Abstract
The RNA-specific adenosine deaminase (ADAR1) and the RNA-dependent protein kinase (PKR) are both interferon-inducible double-stranded (ds) RNA-binding proteins. ADAR1, an RNA editing enzyme that converts adenosine to inosine, possesses three copies of a dsRNA-binding motif (dsRBM). PKR, a regulator of translation, has two copies of the highly conserved dsRBM motif. To assess the functional selectivity of the dsRBM motifs in ADAR1, we constructed and characterized chimeric proteins in which the dsRBMs of ADAR1 were substituted with those of PKR. Recombinant PKR-ADAR1 chimeras retained significant RNA adenosine deaminase activity measured with a synthetic dsRNA substrate when the spacer region between the RNA-binding and catalytic domains of the deaminase was exactly preserved. However, with natural substrates, substitution of the first two dsRBMs of ADAR1 with those from PKR dramatically reduced site-selective editing activity at the R/G and (+)60 sites of the glutamate receptor B subunit pre-RNA and completely abolished editing of the serotonin 2C receptor (5-HT2CR) pre-RNA at the A site. Chimeric deaminases possessing only the two dsRBMs from PKR were incapable of editing either glutamate receptor B subunit or 5-HT2CR natural sites but edited synthetic dsRNA. Finally, RNA antagonists of PKR significantly inhibited the activity of chimeric PKR-ADAR1 proteins relative to wild-type ADAR1, further demonstrating the functional selectivity of the dsRBM motifs.
The RNA-specific adenosine deaminases (ADAR) (1) constitute a multigene family of editing enzymes. They catalyze the C-6 deamination of adenosine to produce inosine in double-stranded (ds) structures present within cellular pre-RNAs and viral RNAs as well as synthetic dsRNA substrates (2–6). So far, two functional RNA adenosine deaminases, denoted ADAR1 and ADAR2, have been described (1, 6). We isolated molecular cDNA clones of ADAR1 as an interferon-inducible enzyme (7, 8). The ADAR1 protein possesses both dsRNA-binding and Z-DNA-binding properties (8–14). The dsRNA-binding domain of the 1,226-aa ORF of the ADAR1 cDNA consists of three copies of the highly conserved dsRNA-binding motif (dsRBM). The prototype dsRBM including the R core amino acid residues was first identified in the interferon-inducible RNA-dependent protein kinase PKR, an enzyme that plays a pivotal role in the antiviral actions of IFN and the control of translation in virus-infected cells (15, 16). PKR possesses two copies of the dsRBM that cannot substitute for each other (17–20). RNAs bound by PKR via the dsRBMs mediate the autophosphorylation and activation of PKR (19, 21), thereby leading to the subsequent phosphorylation of the α subunit of protein synthesis initiation factor eIF-2 (22, 23).
Three naturally occurring splice variant isoforms of human ADAR1 are known that are differentially expressed in different tissues (24). In comparison to the full-length 1,226-aa ADAR1 protein (8–10), denoted ADAR1-a, the variant designated ADAR1-b possesses a deletion of 26 amino acids between the dsRBM RIII core and the catalytic (C) domain. The ADAR1-c variant possesses an additional deletion of 19 amino acids between RII and RIII. The three ADAR1 splice variants exhibit comparable deaminase activity when measured with a synthetic dsRNA substrate (24). However, the ADAR1 splice-site variants display differential editing efficiencies in a site-dependent fashion when tested with two known naturally occurring RNA substrates (25, 26): the pre-RNA encoding ionotropic glutamate receptor channel subunit B (GluR-B) (6, 27) and the pre-RNA encoding serotonin 2C receptor (5-HT2CR) (6, 28).
The sequences obtained for more than 50 dsRBM motifs are highly conserved (29), including those for the three dsRBMs in ADAR1 (8) and two in PKR (17). NMR and crystallography studies of the dsRBM motif from PKR, Escherichia coli RNase III, staufen, and the Xenopus RNA-binding proteins all show a similar α−β−β−β−α protein topology (21, 29–31). Furthermore, at the level of gene organization, the codon phasing is precisely conserved at the junctions of the three exons that specify the three dsRBM copies of ADAR1 as well as at the junctions of the two exons that specify the two dsRBM copies of PKR (24, 32). In spite of the conservation of the dsRBM motifs at the levels of amino acid sequence and exon organization within the Adar and Pkr genes, their functional roles differ in a fundamental manner. With ADAR1, dsRNA functions as the substrate for deamination, and the dsRBM motifs are responsible for the selective recognition of dsRNA structures within the substrate RNAs (11, 25, 33). With PKR, the dsRBM motifs serve to modulate kinase autophosphorylation by binding to RNA regulators, whereby dsRNA functions as an agonist or antagonist rather than a substrate (15, 16).
Although the dsRBMs present in the ADAR and PKR proteins fulfill different functional roles, it is unclear to what extent the specific nature of the dsRBM motifs affects enzymatic activity. For example, it is unknown whether the three dsRBMs of ADAR1 and the two dsRBMs of PKR are functionally exchangeable, independent of their respective C-terminal deaminase and kinase catalytic domains. In an attempt to gain understanding of the functional roles of the dsRBMs of ADAR1 in achieving the site specificity of RNA editing, a domain-swapping approach was used to assess whether there was an obligate requirement for the dsRBM motifs from ADAR1 for catalytic deamination, measured with natural GluR-B and 5-HT2CR neurotransmitter receptor pre-RNAs and with a fully complementary synthetic dsRNA substrate.
