunoprecipitated with anti-hER antibody from the NEv sample than from the NEs sample (lanes 1 and 2). After 0.5 h of MHT treatment, we also see more AIB1 coimmunoprecipitating with hER from the NEv than the NEs sample (lanes 3 and 4). Longer treatment with MHT (3.5 h) results in a significant decrease in the amount of complex formed with MHT compared with that found with E2, as expected for an antagonist (compare lanes 7 and 5). However, we consistently observe a small amount of hER–AIB1 complex in MHT-treated cells. After 3.5 h of E2 treatment, the amount of hER immunoprecipitated from NEv (lane 5) and NEs (lane 6) samples is decreased because of the down-regulation of ER levels by E2 (refs. 15 and 16 and references therein). MHT treatment does not result in down-regulation of hER, consistent with its reported antagonistic properties.
Figure 2.
Coimmunoprecipitation of AIB1 with hER in MCF-7 cells after E2 and MHT treatments. Cells were switched to media with 5% dextran-coated charcoal-stripped FBS for 18 h to withdraw any estrogen. After 0.5 or 3.5 h of ligand treatment, nuclei were extracted with NaCl (NEs) or DV (NEv). Shown is the Western blot analysis for AIB1 (Top) and hER (Bottom) of nuclear extract immunoprecipitations with anti-hER antibody performed as in Fig. 1.
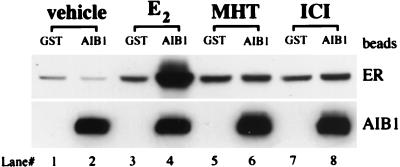
We tested the effects of ER ligands on the ER–AIB1 interaction by using an in vitro GST pulldown assay. Extracts containing a full-length mER prepared from baculovirus-infected Sf9 cells were incubated with ethanol vehicle, E2 (agonist), MHT (partial agonist), or ICI [full antagonist (17)]. Bacterially expressed GST-AIB1 (AIB1 amino acids nos. 605-1294) fusion protein or the GST-only control were purified on glutathione beads and incubated with the unoccupied or occupied mER. Bound proteins were analyzed by Western blot immunostained for mER or AIB1 as shown in Fig. 3. Vehicle treatment did not induce mER binding to the AIB1 fusion protein (lane 2) above background (lane 1). E2 treatment, on the other hand, induced significant binding of mER to GST-AIB1 fusion protein (lane 4). This is not seen after MHT (lane 6) or ICI (lane 8) treatments. Lanes 3, 5, and 7 represent the background binding of mER to GST control beads.
Figure 3.
Interaction of mER with AIB1 in vitro is induced by E2 but not by MHT or ICI. GST alone or GST-AIB1 fusion protein (AIB1) bound to glutathione beads was incubated with unliganded (vehicle), E2, MHT, or ICI-occupied mER. After washing the beads, bound proteins were separated by SDS/PAGE and mER (Top) or AIB1 (Bottom) detected by Western blot.
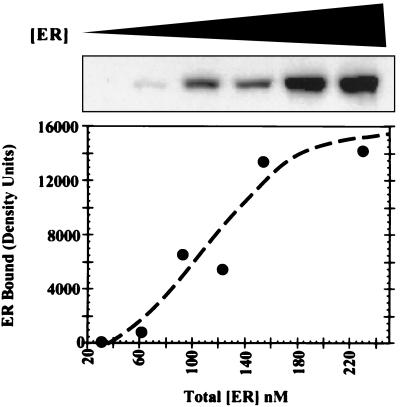
To estimate the affinity of mER for AIB1, a constant limiting amount of bacterially expressed GST-AIB1 fusion protein immobilized on glutathione beads was incubated with increasing concentrations of mER. Bound mER was analyzed by Western blot and the intensity of the mER bands plotted as a function of mER concentration in the reaction. A representative mER Western blot and graphic presentation of total mER concentration vs. amount bound to GST-AIB1 is show in Fig. 4. Three independent experiments were performed and the half-maximal binding estimated from each curve. The estimates of the concentration of mER to give half-maximal complex formation were 120 nM, 75 nM, and 45 nM to give an estimated Kd value of 80 ± 40 nM (mean ± standard deviation).
Figure 4.
Affinity of mER binding to the GST-AIB1 fusion protein. A constant limiting amount of GST-AIB1 immobilized on glutathione beads was incubated with increasing concentrations of mER. Beads were washed and bound mER detected by Western blot as described in Materials and Methods. The intensity of the ER bands was analyzed and graphed as a binding curve as a function of mER concentration.
