Abstract
Myosins, a large family of actin-based motors, have one or two heavy chains with one or more light chains associated with each heavy chain. The heavy chains have a (generally) N-terminal head domain with an ATPase and actin-binding site, followed by a neck domain to which the light chains bind, and a C-terminal tail domain through which the heavy chains self-associate and/or bind the myosin to its cargo. Approximately 140 members of the myosin superfamily have been grouped into 17 classes based on the sequences of their head domains. I now show that a phylogenetic tree based on the sequences of the combined neck and tail domains groups 144 myosins, with a few exceptions, into the same 17 classes. For the nine myosin classes that have multiple members, phylogenetic trees based on the head domain or the combined neck/tail domains are either identical or very similar. For class II myosins, very similar phylogenetic trees are obtained for the head, neck, and tail domains of 47 heavy chains and for 29 essential light chains and 19 regulatory light chains. These data strongly suggest that the head, neck, and tail domains of all myosin heavy chains, and light chains at least of class II myosins, have coevolved and are likely to be functionally interdependent, consistent with biochemical evidence showing that regulated actin-dependent MgATPase activity of Dictyostelium myosin II requires isoform specific interactions between the heavy chain head and tail and light chains.
Myosins, a large family of actin-dependent motors involved in contractile and motile functions in higher and lower animals, plants, and fungi (1), contain one or two (identical) heavy chains of masses ranging from about 110,000 to 250,000 Da and one or more light chains of masses between about 15,000 and 20,000 Da per heavy chain. The heavy chains contain a head (catalytic, motor) domain, generally N-terminal, followed by a neck domain to which the light chains bind, and a C-terminal tail domain, which, in some myosins, dimerizes with an identical heavy chain by forming coiled-coil helical regions and through which some myosin dimers polymerize into filaments. The tail domain of other myosin heavy chains is thought to associate with membranes and organelles or other cargoes that the myosin molecules may transport along actin filaments.
The databases now contain complete DNA sequences of approximately 145 different myosin heavy chains from about 40 different species. Myosin heavy chains have been grouped into 17 different classes (J. Cope and T. Hodge, The Myosin Home Page, http://www.mrc-lmb.cam.ac.uk/myosin/myosin.html) based on the derived amino acid sequences of their head domains. [After completion of this study, a two-member class XVIII was identified (2).] Head domain sequences were used because myosins were defined by the actin-dependent MgATPase activity of the head (3). It was recognized, however, that the initial groupings of myosins by head domain sequences into three (4) and seven (3) classes were tightly correlated with gross structural differences of their tail domains. Although many complete heavy chain sequences are now available, phylogenetic trees of the entire myosin superfamily based on sequences of heavy chain domains other than the head domain have not been developed.
Traditionally, and largely as a result of extensive biochemical studies of the conventional (class II) muscle myosins, myosin catalytic activity and its regulation have been attributed solely to the heavy chain head domain and light chains associated with the neck domain (1). However, there is increasing evidence, especially for the class II myosins from the slime mold Dictyostelium discoideum and the soil amoeba Acanthamoeba castellanii, that contradicts this dogma. Specifically, phosphorylation of serine residues in the tail of Dictyostelium myosin II heavy chain down-regulates the actin-dependent MgATPase activity of the head by increasing the KATPase for F-actin (5) and phosphorylation of serine residues at the C-terminal end of the tail of the heavy chain of Acanthamoeba myosin II down-regulates the activity of its head domain by substantially reducing Vmax (6). In the accompanying paper (7), we show that the actin-dependent MgATPase activity of chimeras in which the head and neck (and associated light chains) domains of Dictyostelium myosin II are fused to the tail domain of either Acanthamoeba myosin II or chicken smooth muscle myosin II is substantially greater than the activity of wild-type Dictyostelium myosin II and essentially unregulated, i.e., in contrast to wild-type Dictyostelium and smooth muscle myosin II, actin-dependent MgATPase activity of the chimeras is only minimally inhibited by unphosphorylated regulatory light chain and, therefore, only minimally activated by phosphorylation of the Dictyostelium regulatory light chain and, in contrast to wild-type Acanthamoeba myosin II, the activity of the chimera with Acanthamoeba myosin II tail is not inhibited by phosphorylation of the Acanthamoeba tail domain. These and other results (8) indicative of strong coupling between the head, neck, and tail domains of the heavy chains of Dictyostelium, Acanthamoeba, and smooth muscle myosin II led me to inquire whether the implied coevolution of the head, neck, and tail domains of these, and possibly all, myosins would be supported by phylogenetic analysis of their amino acid sequences.
Experimental Method
Unrooted phylogenetic trees were constructed by progressive multiple sequence alignment using the default mode of the PC version of the clustal x program (9) with 1,000 bootstrap trials, without correction for gaps or multiple substitutions and without correction for any misalignments (9) that may have occurred. Trees were plotted by using the neighbor joining method (9) and manually adjusted by rotating branches around nodes.