Materials and Methods
Oligonucleotides and Construction of ADAR Expression Vectors.
The following oligonucleotides based on human ADAR1 (8) or human PKR (12) cDNA sequences were used for PCR amplification. Lowercase nucleotides are substitutions introduced to generate either restriction sites as silent mutations or point mutations in dsRBM motifs.
Primers for human ADAR1 (U18121): BclI(+)2563, 5′-CCTTCCATGAtCAGATAGCC-3′ (nt 2563–2582); EagI(−)2652, 5′-GAATCTTGCGGCCGAGCAAG-3′ (nt 2633–2652); RII(+)EagI, 5′-AGATGGCgGCcGAGGAAGCC-3′ (nt 2056–2075); RIII(+)EagI, 5′-AAGCgGCcGATGCGGCTCTCC-3′ (nt 2395–2415).
Primers for human PKR (M85294): R1-K64E(+)210, 5′-GAAGGAAGCAgAAAATGCCGC-3′ (nt 210–230); #4(−)1160, 5′-CATTGTTCCAAGGTCCC-3′ (nt 1144–1160); R2(+)EagI, 5′-GGCgGCcgAtCTTGCATATCTTC-3′ (nt 498–520); R2(−)EagI, 5′-AAGaTcgGCcGCCAATTGTTTTG-3′ (nt 488–510); R1-K60E(+), 5′-GGTAGATCAgAGAAGGAAGC-3′ (nt 199–218); R1-K60E(−), 5′-GCTTCCTTCTcTGATCTACC-3′ (nt 199–218); R2-K150E(+), 5′-GGTTCTACTgAACAGGAAGC-3′ (nt 469–488); R2-K150E(−), 5′-GCTTCCTGTTcAGTAGAACC-3′ (nt 469–488).
All eukaryotic expression vectors (Fig. 1) were constructed in pcDNA I/Neo by subcloning the HindIII-XhoI fragments from the pBluescript SK−(pBS) plasmid (Stratagene) after restriction manipulations as described below. The structures of all final constructions were confirmed by Sanger sequencing.
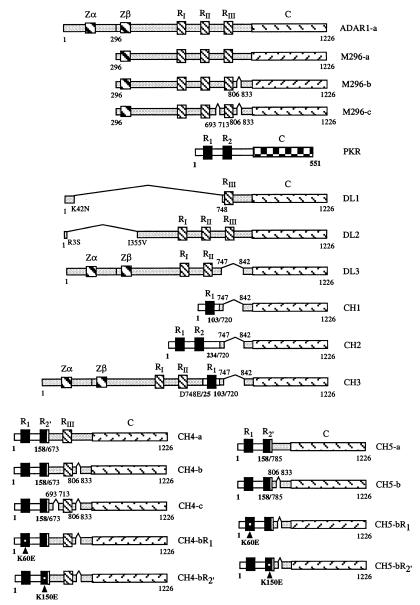
Figure 1.
Schematic illustration of the cDNA structures of human ADAR1 deaminase, PKR kinase, and chimeric PKR-ADAR1 proteins. Zα and Zβ, two Z-DNA-binding domains of ADAR1; RI, RII, and RIII, three copies of the dsRBM of ADAR1; R1 and R2, two copies of the dsRBM of PKR; and C, catalytic domain. The methionine at position 296 used to express the M296 constitutive form of ADAR1 (8, 37) is indicated; a, b, and c refer to the three splice variants of ADAR1 (24). The deletion mutants of ADAR1 (DL1-DL3) and chimeric PKR-ADAR1 constructs (CH1-CH5), including the three CH4 (a, b, c) and two CH5 (a, b) splice variants, are shown. Numbers immediately above or below the schematics refer to the amino acid residue positions, including the positions of the ADAR deletions (DL) and the junctions of the PKR-ADAR1 chimeric (CH) proteins. K60E and K150E, point mutations of the dsRBM motifs from PKR present in CH4-b and CH5-b.
Deletion Mutants.
For the ADAR1 deletion mutants, DL1 was constructed by deletion of the AflII-AccI fragment (nt 168 to 2,286) from ADAR1-a. Similarly, DL2 was generated by deletion of the SacII-NdeI fragment (nt 54 to 1,100) and then insertion of a BglII linker (CAGATCTG) to restore the reading frame. For the DL3 construction, the PCR-derived BclI(+)2563-EagI(−)2652 fragment was used to replace the SalI-EagI fragment (nt 2,286 to 2,652) in pBS-ADAR1-a.
Hybrid Proteins.
For the PKR-ADAR1 chimeric constructs in which dsRBM cores from ADAR1 and PKR were swapped, CH1 was generated by replacing the HindIII(pBS site)-SacI(nt 2,198) fragment of pBS-DL3 with the HindIII(pBS site)-StuI(nt 308) fragment from pBS-PKR. CH2 was likewise constructed by replacing the HindIII(pBS site)-SacI(nt 2,198) fragment of pBS-DL3 with the HindIII(pBS site)-BsaI(nt 721) fragment from pBS-PKR. CH3 was constructed by replacing the SalI(nt 2286)-ApaI(pBS site) fragment in pBS-ADAR1-a with the ScaI(nt 99 in PKR)-ApaI(pBS site) fragment of pBS-CH1.
