Abstract
The centromere–kinetochore complex of Saccharomyces cerevisiae is a specialized chromosomal substructure that mediates attachment of duplicated chromosomes to the mitotic spindle by a regulated network of protein–DNA and protein–protein interactions. We have used in vitro assays to analyze putative molecular interactions between components of the yeast centromerekinetochore complex. Glutathione S-transferase pull-down experiments showed the direct interaction of in vitro translated p110, p64, and p58 of the essential CBF3 kinetochore protein complex with Cbf1p, a basic region helix-loop-helix zipper protein (bHLHzip) that specifically binds to the CDEI region on the centromere DNA. Furthermore, recombinant p64 and p23 each stimulated the in vitro DNA binding activity of Cbf1p. The N-terminal 70 amino acids of p23 were sufficient to mediate this effect. P64 could also promote the multimerization activity of Cbf1p in the presence of centromere DNA in vitro. These results show the direct physical interaction of Cbf1p and CBF3 subunits and provide evidence that CBF3 components can promote the binding of Cbf1p to its binding site in the yeast kinetochore. A functional comparison of the centromere binding proteins with transcription factors binding at MET16 promoters reveals the strong analogy between centromeres and the MET16 promoter.
Eukaryotic centromeres/kinetochores are unique regions on each chromosome that are required for high-fidelity chromosome segregation during cell division. They serve as attachment sites for the kinetochore microtubules facilitating proper sister chromatid separation during anaphase (reviewed in refs. 1 and 2). Although the centromeres of higher eukaryotes are complex structures often spanning several megabases on the DNA (2, 3), the minimal functional centromere of Saccharomyces cerevisiae (yeast) consists only of a 125-bp DNA sequence (CEN) and is organized into three centromere DNA elements (CDEs): CDEI, CDEII, and CDEIII (4). The central element, CDEII, is a highly dAT-rich region of 78–86 bp, flanked by two palindromic sequences, CDEI (8 bp) and CDEIII (25 bp). Several centromere binding proteins have been identified in budding yeast that bind to this region in vivo and/or in vitro and are required for mitotic plasmid stability. Centromere binding factor 1 (Cbf1p) and the multiprotein-complex CBF3, consisting of the four proteins p110Cbf3a/Cbf2p/Ndc10p/Ctf14p, p64Cbf3b/Cep3p, p58Cbf3c/Ctf13p, and p23Cbf3d/Skp1p, were identified by their in vitro binding to CDEI and CDEIII, respectively (reviewed in ref. 2; see also refs. 5 and 6). Cse4p and Mif2p, the yeast counterparts to the mammalian centromere proteins CENP-A and CENP-C, respectively, have been identified by their genetic interactions with other S. cerevisiae centromere proteins and by in vivo crosslinking to centromeric DNA (7–10). One-hybrid studies have further identified the kinetochore proteins Okp1, Mcm21, and Ctf19 (11, 12), which need the CDEIII–CBF3 complex for centromere localization (11).
Cbf1p (also known as Cpf1 or CP1) is an abundant, nonessential basic helix-loop-helix leucine zipper (bHLHzip) protein from S. cerevisiae. It binds to the degenerate octanucleotide RTCACRTG (R = purine) in centromeres (where it is known as CDEI) and in several promoters (5, 13). Deletion of the Cbf1p encoding gene (CEP1) leads to a 10–30-fold increase in mitotic chromosome nondisjunction and a high rate of sister chromatid separation in meiosis (14–16). Furthermore, cep1 null mutants are methionine auxotrophs, and the role of Cbf1p in promoters of the methionine biosynthetic pathway has been studied extensively (17, 18). In MET16, a transcription activation complex is formed by Cbf1p, Met4p, and Met28p, and it has been shown that the formation of the complex on the DNA is regulated by Met28p stimulation of Cbf1p DNA binding (19, 20). Thus, it is evident that the association of other factors with Cbf1p allows the discrimination between different RTCACRTG sequences throughout the genome.
A current model predicts that the centromere DNA is bent around a specialized centromeric nucleosome that includes the histone H3-like protein Cse4 and involves the CDEII region (9–11, 21, 22). This model is supported by data showing the intrinsic DNA curvature of the CDEII region (21, 23, 24). Bending of CEN DNA can also be induced by binding of Cbf1p and CBF3, respectively, to CDEI and CDEIII (25–27). The wrapping of the DNA around a central nucleosome brings the CDEI and CDEIII regions of the centromere into close proximity with each other, making a direct physical interaction of Cbf1p and CBF3 very likely. Based on two-hybrid assays, protein–protein and protein–DNA immunoprecipitations, a network of physical interactions has been described that links the CBF3 complex to Cbf1, Mif2, and Cse4 via a complex formed by Okp1, Mcm21, and Ctf19 (11). However, no direct interaction between Cbf1p and components of the CBF3 complex could be found by two-hybrid experiments (J.L., unpublished data). Recent studies have demonstrated that in vivo Cbf1p cannot bind to the CDEI region of the centromere without a functional CBF3 complex being present (9, 28). It was proposed that the CBF3 complex might form a nucleation site on the CDEIII region, around which a fully functional centromere is assembled if CDEI and CDEII are adjacent (9).