Normally, sequences were entered in alphabetical order but, in some instances, the sequence order was randomized, and 10,000 bootstrap trials were performed. Complete heavy chain sequences were divided into head domains and combined neck/tail domains or, for class II myosins, separate head, neck, and tail domains, as described below. The myosins that were analyzed, their accession numbers, and the residues comprising the head and neck/tail domains or head, neck, and tail domains are enumerated in the figure legends.
The first step was to establish the boundaries between the head, neck, and tail domains by multiple alignments within each of the 17 myosin classes defined by the phylogenetic tree (http://www.mrc-lmb.cam.ac.uk/myosin/myosin.html) of the core motor domains. For class II myosins, this was straightforward as all of the head domains terminate with amino acids FFK or FFR, the neck domains are the next 71 amino acids (except for ScMYO1, which has 72 residues in its neck domain) and all of the tail domains begin with a Pro residue. It also was not difficult to define the end of the head domains of the other myosins; for example, class I myosin heads terminate with FIR or FIK and, occasionally, FVK. However, it proved very difficult, if not impossible, to define unequivocally the junctions between the neck and tail domains of the non-class II myosins. Although neck domains characteristically are defined by the presence of one or more IQ motifs (10), to which the calmodulin-like light chains bind, these motifs can be sufficiently degenerate or incomplete that reasonable people can reach different conclusions on the number of IQ motifs in a given heavy chain, especially as there is no independent biochemical information about the nature or number of light chains of most myosins. For this reason, separate phylogenetic analyses of the neck domains for myosins other than class II myosins were not attempted. Instead, phylogenetic trees of the head domains were compared with phylogenetic trees of the combined neck and tail domains. This almost certainly did not affect the conclusions (other than the absence of explicit information about the phylogeny of the neck domains) because the neck domains are substantially shorter than the tail domains and, thus, contribute very little to the combined neck/tail sequences. Also, combining the neck domains of class II myosins with either the head or tail domains did not change the phylogenetic trees obtained for the head and tail domains alone.
Phylogenetic trees can be presented interchangeably as radial (Fig. 1 and see Figs. 4–6) or vertical (Fig. 2 and 3) plots. The sum of the lengths of the lines connecting any two members of a radial tree and the sum of the lengths of the horizontal lines between any two members of a vertical tree is proportional to the divergence between their amino acid sequences; the standard bar with each tree represents 10% sequence divergence. The vertical distances between members of a vertical tree, the angles between branches in a radial tree, and the positions of branches around the nodes of both radial and vertical trees are arbitrary and have no significance.
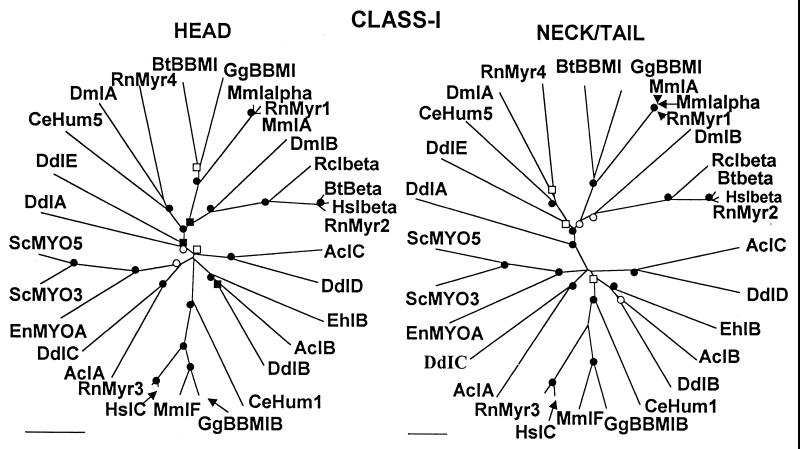
Figure 1.
Comparison of unrooted phylogenetic trees of the head domains and neck/tail domains of the heavy chains of class I myosins. The standard scale bars of percent divergence are different lengths for the two trees. ●, Nodes found in 100% of bootstrap trials; ○, nodes found in >95% of bootstrap trials; ■, nodes found >90% of bootstrap trials; □, nodes found in >85% of bootstrap trials. The GenBank accession numbers for each myosin and, in parentheses, the residue numbers used to define the head and neck/tail domains are: AcIA, AAC35357 (1–674, 675-1215); AcIB, P19706 (1–664, 665-1147); AcIC, AAC98089 (1–671, 672-1186); BtBBMI, P10568 1–681 (682–1043); BtIbeta, Z22825 (1–683, 684-1028); CeHum1, U52515 (1–614, 615-1026); CeHum5, X75564 (687, 688-1017); DdIA, P22467 (, ); DdIB, A33284 (1–678, 679-1111); DdIC, AAC37427 (1–685, 686-1181); DdID, P34109 (1–678, 679-1113); DdIE, Q03479 (1–678, 679-1003); DmIA, U05795 (1–677, 678-1011); DmIB, U07596 (1–681, 689-1026); EhIB, AAC47535 (1–700, 701-1049); EnMYOA, U12427 (706, 707-1249); GgBBMI, U04049 (1–684, 685-1045); GgBBMIB, X70400 (1–677, 678-1099); HsIbeta, X98507 (1–679, 680-1109); HsIC, U14391 (1–683, 684-1028); MmIA, S32404 (1–675, 676-1094); MmIalpha, L00923 (1–689, 690-1079); MmIF, X97650 (1–677, 678-1099); RcIbeta, U14549 (1–683, 684-1028); RnMyr1, A45439 (1–687, 688-1136); RnMyr2, X74800 (1–683, 684-1028); Rnmyr3, X74815 (1–679, 680-1107); Rnmyr4, X71997 (1–682, 683-1006); ScMYO3, P36006 (1–702, 703-1273); ScMYO5, AAB37419 (1–702, 703-1219). Ac, Acanthamoeba castellanii; Bt, Bos taurus (cow); Ce, Caenorhabditis elegans (nematode); Dd, Dictyostelium discoideum; Dm, Drosophila melanogaster; Eh, Entamoeba histolytica; En, Emiricella nidulans (Aspergillus); Gg, Gallus gallus (chicken); Hs, Homo sapiens; Mm, Mus musculus (mouse); Rc, Rana culesbelana (bullfrog); Rn, Rastus norvegicus (rat); Sc, Saccharomyces cerevisiae; BB, brush border.
