Abstract
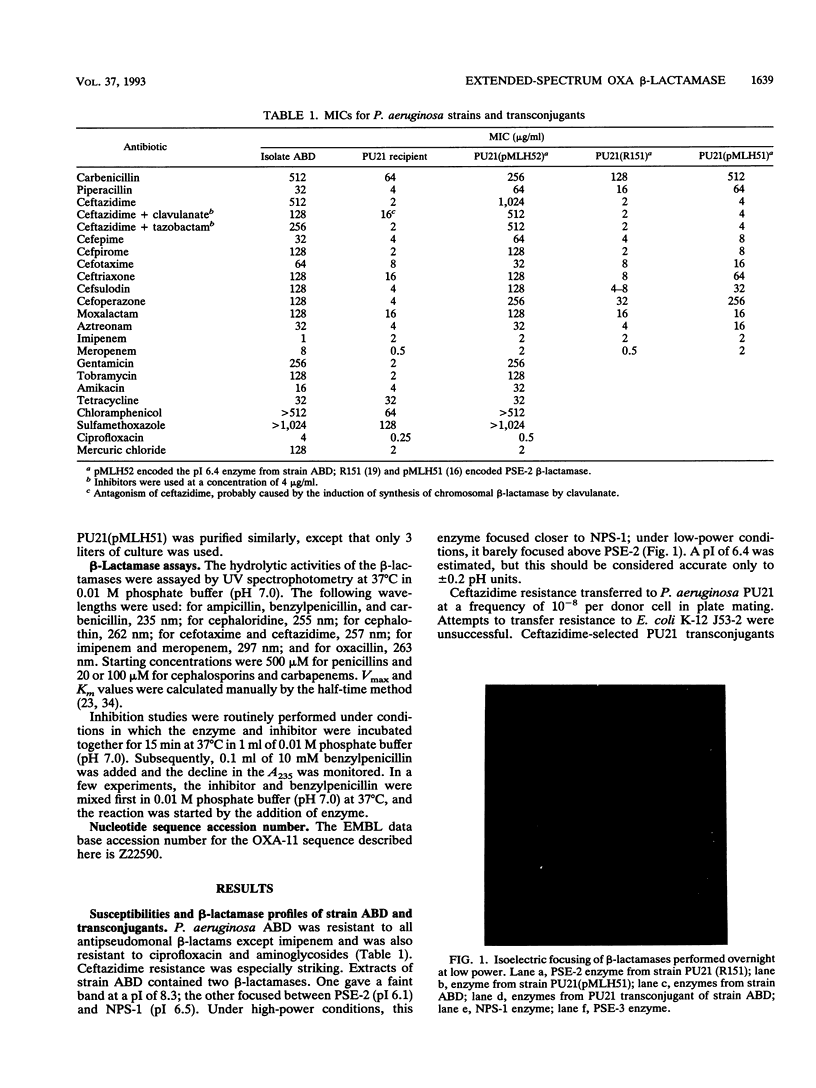
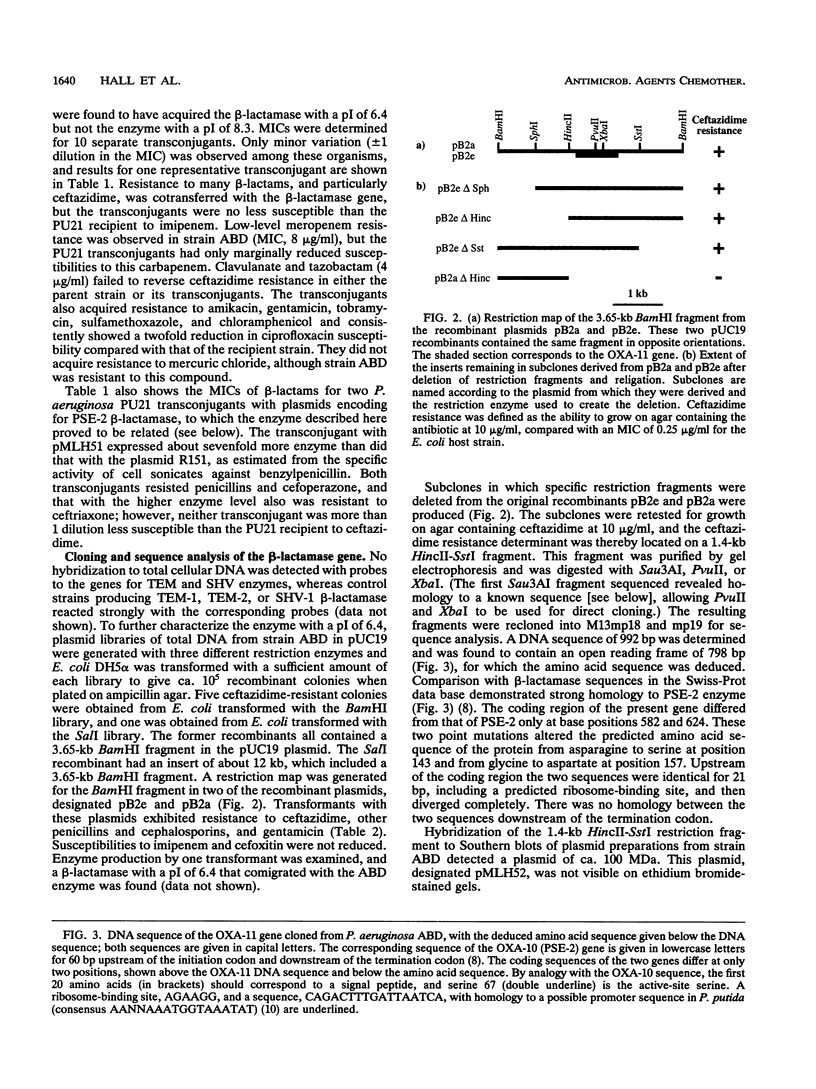
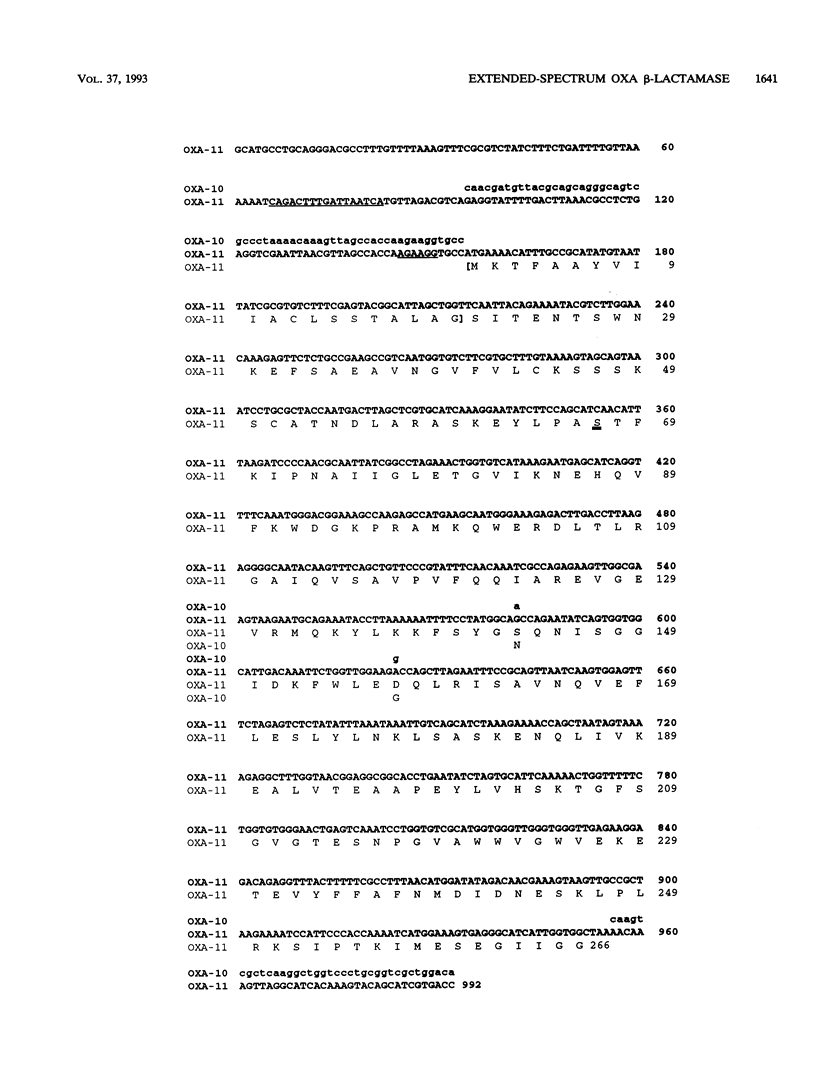
Pseudomonas aeruginosa ABD, which was isolated in October 1991 from blood cultures of a burn patient in Turkey, was resistant to cephalosporins, particularly ceftazidime (MIC, 512 micrograms/ml), penicillins, aztreonam, and meropenem, but not to imipenem. Cephalosporin and penicillin resistance transferred to P. aeruginosa PU21 and was associated with a beta-lactamase with a pI of 6.4 encoded by a 100-MDa plasmid designated pMLH52. Like extended-spectrum TEM and SHV beta-lactamases, this enzyme hydrolyzed penicillins and newer cephalosporins but did not hydrolyze cefoxitin or carbapenems. However, it differed from TEM and SHV derivatives in being a potent oxacillinase, and its encoding gene did not hybridize with probes to TEM and SHV genes. To characterize the enzyme, libraries of total DNA were cloned into plasmid pUC19 and were transformed into Escherichia coli DH5 alpha. Recombinant plasmids that gave ceftazidime resistance all contained a 3.65-kb BamHI fragment. Deletions from this fragment allowed the beta-lactamase gene to be located on a 1.4-kb section of DNA, which contained an open reading frame of 798 bases. This encoded a protein that was deduced to differ from PSE-2 beta-lactamase only in having serine instead of asparagine at position 143 and aspartate instead of glycine at position 157. It is concluded that the resistance of isolate ABD dependent on an extended-spectrum variant of the PSE-2 enzyme. The ability of this enzyme to cause ceftazidime resistance dependent primarily on a low Km for the compound; Vmax remained low. It is proposed that PSE-2 should be transferred to the OXA group as OXA-10 and that the new enzyme be designated OXA-11.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett P. M., Heritage J., Hawkey P. M. An ultra-rapid method for the study of antibiotic resistance plasmids. J Antimicrob Chemother. 1986 Sep;18(3):421–424. doi: 10.1093/jac/18.3.421. [DOI] [PubMed] [Google Scholar]