In this study, we addressed the question whether Cbf1p can interact physically with CBF3 components. We provide evidence that CBF3 components can directly bind to Cbf1p in the absence of CEN DNA. Furthermore, we show that the p64 and p23 CBF3 subunits specifically stimulate the DNA binding activity of Cbf1p on the centromere. These results shed further light on the molecular architecture of the S. cerevisiae centromere complex and provide insight into the molecular interactions that take place between its components. The evolutionary relationship between yeast centromeres and certain methionine promoters is also discussed.
Materials and Methods
Plasmids.
Plasmids pWJ110P, pJL33, pJL36, and pOS233 containing cDNAs from CBF3 components p110, p64, p58, and p23, respectively, have been described (29, 30). Plasmid pMBO27, containing the Mif2 protein cDNA (31), was kindly provided by M. Brown (Fred Hutchinson Cancer Research Center, Seattle). Plasmids pET28-Cbf1p and pET28-Cbf1pΔN209 for bacterial expression of His6-tagged Cbf1 fusion proteins were generous gifts from D. Thomas and are described in detail in ref. 20. To obtain glutathione S-transferase (GST)-Cbf1 fusion proteins, the Cbf1p ORFs of plasmids pET28-Cbf1p and pET28-Cbf1pΔN209 were PCR-amplified with 5′ and 3′ primers containing BamHI and EcoRI restriction sites, respectively, and the resulting PCR products cloned into pGEX2T (32). For expression of a Pho4 protein derivative, a 264-bp PCR product representing the bHLH domain of Pho4 (amino acids 249–311) was generated from genomic yeast DNA using 5′ and 3′ Pho4 sequence-specific primers containing BamHI and EcoRI sites, respectively. PCR products were first inserted into the pCR-Script vector (Stratagene) to verify correctness by sequencing and further subcloned into BamHI/EcoRI-prepared expression vector pGEX 2T (32).
CEN DNAs.
The 300-bp DNA fragments carrying the CEN DNAs from individual yeast chromosomes are described in detail in ref. 25. For Pho4p band shifts, a CEN/PHO5 upstream activating sequence (UAS) hybrid DNA was generated by PCR on pBS-CEN7 (25) using 5′ primer (5′-tcaaatctcacacgtgttatat-3′) and 3′ primer (5′-cgctctagaactagtggatc-3′) resulting in a 213-bp-long DNA fragment. Fragments were diluted to a concentration of 5 nM and stored at −20°C.
Recombinant Proteins.
Full-length Cbf1p and Cbf1pΔN209 were expressed as His6-tagged fusion proteins in Escherichia coli BL21(DE3) from plasmids pET-Cbf1p and pET-Cbf1pΔN209, respectively, and purified according to protocols detailed in Kuras et al. (20). CBF3 proteins were expressed and purified as described previously (30, 31). GST-fusion proteins were expressed in E. coli BL21(DE3) and purified according to published protocols (33). Protein concentrations were determined using the micro BSA protein assay kit (Pierce).
Band Shift Assays.
Protein–DNA binding reactions (20 μl) contained protein(s) at the concentration indicated, 0.5 nM CEN DNA, 25 mM Hepes buffer at pH 7.6, 50% glycerine, and 10 mg/ml BSA (binding buffer). Reaction mixtures were kept on ice for 20 min. The reaction mixture was loaded on 4% native polyacrylamide gels pre-electrophoresed for 1 h. Gels were run in 0.5 TBE (1 × TBE contains 100 mM Tris/83 mM borate/0.1 mM EDTA at pH 8.0) at 20 mA at room temperature and stained in SYBR Gold nucleic acid stain solution (Molecular Probes) for 10 min in the dark. The fluorescence of the gel bands was visualized by UV light. Digital pictures were taken by a high resolution CCD camera and stored as tag image files (TIF) (Cybertec, Berlin). Band shift assays detecting cAMP binding protein–DNA complexes were performed as described (34).
GST Pull-Down Assays.