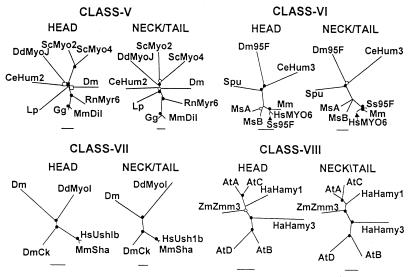
Figure 4.
Comparisons of unrooted phylogenetic trees of the head domains and neck/tail domains of the heavy chains of myosins in classes V, VI, VII, and VIII. Note that scale bars for percent divergence are somewhat different for each tree. ●, Nodes found in 100% of bootstrap trials; ○, nodes found in >95% of bootstrap trials; ■, nodes found >80% of bootstrap trials; □, nodes found in >70% of bootstrap trials. The GenBank accession numbers for each myosin and, in parentheses, the residue numbers used to define the head and neck/tail domains are: class V—CeHum2, AAA97926 (1–792, 793-1839); Dm, AAC99496 (1–759, 760-1792); DdMyoJ, P54697 (1–809, 810-2245); Gg, CAA77782 (1–752, 753-1828); Lp, AAF12809 (1–740, 741-1848); MmDil, CAA40651 (1–751, 752-1853); RnMyr6, AAB38840 (1–751, 752-1846); ScMyo2, NP014971 (1–769, 770-1574); ScMyo4, NP009373 (1–765, 766-1471); class VI—CeHum3, AAC67447 (1–762, 763-1219); Dm95F, CAA47462 (1–754, 755-1253); HsMYO6, AAC516564 (1–759, 760-1262); Mm, NP 032688 (1–762, 763-1265); MsA, AAD52005 (1–759, 7670–1304); MsB, AAD52006 (1–758, 759-1270); Spu, AAF72176 (1–757, 758-1267); Ss95F, CAA84559 (1–760, 751-1254); class VII—DdMyoI, AAF06035 (1–696, 697-2457); Dm, AAF34810 (1–725, 726-2121); DmCr; AAF44915 (1–758, 759-2167); HsUsh1b, AAB03679 (1–729, 730-2215); MmSha, AAB40708 (1–766, 767-2215); class VIII—AtA, CAB61875 (1–733, 734-1166); AtB, CAA84065 (1–656, 657-1101); AtC, AAD50052 (1–727, 728-1155); AtD, CAA19731 (1–728, 729-1126); HaHamy1, AAB71526 (1–690, 691-1120); HaHamy3, AAB93521 (, ); ZmZmm3, AAD31926 (1–665, 666-1099). Abbreviations are as in previous figures and At, Arabidopsis thaliana; Ha, Helianthus annuus (sunflower); Lp, Loigo pealei (squid); Ms, Marone saxatilis (striped bass); Spu, Strongylocentrotus purpuratus (sea urchin).
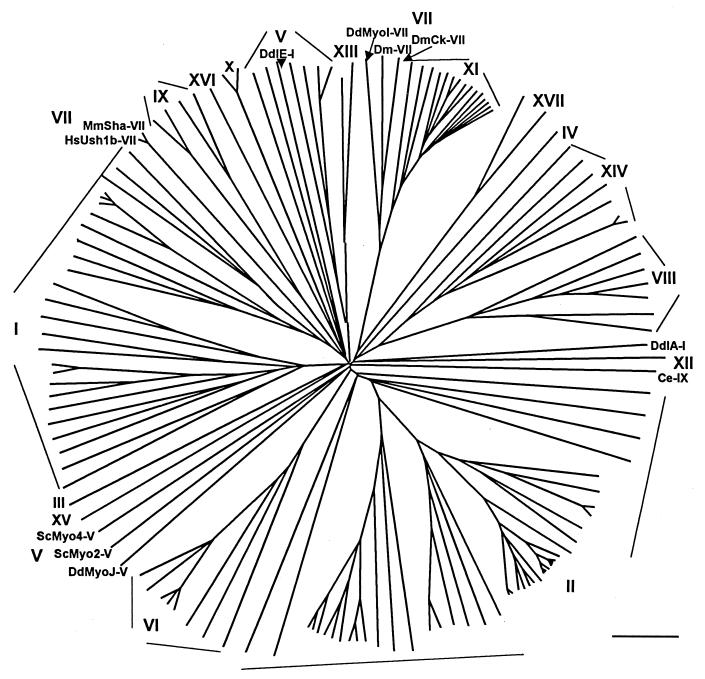
Figure 6.