The CH4 and CH5 sets of PKR-ADAR1 chimeric constructs were engineered to retain an intact spacer region between the dsRBM motifs and the catalytic domain of ADAR1. First, an EagI site in the R2 motif of PKR was generated by two successive rounds of PCR amplification. Primer pairs R1-K64E(+)210 and R2(−)EagI, and R2(+)EagI and #4(−)1160 generated two PKR fragments possessing 13 bp of overlapping sequence, which were used as templates in a second round of PCR with the two distal primers K64E(+)210 and #4(−)1160. The NcoI-AccI (nt 320–798) fragment derived from the resultant 941-bp PCR product was used to replace the corresponding fragment in pBS-PKR, thus generating an EagI site in R2 of PKR (nt 500). By using the HindIII(pBS site)-EagI(nt 500) fragment of pBS-PKR to replace the HindIII(pBS site)-EagI(nt 2,652) fragment of pBS-ADAR1-a, an intermediate chimeric vector pBS-CH was obtained. EagI-EagI PCR fragments of ADAR1, amplified from either ADAR1-a, -b, or -c templates by using the primer pair RII(+)EagI and EagI(−)2,652, were subcloned into the EagI site of pBS-CH to generate the three CH4 constructs, CH4-a, -b, and -c, respectively. Similarly, CH5-a and -b constructs were obtained by inserting the PCR fragments amplified with the primer pair RIII(+)EagI and EagI(−)2,652 from ADAR-a and -b, respectively, into the EagI site of pBS-CH.
RNA-Binding Mutants.
A PCR-based site-directed mutagenesis strategy was used to mutate the highly conserved and critical lysine residue required for RNA binding (11). For mutants CH4-bR1 and CH5-bR1, primer pairs T3(+) and R1-K60E(−) and R1K60E(+) and T7(−) were used to amplify two overlapping products from either the pBS-CH4-b or the pBS-CH5-b templates; these overlapping products were then used as templates in a second round of PCR with the two distal primers T3(+) and T7(−). The HindIII-XhoI fragments of the resultant PCR products, containing the K60E mutation in R1 (11), were subsequently subcloned into pcDNA I/Neo. Mutants CH4-bR2′ and CH5-bR2′ were generated by using a similar strategy, but with the primer pairs T3(+) and R2-K150E(−), and R2-K150E(+) and T7(−).
Expression of the Recombinant ADAR1 and PKR-ADAR1 Chimeric Proteins.
The prototype ADAR1 in the N-terminally truncated (M296) form as well as the deletion-mutant ADAR1 and chimeric PKR-ADAR1 proteins (Fig. 1) were expressed in transfected monkey kidney COS-1 cells (8, 24). The cytoplasmic fractions, which are free of the endogenous COS nuclear ADAR enzymes, were prepared and used as the source of expressed recombinant proteins as previously described in detail (14, 24). Western immunoblot analysis was carried out to quantitate protein levels by using antiserum generated against the catalytic domain of recombinant ADAR1 protein (8). Antibody–antigen complexes were detected with 125I-labeled protein A by autoradiography, followed by quantitation with a Bio-Rad GS-525 molecular imager.
Analysis of A-to-I RNA Editing Activity in Vitro.
Two types of assays were used to measure RNA-specific adenosine deamination. All quantitations were done by using a GS-525 imager system. Error bars shown in figures were determined from two to three independent experiments.
Synthetic dsRNA substrate.
The dsRNA-specific adenosine deaminase activity was measured with a 32P-labeled synthetic dsRNA substrate as previously described (8, 14, 24). To test for antagonism, either synthetic aptamer clone 3 RNA (34) or wild-type adenovirus VAI RNA transcribed in vitro (35) was included in the reaction mixture. Inosine formation from adenosine was analyzed by TLC after hydrolysis with nuclease P1 (Pharmacia).
Natural RNA substrates.
Recombinant proteins were also analyzed for site-selective A-to-I editing activity with natural pre-RNA substrates. For GluR-B RNA, measurement of editing efficiency at the intronic hotspot (+)60 and exon 13 R/G sites was done by using RT-based poisoned primer extension assays as previously described (25); for 5-HT2CR RNA, a poisoned primer extension strategy was also used to determine the editing efficiency at the A and C sites by using specific 32P end-labeled primers (26). Extended products were resolved on denaturing 16% PAGE gels.
Results
dsRBMs from PKR Kinase Can Substitute for Those of the ADAR1 Deaminase in Mediating dsRNA-Specific Adenosine Deamination.