- Bernard H., Tancrede C., Livrelli V., Morand A., Barthelemy M., Labia R. A novel plasmid-mediated extended-spectrum beta-lactamase not derived from TEM- or SHV-type enzymes. J Antimicrob Chemother. 1992 May;29(5):590–592. doi: 10.1093/jac/29.5.590. [DOI] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 2c, 2d, 2e, 3, and 4. Antimicrob Agents Chemother. 1989 Mar;33(3):271–276. doi: 10.1128/aac.33.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Hall R. M., Brookes D. E., Stokes H. W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991 Aug;5(8):1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Huovinen P., Huovinen S., Jacoby G. A. Sequence of PSE-2 beta-lactamase. Antimicrob Agents Chemother. 1988 Jan;32(1):134–136. doi: 10.1128/aac.32.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huovinen P., Jacoby G. A. Sequence of the PSE-1 beta-lactamase gene. Antimicrob Agents Chemother. 1991 Nov;35(11):2428–2430. doi: 10.1128/aac.35.11.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Asai Y., Nakazawa A., Nakazawa T. Nucleotide sequence of a DNA segment promoting transcription in Pseudomonas putida. J Bacteriol. 1986 Jun;166(3):739–745. doi: 10.1128/jb.166.3.739-745.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Carreras I. Activities of beta-lactam antibiotics against Escherichia coli strains producing extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1990 May;34(5):858–862. doi: 10.1128/aac.34.5.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Medeiros A. A. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991 Sep;35(9):1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A. Properties of R plasmids determining gentamicin resistance by acetylation in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1974 Sep;6(3):239–252. doi: 10.1128/aac.6.