Unrooted phylogenetic tree of the neck/tail domains of the heavy chains of 144 myosins in classes I–XVII. The myosins are identified by their classes as defined by previous phylogenetic trees of their head domains and/or by the authors of the original papers describing the myosin. Because of limited space, with a few exceptions, the positions of individual myosins are not identified, and the resolution is insufficient to show clearly all 47 myosin II branches. As in Fig. 1, the distances between any two myosins is proportional to the divergence between their amino acid sequences. The GenBank accession numbers for each myosin and, in parentheses, the residue numbers used to define the neck/tail domains, in addition to those given in the previous figure legends are: class III—DmNINAC, P10676 (1007–1501); class IV—AcHMWM, IV, P47808 (740–1577); class X—Bt, AAB39486 (728–2052); Mm, CAB56466 (728–2061); class XII—CeHum4, CAA91469 (925–2810); class XIII—AclMyo1, AAB53061 (); AclMyo2, AAB53062 (800–1145); class XV—Mm, AAC40124 (703–1783); class XVI—RnMyr8, AAF20150 (1133–1322); class XVII—EnCsmA, BAA21714 (756–1852); PgCsm, BAA74449 (767–1869). Abbreviations are as in previous figures and Acl, Acetabularia cliftonii (alga); Pg, Pyricularia grisea (blast fungus).
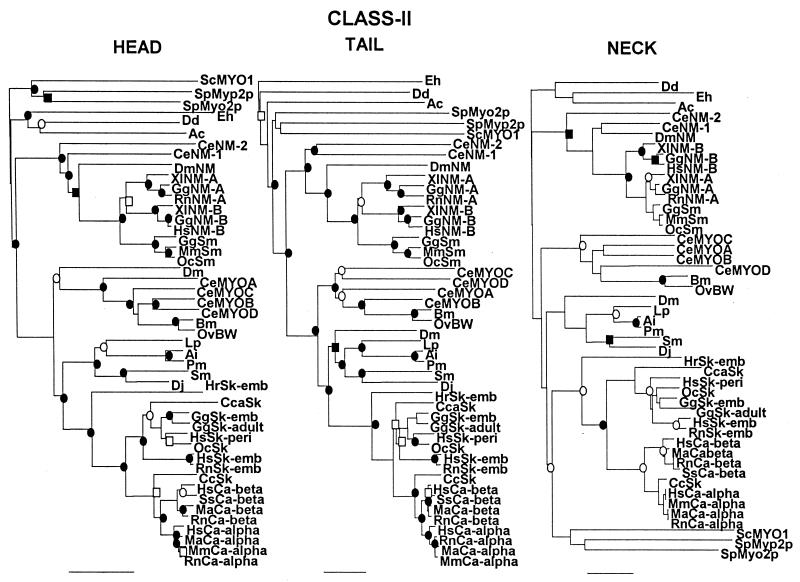
Figure 2.