Discussion
Very few data have been published on interactions between endogenous coactivators and nuclear receptors in whole cells. Our work reported here details the endogenous interaction of hER with AIB1, but not SRC-1, in the MCF-7 human breast cancer cell line. Recently the interaction of endogenous progesterone receptor with SRC-1 in T-47D cells was also reported (18). However, most studies have looked at these interactions after transfecting either the receptor and/or coactivator of interest. These studies demonstrate the dramatic potential these molecules have for interaction, yet they do not give information on what interactions occur and are important with endogenous concentrations of receptor and comodulator in a specific cell type. To date, over 30 comodulators have been identified, and many of them have been shown to bind nuclear receptors and several other proteins in vitro (ref. 19 and references therein). The challenge is to evaluate which endogenous comodulators are important in mediating cell-specific responses to specific ligands.
In this paper, we show a ligand-specific interaction between endogenous hER and AIB1 in the MCF-7 human breast cancer cell line. Coimmunoprecipitation of AIB1 with anti-hER antibody shows a significant amount of AIB1 associated with E2-occupied hER. We were also able to coimmunoprecipitate hER with anti-AIB1 antibody from MCF-7 cell nuclear extracts (data not shown). Comigration of hER and AIB1 in sucrose gradients supports this observation (data not shown). The MHT-occupied mER did not bind GST-AIB1 in vitro. However, after MHT treatment some interaction between hER and AIB1 was detected in vivo, although consistently less than that seen with E2. We are considering several possibilities for these observations. First, MHT may induce ER–AIB1 complexes with lower affinity than E2, but because of the high concentrations of AIB1 in MCF-7 cells, formation of some complex still occurs. It should be noted that the in vitro assays used a truncated AIB1. Second, other interactions that occur in MCF-7 cells may stabilize this complex. For example, a phosphorylated and/or activated ER subpopulation, which is able to bind to AIB1 regardless of MHT treatment, may exist. This hypothesis is based on work with other receptors, where it has been shown that phosphorylation of the orphan nuclear receptor SF-1 recruits coactivators, even in the absence of ligand (20).
Work from other laboratories suggests that the AIB1 coactivator has a major role in mediating estrogen effects in breast tissue. The O'Malley laboratory has recently generated the AIB1-null mouse, which displays a phenotype of greatly reduced sensitivity of breast tissue to estrogen and progesterone administration. The study also showed that AIB1 expression is particularly high in the mammary epithelial cells (5).Western blot analysis has shown that AIB1 protein expression is abundant in MCF-7 cells compared with several other human cancer cell lines (21). Another study supported this observation by showing that AIB1 mRNA was overexpressed in MCF-7 cells, whereas in other breast or endometrial cell lines, expression was low (22). AIB1 mRNA was also expressed at higher levels than other SRC-family members in MCF-7 cells, suggesting a specific role for this coactivator (7). The highest expression of AIB1 mRNA was seen in ER-positive breast cancer cell lines in this study. This is consistent with the fact that we could easily detect AIB1–hER complexes from MCF-7 cells, whereas SRC-1–hER complexes were undetectable by using the same methods. Immunohistochemical localization of SRC-1 and ER in the mouse mammary gland by the Jeng laboratory showed that these two proteins are expressed in distinctly different epithelial cells (6). The potential for ER to interact with SRC-1 has been well established in a number of assays (ref. 23 and references therein). However, our data clearly suggest that AIB1–hER complexes predominate in MCF-7 cells, which is the classic cell model for estrogen-dependent breast cancer cell proliferation. Taken together, our work and studies from other laboratories suggest that AIB1 may be the major SRC family coactivator for ERα in breast epithelial cells.
We used GST pulldown assays to estimate a Kd value for the AIB1–mER interaction of 80 ± 40 nM. The validity of this estimated Kd is limited by the quality of the mER and AIB1 proteins used in the assay and the quantitation of bands from chemiluminescent reagents. We estimate that the majority of our baculovirus-expressed mER binds estradiol based on comparison of Western blot signal from rat uterine and Sf9 cell ER preparations that have been quantitated for 3H-E2 binding capacity (data not shown). However, we do not have an independent method to verify the biological activity of our GST-AIB1 fusion protein preparation, and the partial AIB1 protein fused to GST may behave differently from full length AIB1. However, there are few other studies that have estimated the affinity of nuclear receptor–coactivator interactions, and our estimate provides a reference point for ER interaction with other coactivators. Peptides containing three nuclear receptor boxes (NR boxes) from the coactivator GRIP-1 were used to estimate binding to the thyroid hormone receptor β. The Kd for this interaction was in the micromolar range (24). The higher affinity we observe most likely is the result of our use of a much larger fragment of AIB1 instead of a small polypeptide. High-affinity binding for an ER–AIB1 interaction (Kd ≈ 1 nM) was reported by using a BIAcore method (21). This method involves immobilization of an AIB1 fragment to dextran, and ER dissociation rates were very slow, which could account for the very low Kd observed. The affinity we observe is in a range that would be responsive to changing ER and/or AIB1 protein concentrations. We hypothesize that increasing the concentration of ER–AIB1 complexes may increase transcription of certain estrogen-regulated genes, even in response to low levels of estrogen, therefore promoting cellular proliferation and tumorigenesis.