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. Y., Gur D., Hall L. M., Livermore D. M. Survey of the prevalence of beta-lactamases amongst 1000 gram-negative bacilli isolated consecutively at the Royal London Hospital. J Antimicrob Chemother. 1992 Oct;30(4):429–447. doi: 10.1093/jac/30.4.429. [DOI] [PubMed] [Google Scholar]
- Livermore D. M. Clinical significance of beta-lactamase induction and stable derepression in gram-negative rods. Eur J Clin Microbiol. 1987 Aug;6(4):439–445. doi: 10.1007/BF02013107. [DOI] [PubMed] [Google Scholar]
- Livermore D. M. Do beta-lactamases 'trap' cephalosporins? J Antimicrob Chemother. 1985 May;15(5):511–514. doi: 10.1093/jac/15.5.511. [DOI] [PubMed] [Google Scholar]
- Livermore D. M., Maskell J. P., Williams J. D. Detection of PSE-2 beta-lactamase in enterobacteria. Antimicrob Agents Chemother. 1984 Feb;25(2):268–272. doi: 10.1128/aac.25.2.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M., Yang Y. J. Beta-lactamase lability and inducer power of newer beta-lactam antibiotics in relation to their activity against beta-lactamase-inducibility mutants of Pseudomonas aeruginosa. J Infect Dis. 1987 Apr;155(4):775–782. doi: 10.1093/infdis/155.4.775. [DOI] [PubMed] [Google Scholar]
- Mathew A., Harris A. M., Marshall M. J., Ross G. W. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J Gen Microbiol. 1975 May;88(1):169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- Nichols W. W., Hewinson R. G. Rapid and automated measurement of Km and specific Vmax values of beta-lactamases in bacterial extracts. J Antimicrob Chemother. 1987 Mar;19(3):285–295. doi: 10.1093/jac/19.3.285. [DOI] [PubMed] [Google Scholar]