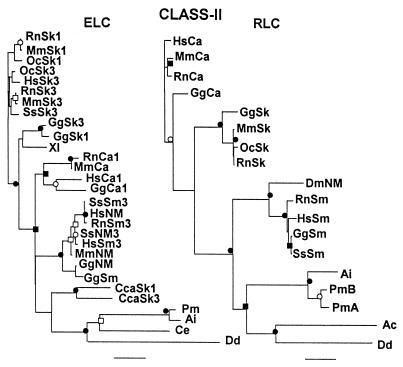
Comparison of unrooted phylogenetic trees of the head, neck, and tail domains of the heavy chains of class II myosins. These trees are not displayed in radial form (as in Fig. 1) because some of the subgroups are too tightly clustered to be readily visualized in that format. The scale bars of percent divergence are different lengths for the three trees. ●, Nodes found in 100% of bootstrap trials; ○, nodes found in >95% of bootstrap trials; ■, nodes found >90% of bootstrap trials; □, nodes found in >80% of bootstrap trials. The same results were obtained for the neck domains after 10,000 bootstrap trials and when the order of sequence entry was changed. The GenBank accession numbers for each myosin and, in parentheses, the residue numbers used to define the head and neck/tail domains are: Ac, P05659 (1–775, 776–846, 847-1509); Ai, P24733 (1–763, 764–835, 836-1938); Bm, M74000 (1–773, 774–844, 845-1957); CcSk, U53862 (1–761, 762–832, 833-1931); Cca, BAA22069 (1–767, 768–838, 839-1935); CeMYOA, P12844 (1–79, 780–850, 851-1969); CeMYOB, P02566 (1–778, 779–849, 850-1966); CeMYOC, P12845 (1–782, 783–853, 854-1947); CeMYOD, P02567 (1–773, 774–844, 845-1938); CeNM-1, U41990 (1–763, 764–834, 835-1956); CeNM-2, U49263 (1–781, 782–852, 853-2003); Dd, A26655 (1–747, 748–818, 819-2116); Dj, BAA34955 (1–557, 558–628, 629-1743); Dm, A32491 (1–1198, 1199–1259, 1260–2385); DmNM, A36014 (1–815, 816–886, 887-2017); Eh, L03534 (1–780, 781–851, 852-2139); GgNM-A, P14105 (1–764, 765–835, 836-1959); GgNM-B, M93676 (1–802, 803–873, 874-2007); GgSk-emb, P02565 (1–771, 772–842, 843-1940); GgSk-adult, P13538 (1–768, 769–839, 840-1938); GgSm, P10587 (1–777, 778–848, 849-1979); HrSk-emb, D45163 (1–766, 767–837, 838-1927); HsCa-alpha, D00943 (1–768, 769–839, 840-1939); HsCa-beta, P12883 (1–766, 767–837, 838-1935); HsNM-B, M69181 (1–771, 772–842, 843-1976); HsSk-emb, P11055 (1–767, 768–838, 839-1940); HsSk-peri, Z38133 (1–769, 770–840, 841-1937); Lp, AAC24207 (1–765, 766–836, 83701935); MaCa-alpha, L15351 (1–768, 769–839, 840-1939); MaCa-beta, L12104 (1–765, 766–836, 837-1934); MmCa-alpha, M76598 (1–768, 769, 839, 840- 1938); MmSm, D85924 (1–771, 772–842, 843-1938); OcSk, U32574 (1–769, 770–840, 841-1948); OcSm, M77812 (1–771, 772–842, 843-1972); OvBW M74066 (1–773, 774–844, 845-1957); Pm, AAB03660 (1–766, 767–837, 8938–1941); RnCa-alpha, S06005 (1–767, 768–838, 839-1938); RnCa-beta, P02564 (1–766, 767–837, 838-1935); RnNM-A, U41463 (1–764, 765–835, 836-1961); RnSk-emb, P12847 (1–767, 768–838, 835-1940); ScMYO1, NP 011888 (1–779, 780–851, 852-1928); Sm, L01634 (1–752, 753–823, 824-1940); SpMyo2p, AAC04615 (1–755, 756–826, 827-2104); SpMyp2p, U75357 (1–743, 744–814, 815-1526); SsCa-beta, U75316 (1–766, 767- 837, 838-1935); XlNM-A, AAC83556 (1–1-764, 765–835, 836-1964); XlNM-B, A47297 (1–787, 788–858, 859-1992). Abbreviations: As in Fig. 1 and Bm, Btugia malayi (nematode); Cc, Coturnix coturnix (quail); Cca, Cyprinus carpio (carp); Dj, Dugesia japonica (planaria); Hr, Halocynthia roretzi (ascidia); Ma, Mesocritus aureus (hamster); Oc. Oryctolague cuniculus (rabbit); Ov, Onchocerca volvulus (nematode); Pm, Placopecten magellanicus (mollusc); Sm, Schistosoma mansoni; Sp, Schizosaccharomyces pombe; Ss, Sus scrufo (pig); Xl, Xenopus laevis; BW, body wall; Ca, cardiac; emb, embryonic; NM, nonmuscle; peri, perinatal; Sk, skeletal muscle.
Figure 3.
Comparison of unrooted phylogenetic trees of essential and regulatory light chains of class II myosins. The scale bars of percent divergence are different lengths for the two trees. ●, Nodes found in 100% of bootstrap trials; ○, nodes found in >95% of bootstrap trials; ■, nodes found >90% of bootstrap trials; □, nodes found in >80% of bootstrap trials. The same results were obtained for the neck domains after 10,000 bootstrap trials and when the order of entry of the sequences was changed. The GenBank accession numbers for each myosin and, in parentheses, the residue numbers used to define the head and neck/tail domains are: essential light chains—Ai, P07291 (); CcaSk1, BAA12732 (); CcaSk3, BAA12733 (); Ce, P19626 (); Dd, P09402 (); GgCa1, P02606 (); GgNM, P08296 (); GgSk1, P02604 (); GgSk3, P02605 (); GgSm, P02607 (); HsCa1, P08590 (); HsNM, MOHU6N (); HsSk3, P06741 (); HsSm3, AAA59853 (); MmCa, AAA39720 (); MmNM, Q60605 (); MmSk1, P05977 (); MmSk3, P05978 (); OcSk1, P02603 (); OcSk3, P02603 (); Pm, AAB02928 (); RnCa1, P17209 (); RnSm3, Q64119 (); RnSk1, AAA98533 (); RnSk3, P02601 1–150); SsNM3, P16475 (); SsSk3, X94689 (); SsSm3, P24572 (); Xl., AAB00988 (); regulatory light chains (18)—Ac, 1810397A (); Ai, P13543 (); Dd, P13833 (); DmNM, AAA28576 (); GgCa, P02611 (); GgSk, P02609 (); GgSm, P02612 (); HsCa, P10916 (); HsSm, P24844 (); MmCa, P51667 (); MmSk, P97457 (); OcSk, P24732 (); PmA, AAB02931 (); PmB, AAB02930 (); RnCa, P08733 (); RnSk, P04466 (); RnSm, Q64122 (); SsSm, P29269 (). Abbreviations are as in previous figures.