The core sequences of the dsRBM motifs from ADAR1 (RI, RII, RIII) and PKR (R1, R2) are highly conserved (Table 1). To investigate the functional role of the dsRBMs of ADAR1 in determining the site selectivity of A-to-I editing, we examined the ability of dsRBMs from PKR to substitute for those of ADAR1 in the editing of synthetic and natural substrates. Three deletion mutants designated DL1, DL2, and DL3 were derived from ADAR1-a as intermediates in our initial cloning strategy (Fig. 1). With a synthetic dsRNA substrate, neither DL1 that lacked both the RI and RII motifs as well as the Zα and Zβ domains, nor DL3 which lacked only the RIII motif, exhibited detectable deaminase activity (Table 2). By contrast, the DL2 deletion mutant that lacked the N-terminal region including the Zα and Zβ domains but retained the three dsRBM motifs showed deaminase activity comparable to that of the full-length and M296 forms of ADAR1-a (Table 2). The chimeric PKR-ADAR1 protein CH3, which included the dsRBM core sequence R1 from PKR inserted into the spacer region of DL3 between the RII motif and catalytic domain (Fig. 1), lacked deaminase activity. Furthermore, when all three dsRBMs of ADAR1 were replaced, either with the single dsRBM motif R1 from PKR (chimera CH1) or with both dsRBM motifs of PKR (chimera CH2), not unexpectedly, no enzyme activity was detectable (Table 2). Even though no deaminase activity was observed for the DL1, DL3, CH1, CH2, and CH3 proteins under any assay condition examined, these recombinant mutant proteins were of the appropriate size as analyzed by SDS/PAGE (data not shown). Presumably the conformation of these inactive deletion and chimeric mutant proteins was perturbed in a manner that precluded either substrate binding or catalysis.
Table 1.
Homology between the core (R) sequences of the dsRBM motif from human and rat ADAR1 and PKR proteins
| †Rn \ *Hs | ADAR1
|
PKR
|
CH4
|
CH5
|
||||
|---|---|---|---|---|---|---|---|---|
| RI | RII | RIII | R1 | R2 | ‡R2′ | R2′ | ||
| RI | — | 60 | 67 | 67 | 33 | — | — | |
| ADARI | RII | 60 | — | 62 | 48 | 33 | 45 | — |
| RIII | 64 | 62 | — | 57 | 36 | — | 48 | |
| PKR | R1 | 60 | 45 | 52 | — | 43 | — | — |
| R2 | 31 | 38 | 38 | 40 | — | 88 | 88 | |
The core (R) sequences of the dsRBM motif from human (Hs) ADAR1 (42 aa, U18121) correspond to residues 528–569 for RI, 639–680 for RII and 751–792 for RIII; and from PKR (42 aa, M85294), residues 34–75 for R1 and 124–165 for R2. The values in boldface indicate relatively high homology between dsRBMs of different proteins.
The underlined values indicate the similarity percentage for rat (Rn) proteins. The dsRBM core sequences from ADAR1 (42 aa, U18942) correspond to residues 478–519 for RI, 589–630 for RII and 697–738 for RIII; and from PKR (42 aa, L29281), residues 33–74 for R1 and 119–160 for R2.
The fusion R core (R2′) that consists of the N-terminal 35 residues of the R2 of PKR and the C-terminal 7 residues of the RII in CH4 or RIII in CH5 of ADAR1 are compared with the composite R core sequences, respectively.
Table 2.
Ability of engineered recombinant ADAR1 proteins to deaminate synthetic dsRNA
| Construction | A → I Deaminase activity |
|---|---|
| Wild-type proteins | |
| ADAR1-a | 1.07 |
| M296-a | ≡1.00 |
| Mutant proteins | |
| DL1 | None |
| DL2 | 1.13 |
| DL3 | None |
| CH1 | None |
| CH2 | None |
| CH3 | None |
| CH4-a | 0.79 |
| CH5-a | 0.12 |
The relative specific deaminase activity of the recombinant wild-type M296-a protein measured with a synthetic dsRNA substrate under standard conditions is defined as 1.00. None, no deaminase activity was detectable. cDNA structures of wild-type and mutant versions of RNA-specific deaminase proteins are as shown in Fig. 1. DL, deletion mutant; CH, chimeric hybrid mutant protein.
Editing activities observed with the three naturally occurring splice variants of ADAR1 (24), illustrated by M296-a, -b, and -c (Fig. 1), indicated that the spacer regions between the dsRBMs and the catalytic domain might be functionally important, because the A-to-I editing at certain sites of the GluR-B and 5-HT2CR pre-RNA substrates differed about 2-fold between ADAR1 variants (25, 26). Therefore, chimeric PKR-ADAR1 proteins were engineered that precisely retained the distance between the dsRBM motifs and the catalytic domain, identical to that of the natural splice variants. Three variants of the CH4 chimeric protein (CH4-a, -b, and -c) were constructed by substituting the RI and RII dsRBM motifs of each splice-site variant of ADAR1 with the R1 and R2 dsRBMs of PKR (Fig. 1). Likewise, two variants of the CH5 chimera (CH5-a and -b) were generated by replacing all three dsRBMs of ADAR1-a and -b (Fig. 1). The fusion R2′ core sequence in the CH4 or CH5 constructs consisted of 35 amino acids from the N-terminal R2 core sequence of PKR and only 7 amino acids from the C-terminal RII or RIII core sequences of ADAR1; R2′ retained high homology (88%) with the native R2 core of PKR (Table 1).
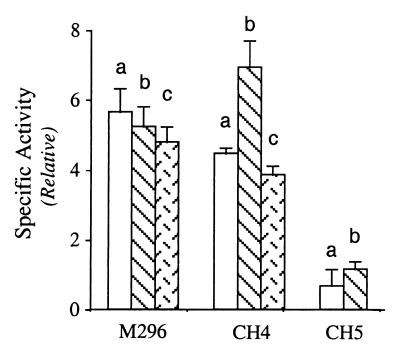
Chimeric CH4 proteins possessed high RNA-specific adenosine deaminase activity with a synthetic dsRNA substrate (Fig. 2). Furthermore, all three of the splice variants, CH4-a, -b, and -c, showed comparable deaminase activity, and their activities were similar to those of the three splice variants of ADAR1 in the M296 form (Fig. 2). The two CH5 chimeras that contained only the two dsRBMs of PKR as a unit also exhibited significant enzymatic activity (Fig. 2). However, the specific deaminase activity of the CH5 chimeric proteins was significantly lower than that of the corresponding CH4 chimeric variants and the M296 variants.