Acknowledgments
We thank Candi Jones for technical assistance and Karen Williams (Uniformed Services University of the Health Sciences, Bethesda, MD) for all aspects of project support. We also thank Robert Lechleider and Mike Fritsch for critically reading the manuscript, Aviva Symes for discussions, and Todd Martensen for editorial assistance. The gift of the mER-expressing baculovirus construct from Iain Anderson and Jack Gorski (University of Wisconsin, Madison) is gratefully acknowledged. This work was supported by a grant from the National Institutes of Health (DK50028) awarded to F.E.M.
Abbreviations
- ER
estrogen receptor
- hER
human ER
- mER
mouse ER
- E2
estradiol
- MHT
monohydroxytamoxifen
- ICI
ICI 182,780
- DV
decavanadate
- NEs
nuclear extract prepared with salt
- NEv
nuclear extract prepared with decavanadate
- CE
cytosolic extract
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220427297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220427297
References
- 1.Warner M, Nilsson S, Gusfafsson J A. Curr Opin Obstet Gynecol. 1999;11:249–254. doi: 10.1097/00001703-199906000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Bevan A P, Burgess J W, Yale J-F, Drake P G, Lachance D, Baquiran G, Shaver A, Posner B I. Am J Physiol. 1995;268:E60–E66. doi: 10.1152/ajpendo.1995.268.1.E60. [DOI] [PubMed] [Google Scholar]
- 3.Chen J D, Li H. Crit Rev Eukaryot Gene Expr. 1998;8:169–190. doi: 10.1615/critreveukargeneexpr.v8.i2.40. [DOI] [PubMed] [Google Scholar]
- 4.Leo C, Chen J D. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O'Malley B W. Proc Natl Acad Sci USA. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shim W S, Di Renzo J, De Caprio J A, Santen R J, Brown M, Jeng M H. Proc Natl Acad Sci USA. 1999;96:208–213. doi: 10.1073/pnas.96.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 8.Bautista S, Valles H, Walker R L, Anzick S, Zeillinger R, Meltzer P, Theillet C. Clin Cancer Res. 1998;4:2925–2929. [PubMed] [Google Scholar]
- 9.Bevan C, Parker M. Exp Cell Res. 1999;253:349–356. doi: 10.1006/excr.1999.4719. [DOI] [PubMed] [Google Scholar]
- 10.Fritsch M, Aluker M, Murdoch F E. Biochemistry. 1999;38:6987–6996. doi: 10.1021/bi982190k. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Azorsa D O, Meltzer P S. Hybridoma. 1999;18:281–287. doi: 10.1089/027245799315943. [DOI] [PubMed] [Google Scholar]
- 13.Anderson I, Bartley C R, Lerch R A, Gray W G, Friesen P D, Gorski J. Biochemistry. 1998;37:17287–17298. doi: 10.1021/bi981079b. [DOI] [PubMed] [Google Scholar]
- 14.Gorski J, Welshons W, Sakai D. Mol Cell Endocrinol. 1984;36:11–15. doi: 10.1016/0303-7207(84)90079-0. [DOI] [PubMed] [Google Scholar]
- 15.Alarid E T, Bakopoulos N, Solodin N. Endocrinology. 1999;13:1522–1534. doi: 10.1210/mend.13.9.0337. [DOI] [PubMed] [Google Scholar]
- 16.Nawaz Z, Lonard D M, Dennis A P, Smith C L, O'Malley B W. Proc Natl Acad Sci USA. 1999;96:1858–1862. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakeling A E, Bowler J. J Steroid Biochem Mol Biol. 1992;43:173–177. doi: 10.1016/0960-0760(92)90204-v. [DOI] [PubMed] [Google Scholar]
- 18.McKenna N J, Nawaz Z, Tsai S Y, Tsai M J, O'Malley B W. Proc Natl Acad Sci USA. 1998;95:11697–11702. doi: 10.1073/pnas.95.20.11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonnell D P. J Soc Gynecol Invest. 2000;7:10–15. doi: 10.1016/s1071-5576(99)00055-6. [DOI] [PubMed] [Google Scholar]
- 20.Hammer G D, Krylova I, Zhang Y, Darimont B D, Simpson K, Weigel N L, Ingraham H A. Mol Cell. 1999;3:521–526. doi: 10.1016/s1097-2765(00)80480-3. [DOI] [PubMed] [Google Scholar]
- 21.Suen C S, Berrodin T J, Mastroeni R, Cheskis B J, Lyttle C R, Frail D E. J Biol Chem. 1998;273:27645–27653. doi: 10.1074/jbc.273.42.27645. [DOI] [PubMed] [Google Scholar]
- 22.Thenot S, Charpin M, Bonnet S, Cavailles V. Mol Cell Endocrinol. 1999;156:85–93. doi: 10.1016/s0303-7207(99)00139-2. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Qiu Y, De Mayo F J, Tsai S Y, Tsai M J, O'Malley B W. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 24.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]