Results and Discussion
Class I Myosins.
Thirty myosins from 13 species from amoeba to human were analyzed (Fig. 1). Except for the difference in scale (see standard bar on each figure), the phylogenetic trees of the neck/tail domains and head domains are virtually superimposable, and the two trees are equally robust with 18 of 22 nodes obtained in >95% of bootstrap trials. Within branches, the same myosins have essentially the same relationships in the two trees. For example, Btbeta, HsIbeta, and RnMyr2 are very close together, and EhIB, AcIB, and DdIB are rather distant from each other in both trees. Also, in both trees myosins from the same species are on separate, often quite distant, branches, except for ScMYO3 and ScMYO5 and MmIalpha and MmIA, which are together on both trees.
Class II Myosins.
Trees were developed for the head, neck, and tail domains of 47 myosins from 25 species (Fig. 2). The trees for the head and tail domains are very similar. In both trees, vertebrate striated muscle myosins form a subgroup with separate subgroups for cardiac alpha, cardiac beta, and skeletal muscle myosins; the two cardiac myosin subgroups are closer together than either is to vertebrate skeletal muscle myosin; invertebrate striated muscle myosins are a somewhat more distant subgroup; and vertebrate smooth muscle myosins are closer to vertebrate and invertebrate nonmuscle myosins than to striated muscle myosins. Yeast myosins and protozoal myosins form two separate groups in both trees but with an inversion of their relationship to smooth muscle and nonmuscle myosins. With a few exceptions, the relationships of individual myosins within each group and subgroup are quite similar in the two trees—the position of Drosophila muscle myosin II (Dm) is slightly different in the two trees. The tail tree is essentially as robust as the head tree; 33 nodes were found in >90% of the bootstrap trials of the tail tree compared with 36 nodes in the head tree. Bezanilla and Pollard (11) reported similar results in comparing phylogenetic trees of the combined head/neck domains (as defined here) and tail domains of 34 class II myosins.
The phylogenetic tree for the neck domains (Fig. 2) is very similar to the trees for the head and tail domains with similar groupings of cardiac and skeletal striated muscle myosins, smooth muscle myosins, and nonmuscle myosins of vertebrate and invertebrate species. Protozoal myosins also have a similar position in the three trees but yeast myosins are closer to striated muscle myosins in the neck tree rather than to the smooth muscle and nonmuscle myosins as in the head and tail trees. Fewer nodes (17 of them) were found in >90% of bootstrap trials of the neck tree, than in the other two trees, perhaps because the necks contain only 71 amino acids. Generally, the distances between head domains are less than the distances between neck domains and tail domains have the greatest sequence divergence (note the different lengths of the scale bars for the three trees). However, the relative distances between any two myosins are very similar in the three trees.
Because fewer myosins were analyzed, the trees for the 29 essential light chains and 18 regulatory light chains (Fig. 3) cannot be compared precisely either to each other or to the trees for the three heavy chain domains. However, as was found for the heavy chain domains, the essential and regulatory light chains of smooth muscle myosins group with nonmuscle myosin light chains, invertebrate and vertebrate striated myosin light chains fall into separate groups, and cardiac and skeletal muscle myosin light chains also form subgroups. Although the light chain trees are less robust than the heavy chain trees, i.e. a smaller fraction of nodes were found in >90% of the bootstrap trials, the same trees were obtained for the light chains with 10,000 trials as with 1,000 trials and the order of entry of the sequences made no difference.
Myosin Classes V, VI, VII, VIII, IX, XI, and XIV.
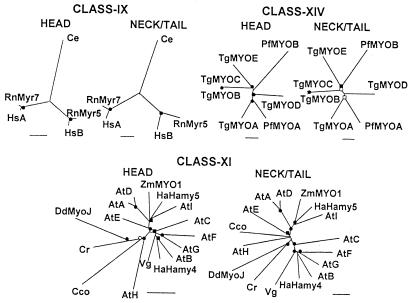
Although there may be more myosins yet to be discovered, at this time these seven classes contain only from five (class IX) to 15 (class XI) members from a limited number of species. Class VIII is limited to higher plants and class XIV to two parasitic protozoa. AtMYA1 (GenBank accession number CAA82234), AtMYA2 (GenBank accession number CAA84067), HaHamy2 (GenBank accession number AAB71527), and ZmMyo2 (GenBank accession number AAD34597) were not included in the class XI tree because the available sequences were too short at either the N-terminal or C-terminal end. Note that DdMyoJ was included in both the class V and class XI trees because in the two papers describing this myosin (12, 13) some uncertainty was expressed as to which of these two classes it belongs or if, indeed, these are separate classes.