Figure 2.
Analysis of dsRNA adenosine deaminase activity of recombinant chimeric PKR-ADAR1 proteins. Relative specific deaminase activities were calculated based on the A-to-I conversion with a synthetic 32P-labeled dsRNA, catalyzed by three ADAR1 splice variant proteins (a, b, and c) of the M296 form (8, 24); three PKR-ADAR1 chimeric CH4 variant proteins (a, b, and c); and two chimeric CH5 proteins (a and b).
Both dsRBMs of PKR Are Essential for Deaminase Activity of the Chimeric Proteins.
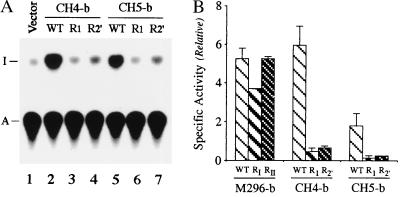
Prior analyses of single substitution ADAR1 mutants established that the RIII motif is essential for deaminase activity, whereas substitution mutants at either the RI or RII dsRBM motifs retain significant enzyme activity with dsRNA (11, 24). The equivalent single site-directed mutation (K60E or K150E) was therefore introduced into the R1 or R2 dsRBM motifs of PKR to generate the mutant versions of chimeric CH4-b (CH4-bR1 and CH4-bR2′) and CH5-b (CH5-bR1 and CH5-bR2′) proteins (Fig. 1). Mutation of either R1 or R2 within the chimeric CH4-b and CH5-b proteins almost completely destroyed the A-to-I editing activity, as measured with a synthetic dsRNA substrate (Fig. 3A). The specific deaminase activities of the CH4-b and CH5-b dsRBM mutants were greatly reduced relative to the wild type (Fig. 3B). By contrast, the corresponding mutations when present in the M296 form of the prototype ADAR1 did not significantly affect activity (Fig. 3B), consistent with earlier observations (24).
Figure 3.
Both RNA-binding motifs of chimeric PKR-ADAR1 proteins are essential for dsRNA adenosine deaminase activity. (A) Autoradiogram showing dsRNA-specific adenosine deaminase activity catalyzed by wild-type (WT; lanes 2 and 5) and mutant (lanes 3, 4, 6, and 7) chimeric CH4 and CH5 proteins. The K60E or K150E mutation was introduced into the R1 (lanes 3 and 6) and R2 (lanes 4 and 7) dsRBM motifs of PKR within the chimeric CH4-b and CH5-b proteins. The positions to which the adenosine (A) and inosine (I) 5′-nucleoside monophosphates migrated on the TLC plate are indicated. (B) Relative specific deaminase activities were calculated based on percentage of A-to-I conversion with synthetic dsRNA substrate. The relative specific activities of CH4-b and CH5-b chimeric enzymes were compared with those of the M296 version of ADAR1-b.
Site-Selective Editing of GluR-B and 5-HT2CR Pre-RNAs.
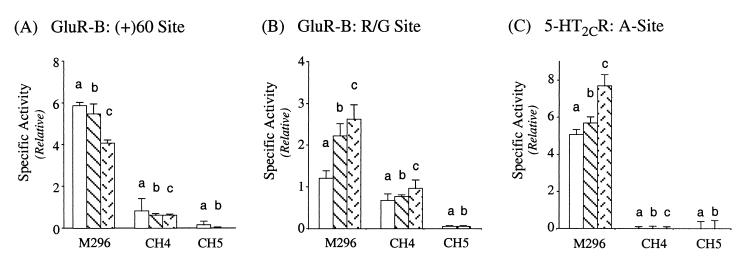
We next examined the ability of the DL2 deletion mutant and the CH4 and CH5 chimeric PKR-ADAR1 recombinant proteins to catalyze site-selective editing of natural RNA substrates as compared with the nonspecific deamination of a synthetic dsRNA. As was seen with synthetic dsRNA (Table 1), DL2 exhibited comparable editing activity to that of M296 for both the R/G and the hotspot + 60 sites of GluR-B RNA (data not shown). The three chimeric CH4 variant enzymes (a, b, and c) were all capable of modifying the two GluR-B RNA substrates in vitro in a site-selective fashion (Fig. 4 A and B). However, as compared with the M296-ADAR1 variants, the CH4 chimeras all showed significantly reduced editing efficiency both for the hotspot (+)60 site (Fig. 4A) and the R/G site (Fig. 4B). This may reflect the effects of the shorter spacer region between R1 and R2 of PKR (49 amino acids) that is very different in primary amino acid sequence from that of the RI and RII spacer region (70 amino acids) of ADAR1. Similar to the results obtained with the synthetic dsRNA substrate (Fig. 3B), mutation of either of the two dsRBMs of PKR present in the chimeric CH4-b enzymes abolished editing activity at the two GluR-B sites (data not shown). The three chimeric CH4 variants (a, b, and c) showed comparable editing efficiency at the R/G site. By contrast, and in full agreement with prior observations (25), the M296-ADAR1-b and -c splice variants edited the R/G site more efficiently than M296-a (Fig. 4B). The two chimeric CH5 variants exhibited extremely low editing activity at both GluR-B sites (Fig. 4 A and B).