- Papanicolaou G. A., Medeiros A. A., Jacoby G. A. Novel plasmid-mediated beta-lactamase (MIR-1) conferring resistance to oxyimino- and alpha-methoxy beta-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990 Nov;34(11):2200–2209. doi: 10.1128/aac.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D. J., Woodford N., Amyes S. G. Characterization of the plasmid mediated beta-lactamase BIL-1. J Antimicrob Chemother. 1992 Aug;30(2):119–127. doi: 10.1093/jac/30.2.119. [DOI] [PubMed] [Google Scholar]
- Philippon A. M., Paul G. C., Jacoby G. A. Properties of PSE-2 beta-lactamase and genetic basis for its production in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1983 Sep;24(3):362–369. doi: 10.1128/aac.24.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A., Labia R., Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989 Aug;33(8):1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi R. J., Ahroon W., Saunders S. S., Arnold S. A. Evoked potentials: computer-automated threshold-tracking procedure using an objective detection criterion. Ear Hear. 1987 Jun;8(3):151–156. [PubMed] [Google Scholar]
- Sanders C. C., Sanders W. E., Jr Clinical importance of inducible beta-lactamases in gram-negative bacteria. Eur J Clin Microbiol. 1987 Aug;6(4):435–438. doi: 10.1007/BF02013106. [DOI] [PubMed] [Google Scholar]
- Seetulsingh P. S., Hall L. M., Livermore D. M. Activity of clavulanate combinations against TEM-1 beta-lactamase-producing Escherichia coli isolates obtained in 1982 and 1989. J Antimicrob Chemother. 1991 Jun;27(6):749–759. doi: 10.1093/jac/27.6.749. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Iyobe S., Inoue M., Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991 Jan;35(1):147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton C. W., Szawelski R. J. Half-time analysis of the integrated Michaelis equation. Simulation and use of the half-time plot and its direct linear variant in the analysis of some alpha-chymotrypsin, papain- and fumarase-catalysed reactions. Biochem J. 1982 May 1;203(2):351–360. doi: 10.1042/bj2030351. [DOI] [PMC free article] [PubMed] [Google Scholar]