In general, the head and neck/tail trees are equally robust within each of the seven classes, i.e., the nodes are as strongly supported in the neck/tail tree as in the head tree for each class in Figs. 4 and 5. As for the class I and class II trees, the trees for the head and neck/tail domains of these seven classes (Figs. 4 and 5) are essentially superimposable, when adjusted for the greater distances between the neck/tail domains, and the relative distances between the head domains and neck/tail domains of any two myosins are very similar. The most obvious exceptions are ScMyo2 and ScMyo4 in class V (Fig. 4), whose neck/tail domains are relatively more divergent than their head domains, and Cr, Cco, and AtH in class XI (Fig. 5), whose relative positions are somewhat different in the head domain and neck/tail domain trees. Also, the node where ScMyo2 and ScMyo4 separate is much closer to the DdMyoJ branch in the class V neck/tail tree than in the head tree (Fig. 4). DdMyoJ is more distant from its closest neighbors in class XI than in class V, especially in the neck/tail trees, consistent with the classification of DdMyoJ as a class-V myosin by Cope and Hodge (http://www.mrc-lmb.cam.ac.uk/myosin/myosin.html). Also, when single head and neck/tail domain trees were prepared containing all of the myosins in class V and class XI, the members of each class segregated at opposite ends of both trees with DdMyoJ (closer to the myosin V group) and Cr (closer to the myosin XI group) in the middle (not shown).
Figure 5.
Comparisons of unrooted phylogenetic trees of the head domains and neck/tail domains of the heavy chains of myosins in classes IX, XI, and XIV. Note that the scale bars for percent divergence are somewhat different for each tree. ●, Nodes found in 100% of bootstrap trials; ○, nodes found in >80% of bootstrap trials; ■, nodes found >70% of bootstrap trials; □, nodes found in >65% of bootstrap trials. The GenBank accession numbers for each myosin and, in parentheses, the residue numbers used to define the head and neck/tail domains are: class IX—Ce, AAC17015 (1–903, 904-1846); HsA, AAD49195 (1–1004, 1005–2548); HsB, AAC50402 (1–941, 942-2022); RnMyr5, CAA54700 (1–942, 943-1980); RnMyr7, CAA04946 (1–1015, 1016–2626); class XI—AtA, AAC14048 (1–762, 763-1583); AtB, AAC16753 (1–733, 734-1477); AtC, AAC04899 (1–734, 735-1611); AtD, AAC64896 (1–766, 767-1556); AtE, AAD32282 (1–653, 654-1490); AtF, AAD21759 (1–729, 730-1502); AtG, CAA22981 (720, 721-1446); AtH, CAB36794 (1–713, 714-1374); AtI, CAA82234 (1–717, 718-1520); Cco, BAA87057 (1–727, 728-2167); Cr, AAC27525 (1–721, 722-1643); HaHamy4 (1–719, 720-1502), AAB71528; HaHamy5, AAB71529 (1–720, 721-1528); Vg, AAF43440 (1–720, 721-1512); ZmMYO1, AAD17931 (1–719, 720-1529); class XIV—PfMYOA, AAD21242 (, ); PfMYOB, AAF25688 (, ); TgMYOA, AAC47724 (, ); TgMYOB, AAC47725 (1–605, 606-1004); TgMYOC, AAC47726 (1–605, 606-1101); TgMYOD, AAD21243 (, ); TgMYOE, AAF25495 (, ). Abbreviations are as in previous figures and Cco, Chara corallina; Cr, Chlamydomonas reinhardtii; Pf, Plasmodium falciparum; Tg, Toxoplasma gondii; Vg, Vallisneria gigantea.
Myosin Classes I–XVII.
Phylogenetic trees were produced for the head domains and neck/tail domains of 144 myosins including those analyzed in Figs. 1–5 and members of the eight classes not analyzed in the previous figures: classes III, IV, XII, XV, and XVI, comprising one myosin each (class III contains two myosins but Limulus polyphemus myosin was not included because the sequence in the database (GenBank accession number AAC16332) has a tail of only 46 residues compared with 495 for DmIII), and classes X, XIII, and XVII, each of which has two members.
Because there is much greater sequence identity among the head domains, not surprisingly, the neck/tail domain tree (Fig. 6) is less robust than the head domain tree (not shown), 80 vs. 102 nodes supported by >90 of bootstrap trials. Nonetheless, with a few exceptions, the neck/tail domains segregated into the 17 classes previously defined by analysis of the head domains. Two of the five Dictyostelium class I myosins, DdIA and DdIE, separated from the 28 other class I myosins and from each other; Ce-IX did not segregate with the four other class IX myosins; class V myosins divided into two groups, six vertebrate and invertebrate myosins and three Dictyostelium and yeast myosins; and class VII myosins separated into two groups, three Drosophila and Dictyostelium myosins and two mammalian myosins. The divisions within class V, class VII, and class XI parallel the alignments within the individual class trees (Figs. 4 and 5), but the separations of the two Dictyostelium class I myosins do not. Although not shown in detail, the alignments within each class in Fig. 6 are otherwise essentially the same as the alignments within the nine individual class trees. Interestingly, in the 144-head domain tree (not shown) DdMyoJ and DdMyoI also separated from the other class V and class VII myosins, respectively.