Figure 4.
Analysis of site-selective editing of GluR-B and 5-HT2CR receptor RNAs by chimeric PKR-ADAR1 proteins in vitro. A-to-I editing catalyzed by the chimeric CH4 and CH5 proteins as compared with the M296 version of ADAR1 was determined for: (A) GluR-B RNA intronic hotspot (+)60 site; (B) GluR-B RNA R/G site; and (C) 5-HT2CR RNA A site.
In contrast to the GluR-B RNA substrate, none of the chimeric CH4 or CH5 deaminases were able to edit the 5-HT2CR RNA substrate, either at the A site (Fig. 4C) or the C site (data not shown). The A site was efficiently edited by all three splice variants of M296 (Fig. 4C). Furthermore, DL2 that lacks the two Z-DNA-binding domains (Fig. 1) edited the A and C sites with comparable efficiency to that of the full-length or M296 form of ADAR1-a (data not shown; ref. 26).
The dsRBMs of PKR and ADAR1 Are Functionally Different in Recognizing Antagonistic RNAs.
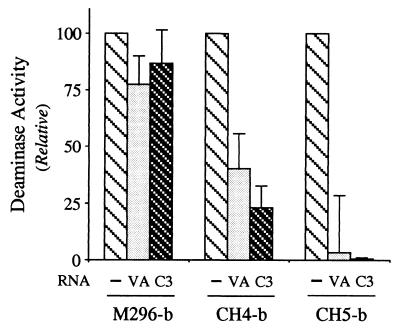
The highly structured VAI RNA of adenovirus (36) and the synthetic clone 3 aptamer identified by RNA selection with PKR dsRBM motifs (34) both antagonize the activation of PKR kinase by binding to its dsRBMs. The ability of wild-type M296 ADAR1 to deaminate synthetic dsRNA was not significantly affected by the addition of either 1 μM VAI RNA or 10 μM C3 RNA, at which concentrations they significantly antagonize the activation of PKR (34, 36). By contrast, the activities of the CH4 and CH5 chimeras were impaired by both VAI and C3 RNAs at 1 μM, even though the activity of M296 was not (Fig. 5). The inhibition of CH5, which possesses the R1 and R2 dsRBM motifs from PKR in place of the three dsRBMs of ADAR1 (Fig. 1), was somewhat greater than that of CH4, which retains the RIII dsRBM from ADAR1 (Fig. 5).
Figure 5.
Effect of adenovirus VA RNA and C3 aptamer RNA on RNA-specific adenosine deaminase activity catalyzed by the M296 version of ADAR1-b and the chimeric CH4-b and CH5-b proteins. A synthetic 32P-labeled dsRNA substrate was used in the standard TLC-based adenosine deaminase assay, modified to contain either adenovirus VAI RNA or selected C3 aptamer RNA at a final concentration of ≈1 μM.
Discussion
To gain understanding of the functional specificity of the dsRBM motifs of the ADAR1 deaminase, we asked whether the dsRBM motifs of the PKR kinase could substitute for those of ADAR1 as measured by retention of A-to-I editing activity. The answer is yes, depending on the nature of the substrate RNA. Several important points emerged from our functional analysis of PKR-ADAR1 chimeric and ADAR1 deletion mutant proteins. First and foremost, chimeric proteins surprisingly could be engineered that retained RNA-specific deaminase activity. The two dsRBM motifs from PKR as a module could replace those from ADAR1 in a chimeric enzyme capable of catalyzing nonspecific deamination of a fully complementary synthetic dsRNA substrate. Second, the ability of the chimeric ADAR1 hybrids possessing dsRBM motifs from PKR to selectively edit natural RNA substrates such as the GluR-B or 5-HT2CR RNAs varied with the substrate. Third, small structured RNAs known for their ability to antagonize PKR could also antagonize ADAR1 deaminase activity, but their effectiveness as antagonists depended on the origin of the dsRBM motifs, suggesting that specific RNAs target specific dsRBMs. And fourth, deletions within the region between the dsRBM motifs and catalytic domain of ADAR1 affected deaminase activity, whereas deletion of one or both of the Z-DNA-binding motifs from ADAR1 did not impair editing activity.
That the two dsRBM motifs of PKR can functionally substitute for those naturally found in ADAR1 is illustrated by the CH4 and CH5 chimeric PKR-ADAR proteins. The three chimeric splice variants of CH4 (a, b, c), all of which possess the two dsRBMs from PKR and the third from ADAR1, displayed comparable deaminase activity to that of the M296 form of ADAR1 with synthetic dsRNA as the substrate. This suggests that the N-terminal region upstream of the RI motif of ADAR1 and the spacer region between the RI and RII motifs of ADAR1 are not uniquely required for editing activity, in that they can be replaced by the N-terminal region of PKR containing R1 and R2 from the kinase. Likewise, the two chimeric variants of CH5 (a, b), in which all three dsRBMs of ADAR1 were substituted by using the two dsRBMs of PKR, also possessed considerable deaminase activity measured with synthetic dsRNA. That the CH5 chimera was an active enzyme was somewhat unexpected. It is reported that an ADAR1 deletion mutant which lacks the proximal RI dsRBM motif but retains the other two dsRBMs is not active (36), even though site-directed mutagenesis of RI that destroys RNA-binding activity of the dsRBM motif does not eliminate deaminase activity (11, 24). Possibly the ADAR1 dsRBM deletion mutation caused a change in conformation that prevented enzymatic activity (33), whereas the chimeric CH5 protein possessed a favorable conformation from the N-terminal portion of PKR that supports deaminase activity. The CH5 proteins showed <20% deaminase activity relative to the CH4 proteins. This conceivably reflects the inefficiency of having only two dsRBM motifs in CH5, both from PKR, as compared with the three dsRBMs in CH4, one of which (RIII) is functionally the most important dsRBM of ADAR1 (11, 24). Although both CH4 and CH5 are active deaminases, the activity differences seen between these chimeric mutants and those of ADAR1 could reflect alterations in either substrate-binding properties or catalytic rates. Further studies will be necessary to definitively resolve these possibilities.