To obtain in vitro translation products, p110, p64, p58, p23, and Mif2p cDNAs were amplified from above plasmids by PCR using 5′ primers designed to include a T3 promoter and a Kozak consensus dATG codon (5′-gaaattaaccctcactaaagggaaccatggagatg-3′, respective sequences underlined). The PCR products were transcribed using T3 polymerase, and resulting mRNA was translated in a coupled transcription/translation reaction in the presence of [35S]methionine (Amersham Pharmacia) under conditions recommended by the supplier in 50-μl reactions (Promega). Luciferase cDNA for transcription/translation was supplied with the kit (Promega). Binding reactions (100 μl) containing 10 μl of 35S-labeled proteins, 20 μl of GST, GST-Pho4p, or GST-Cbf1pΔN209 bead slurry (containing approximately 500 ng of protein each) were adjusted to 1 × band shift buffer and incubated for 1 h at 4°C with end-over-end tube rotation. Subsequently, beads were washed four times with 500 μl of binding buffer, resuspended in gel loading buffer, and loaded on SDS gels. Gels were dried and analyzed for radioactively labeled binding proteins by autoradiography.
Results
CBF3 Components Bind to Cbf1p in Vitro.
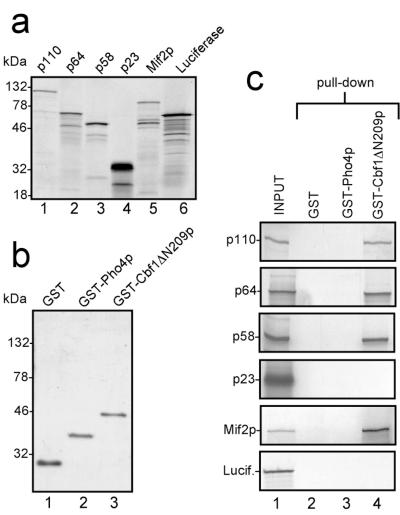
We used GST pull-down assays to identify potential molecular interactions between Cbf1p and components of the CBF3 complex. CBF3 proteins were generated as radioactively labeled in vitro translation products (Fig. 1a). Mif2p was included as a potential positive control as it had been shown previously that Cbf1p and Mif2p interact genetically and that a mif2 temperature-sensitive phenotype can be partially rescued by increased dosage of Cbf1p (8). The full-length translation products of the CBF3 components p110, p64, and p58 migrated as major bands at their expected molecular weights in SDS gels (Fig. 1a, lanes 1–3). The aberrant migration of p23 at 32 kDa (Fig. 1a, lane 4) has been noticed previously for the endogenous protein and results from its highly negative net charge (30). The translation product Mif2p (63 kDa) migrated at approximately 94 kDa (Fig. 1a, lane 5), also consistent with the SDS/PAGE migration behavior of cellular Mif2p obtained from yeast whole protein extracts (9). Luciferase was used as a negative control (Fig. 1a, lane 6). Faster migrating bands were observed in all translation reactions and were presumably attributable to premature termination of translation, initiation at cDNA-internal start codons, or nonspecific degradation in the lysate. GST and GST-fusion proteins containing the bHLH domain of Pho4p (GST-Pho4p) and the bHLHzip domain of Cbf1p (GST-Cbf1pΔN209) were expressed in E. coli and affinity-purified on GST-agarose beads (Fig. 1b). The bacterially expressed GST-Cbf1p contained only the bHLHzip domain of the protein (starting from amino acid 210). This part of the molecule has been shown to be sufficient for providing centromere function and methionine auxotrophy in cbf1 null mutant yeast strains (16). GST-Pho4p contains the bHLH domain of transcription factor Pho4 from S. cerevisiae and served as a control. Both fusion proteins were able to bind to their cognate DNA binding sites at nanomolar concentrations in band shift assays (data not shown), indicating that the bacterially expressed proteins were functionally active in vitro. In GST pull-down assays, the translation products were tested on agarose-immobilized GST, GST-Pho4p, or GST-Cbf1pΔN209 (Fig. 1c, pull-down). This assay showed that CBF3 proteins p110, p64, p58, and Mif2p were bound by GST-Cbf1pΔN209, whereas translation product p23 was not recovered from the beads (Fig. 1c, lane 4). Addition of CEN-DNA or individual CDEs showed no effect on the binding characteristics, indicating that the observed interactions are DNA-independent (data not shown). To rule out that these interactions were attributable to nonspecific binding of the translation products, we tried to bind the translation products to beads that carried the bHLH domain of transcription factor Pho4. None of the translation products bound to those beads (Fig. 1c, lane 3). The translation products also did not bind to GST beads (Fig. 1c, lane 2). Taken together, these results suggest that the CBF3 components p110, p64, and p58 as well as centromere protein Mif2p can directly and specifically interact with Cbf1p in vitro.