Concluding Comments
This analysis demonstrates that, with a few exceptions, phylogenetic analysis divides the neck/tail domains of the heavy chains of 144 myosins into the same 17 classes previously defined by comparison of their head domain sequences. The “mislocation,” when compared with previous alignments, of head domains of DdMyoJ and DdMyoI may have resulted from using total head sequences rather than the much more highly conserved core motor domains previously used, but may reflect some ambiguity in the classification of these two myosins. The additional divergences within the 144-neck/tail domain tree could indicate either a need for further refinement of the classifications or a divergence between the phylogeny of head and neck/tail domains in these few cases. These discrepancies might disappear, however, if the initial sequence alignments were corrected for gaps, multiple substitutions, and possible misalignments before plotting the trees. Thompson et al. (9) have cautioned that misalignments can occur in any automated method, especially when sequences are less than 30% identical as is commonly the case for myosin neck/tail domains in contrast to head domains.
The head domains and neck/tail domains of the individual myosins within each class, including the large myosin I and myosin II classes, segregate very similarly within their respective trees and, at least for class II myosins, light chain phylogeny parallels heavy chain phylogeny. The coalignments of head and neck/tail domains might be even better if the alignment procedure were refined, as discussed above.
The logical inference from these results, that the apparent coevolution of myosin heavy chain head and tail (and possibly neck) domains reflects a functional interdependence of the two (or three) domains, is consistent with the finding that the catalytic properties of the head domain of Dictyostelium myosin II are greatly affected when the tail domain is replaced by the tail domain of either Acanthamoeba, chicken smooth muscle, or chicken skeletal muscle myosin II (7, 8). Whether such tight coupling between head and tail domains can be generalized to all or most myosins needs to be determined. For example, exchange of head and tail domains within or between the cardiac alpha or cardiac beta heavy chain subgroups may have little or no effect because the sequences of the head, neck, and tail domains are 95–99% identical in pairwise comparisons between species within each of these subgroups and 85–90% identical in pairwise comparisons between the subgroups. This contrasts to only about 50% sequence identity between the head domains and 20–25% sequence identity between tail domains of Dictyostelium myosin II and either Acanthamoeba, chicken smooth muscle, or chicken skeletal muscle myosin II.
Acknowledgments
I thank Dr. James R. Sellers (National Heart, Lung, and Blood Institute) for many helpful discussions during the course of this research and Dr. Sellers and Dr. Robert S. Adelstein (National Heart, Lung, and Blood Institute) for suggestions for improving the manuscript.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230441597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230441597
References
- 1.Sellers J R. Myosins. Oxford, U.K.: Oxford Univ. Press; 1999. [Google Scholar]
- 2.Yamashita, R. A., Sellers, J. R. & Anderson, J. B. (2000) J. Muscle Res. Cell Motil., in press. [DOI] [PubMed]
- 3.Cheney R E, Riley M A, Mooseker M S. Cell Motil Cytoskel. 1993;24:215–223. doi: 10.1002/cm.970240402. [DOI] [PubMed] [Google Scholar]
- 4.Goodson H V, Spudich J A. Proc Natl Acad Sci USA. 1993;90:659–663. doi: 10.1073/pnas.90.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truong T, Medley Q G, Côt é. J Biol Chem. 1992;267:9767–9772. [PubMed] [Google Scholar]
- 6.Collins J H, Kuznicki J, Bowers B, Korn E D. Biochemistry. 1982;21:6910–6915. doi: 10.1021/bi00269a045. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Shu S, Yamashita R, Korn E D. Proc Natl Acad Sci USA. 2000;97:12553–12558. doi: 10.1073/pnas.230441497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito K, Liu X, Katayama E, Uyeda T Q P. Biophys J. 1999;76:985–992. doi: 10.1016/S0006-3495(99)77262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheney R E, Mooseker M S. Curr Biol. 1992;4:27–35. doi: 10.1016/0955-0674(92)90055-h. [DOI] [PubMed] [Google Scholar]
- 11.Bezanilla M, Pollard T D. Mol Biol Cell. 2000;11:79–91. doi: 10.1091/mbc.11.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson M D, Urioste A S, Titus M A. J Muscle Res Cell Motil. 1996;17:411–424. doi: 10.1007/BF00123358. [DOI] [PubMed] [Google Scholar]
- 13.Hammer J A, III, Jung G. J Biol Chem. 1996;271:7120–7127. doi: 10.1074/jbc.271.12.7120. [DOI] [PubMed] [Google Scholar]