Recombinant CH4, but not CH5, chimeric proteins displayed an ability to selectively edit natural RNA substrates. In the case of CH4, the two dsRBMs of PKR together with the RIII-containing dsRBM from ADAR1 were capable of supporting site-selective editing of GluR-B RNA at both the intronic hotspot (+)60 and the R/G sites, further suggesting that the spacer between R1 and R2 of PKR does not play a unique role in determining the site specificity. However, the editing of GluR-B by CH4 occurred with significantly reduced efficiency relative to the activity observed for M296 or relative to that seen with CH4 for the synthetic dsRNA substrate. Whereas the three M296 splice variants (a, b, c) possess differential editing activity for the R/G site (25), little difference was observed with the three chimeric CH4 variants. This finding further supports the notion that the two dsRBMs of PKR were not fully equivalent to the dsRBMs of ADAR1 when measured with a natural RNA substrate. However, the possibility that the difference in the spacer region between R1 and R2 of PKR contributes to editing efficiency but not specificity cannot be excluded. Moreover, the two dsRBMs of PKR alone were not sufficient to support site-selective editing of GluR-B RNA, as evidenced by the lack of editing at the (+)60 and R/G sites by CH5 chimeras. In contrast to GluR-B RNA, the 5-HT2CR RNA was not edited by either the CH4 or the CH5 chimeras even at the A site, which is efficiently edited by M296 ADAR1 (26, 28). These results suggest that features of the GluR-B and 5-HT2CR RNA substrates are distinguished differently by the whole RNA-binding domain constituted by the multiple dsRBM motifs and spacers of ADAR1 and PKR. Possibly there is a more stringent requirement for functional interactions between the 5-HT2CR RNA and dsRBD sufficient to achieve editing of the serotonin receptor RNA. The structure of the GluR-B RNA capable of undergoing A-to-I editing may conceivably be more double stranded or more flexible in nature rather than existing per se as an unique higher-ordered single-stranded RNA structure, because the GluR-B RNA can be recognized by dsRBMs of both ADAR1 and ADAR2 (6, 27) as well as PKR in the CH4 chimeras for selective deamination.
Prior mutagenesis studies, in which the dsRBM motifs of ADAR1 were individually inactivated for RNA-binding activity by single site-directed amino acid substitutions, showed that disruption of either the RI or RII dsRBM did not destroy ADAR1 deaminase activity (11, 24). However, we found that the conceptually equivalent mutations introduced into the two dsRBMs of PKR present in the CH4 chimera completely destroyed deaminase activity toward synthetic dsRNA. This was surprising, because the CH4 chimera possessed comparable deaminase activity with synthetic dsRNA, as did wild-type ADAR1. Thus, although the dsRBMs of PKR and ADAR1 appeared functionally equivalent in the background of the wild-type form of the chimeric CH4 protein, mutagenesis results with CH4 (and also CH5) suggest that the dsRBMs from PKR were not functionally identical to those of ADAR1 when examined as mutated dsRBMs in the chimera background.
As an initial strategy to generate engineered ADAR1 chimeric proteins with dsRBM motifs from PKR, restriction sites were used that had the effect of altering the length of the exon 7 spacer region between the dsRBM motifs and the C-terminal catalytic domain of ADAR1. We did not anticipate that this would necessarily be problematic, because the naturally occurring ADAR1-b and -c splice variants possess an alternative exon 7 that lacks the residues amino acids 806 to 833 of ADAR1-a. However, when the deletion was expanded in chimeric proteins CH2 and CH3 to include amino acids 748 to 841, neither chimera possessed detectable deaminase activity. But when the fusion junction was engineered to occur within the highly conserved RII or RIII core sequence of the dsRBM in a manner that consequently did not alter this spacer region, then deaminase activity was preserved. These results thus emphasize the functional importance of the linker region between the dsRNA-binding domain and the catalytic domain of the ADAR deaminase, a notion supported by the existence of the splice variants of ADAR1.
Adenovirus VA RNA is a well established antagonist of the RNA-dependent protein kinase PKR (15, 36) and, at high concentrations, also is able to inhibit ADAR1 (35). Clone 3 (C3) aptamer RNA, selected from a library of ≈1014 different sequences by using the dsRBMs of PKR, is likewise an antagonist of PKR activation (34). Both VA RNA and C3 RNA were able to inhibit the deaminase activity of the CH4 and CH5 chimeric proteins. Interestingly, CH4 and CH5 were significantly more sensitive to inhibition by the VA and C3 RNAs than was the M296 form of ADAR1. It is tempting to speculate that VA and C3 RNAs were capable of distinguishing the dsRBMs of PKR from those of ADAR1. Possibly the chimeric deaminases displayed increased sensitivity because of the presence of the dsRBM domain from PKR, the structure against which the antagonistic RNA is presumably naturally targeted in the biology of the virus (36) or the structure with which the C3 RNA was selected (34).
Acknowledgments
This work was supported in part by Research Grants AI-12520 and AI-20611 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbreviations
- ADAR
RNA-specific adenosine deaminase
- IFN
interferon
- ds
double-stranded
- dsRBM
dsRNA-binding motif
- PKR
RNA-dependent protein kinase
References
- 1.Bass B L, Nishikura K, Keller W, Seeburg P H, Emeson R B, O'Connell M A, Samuel C E, Herbert A. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 2.Bass B L, Weintraub H. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 3.Wagner R W, Smith J E, Cooperman B S, Nishikura K. Proc Natl Acad Sci USA. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar M, Carmichael G G. Proc Natl Acad Sci USA. 1997;94:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattaneo R. Curr Opin Genet Dev. 1994;4:895–900. doi: 10.1016/0959-437x(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 6.Rueter S M, Emeson R B. Modifications and Editing of RNA. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 343–361. [Google Scholar]
- 7.Patterson J B, Thomis D C, Hans S L, Samuel C E. Virology. 1995;210:508–511. doi: 10.1006/viro.1995.1370. [DOI] [PubMed] [Google Scholar]
- 8.Patterson J B, Samuel C E. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connell M A, Krause S, Higuchi M, Hsuan J J, Totty N F, Jenny A, Keller W. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Samuel C E. J Virol. 1996;70:1961–1968. doi: 10.1128/jvi.70.3.1961-1968.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomis D C, Doohan J P, Samuel C E. Virology. 1992;188:33–46. doi: 10.1016/0042-6822(92)90732-5. [DOI] [PubMed] [Google Scholar]
- 13.Herbert A, Alfken J, Kim Y-G, Mian I S, Nishikura K, Rich A. Proc Natl Acad Sci USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Herbert A, Rich A, Samuel C E. Methods Companion Methods Enzymol. 1998;15:199–205. doi: 10.1006/meth.1998.0624. [DOI] [PubMed] [Google Scholar]
- 15.Samuel C E. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 16.Clemens M J, Elia A. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 17.McCormack S J, Thomis D C, Samuel C E. Virology. 1992;188:47–56. doi: 10.1016/0042-6822(92)90733-6. [DOI] [PubMed] [Google Scholar]
- 18.Green S R, Manche L, Mathews M B. Mol Cell Biol. 1995;15:358–364. doi: 10.1128/mcb.15.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCormack S J, Ortega L G, Doohan J P, Samuel C E. Virology. 1994;198:92–99. doi: 10.1006/viro.1994.1011. [DOI] [PubMed] [Google Scholar]
- 20.McMillan N A J, Carpick B W, Hollis B, Toone W M, Zamanian-Daryoush M, Williams B R G. J Biol Chem. 1995;270:2601–2606. doi: 10.1074/jbc.270.6.2601. [DOI] [PubMed] [Google Scholar]
- 21.Nanduri S, Carpick B W, Yang Y, Williams B R, Qin J. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samuel C E. Proc Natl Acad Sci USA. 1979;76:600–604. doi: 10.1073/pnas.76.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathak V K, Schindler D, Hershey J W. Mol Cell Biol. 1988;8:993–995. doi: 10.1128/mcb.8.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, George C X, Patterson J B, Samuel C E. J Biol Chem. 1997;272:4419–4428. doi: 10.1074/jbc.272.7.4419. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Samuel C E. J Biol Chem. 1999;274:5070–5077. doi: 10.1074/jbc.274.8.5070. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Emeson R B, Samuel C E. J Biol Chem. 1999;274:18351–18358. doi: 10.1074/jbc.274.26.18351. [DOI] [PubMed] [Google Scholar]
- 27.Seeburg P H, Higuchi M, Sprengel R. Brain Res Brain Res Rev. 1998;26:217–229. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- 28.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Nature (London) 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 29.Kharrat A, Macias M J, Gibson T J, Nilges M, Pastore A. EMBO J. 1995;14:3572–3584. doi: 10.1002/j.1460-2075.1995.tb07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bycroft M, Grunert S, Murzin A G, Proctor M, St. Johnston D. EMBO J. 1995;14:3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryter J M, Schultz S C. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhen K L, Shen X, Carlisle E R, Richardson A L, Weier H U, Tanaka H, Samuel C E. Genomics. 1996;36:197–201. doi: 10.1006/geno.1996.0446. [DOI] [PubMed] [Google Scholar]
- 33.Lai F, Drakas R, Nishikura K. J Biol Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 34.Bevilacqua P C, George C X, Samuel C E, Cech T R. Biochemistry. 1998;37:6303–6316. doi: 10.1021/bi980113j. [DOI] [PubMed] [Google Scholar]
- 35.Lei M, Liu Y, Samuel C E. Virology. 1998;245:188–196. doi: 10.1006/viro.1998.9162. [DOI] [PubMed] [Google Scholar]
- 36.Mathews M B, Shenk T. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.George C X, Samuel C E. Proc Natl Acad Sci USA. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]