Figure 1.
CBF3 components interact directly with Cbf1p. (a) In vitro translated CBF3 components, Mif2p and luciferase. Rabbit reticulocyte lysates containing T3-RNA polymerase and [35S]methionine were programmed with PCR products representing the open reading frames of p110, p64, p58, p23, Mif2p, and luciferase, respectively. After incubation for 60 min, aliquots of each reaction mixture were applied to an SDS-gel, and the gel was autoradiographed. (b) Recombinant GST-fusion proteins. GST alone, GST-Pho4p, and GST-Cbf1ΔN209p were expressed in E. coli and affinity-purified. Aliquots of each protein were analyzed in a Coomassie-stained SDS-gel. (c) GST pull-down assays. Aliquots of the indicated translations were used for GST pull-down assays as described in Materials and Methods. After washing, the beads were loaded on SDS-gels, and bound proteins were identified by autoradiography. One-fifth of the translation product used in each pull-down assay was also run on the respective gels (INPUT). Numbers on the left side of the gels in a and b indicate the positions of the molecular weight standards.
CBF3 Components p64 and p23 Stimulate the DNA Binding Activity of Cbf1p.
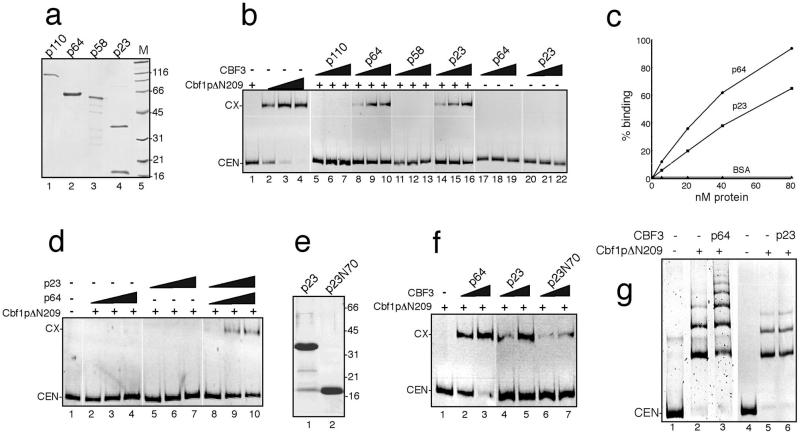
It had previously been shown that binding of Cbf1p to its binding motif in the methionine promoter can be stimulated by the transcription factor Met28 (20) and that Met28p directly interacts with Cbf1p in two-hybrid experiments. Therefore, our finding that CBF3 subunits can directly bind to Cbf1p prompted us to investigate the effect of the four CBF3 subunits on the Cbf1p–CDEI complex formation using electrophoretic mobility shift assays (EMSA). To avoid possible effects caused by the GST moiety of the GST-Cbf1pΔN209 fusion protein, a His-tagged Cbf1pΔN209 fusion protein was used for these assays (Fig. 2b, lanes 1–4). To apply purified proteins, we recombinantly expressed the CBF3 subunits in E. coli as histidine-tagged fusion proteins and purified them by metal-chelate chromatography (Fig. 2a). After establishing a Cbf1p concentration that resulted in a very low level of DNA binding activity, increasing amounts of CBF3 proteins were added to the binding reactions (Fig. 2b, lanes 5–22). Whereas p110 (lanes 5–7) and p58 (lanes 11–13) did not increase the binding of Cbf1p to the centromere, addition of p64 and p23 to the binding reaction resulted in the appearance of Cbf1p–DNA complexes at Cbf1p concentrations that previously had not resulted in any detectable shifted band (Fig. 2b, lanes 8–10 and 14–16, respectively). P64 and p23 did not alter the electrophoretic mobility of the Cbf1p–CEN complex. This can be explained by assuming that the proteins are initially components of the Cbf1p–CEN complex but dissociate during EMSA. This transient binding has been reported previously for Cbf1p and other protein–DNA complexes (ref. 20 and references therein).
Figure 2.
CBF3 components increase the DNA binding of Cbf1p. (a) Recombinantly expressed CBF3 subunits. P110 was expressed in Pichia pastoris, and p64, p58, and p23 were expressed in E. coli (see Materials and Methods for details). After metalchelate-affinity purification, aliquots of each protein sample were analyzed by SDS/PAGE. Numbers on the right indicate the molecular weights of the marker proteins run in lane 5 (M). (b) P64 and p23 exhibit DNA-binding stimulation activity. A shift complex between Cbf1p and CEN-DNA was established by incubating CEN-DNA with increasing amounts (0.5, 5, 10, and 20 nM) of recombinant, His6-tagged Cbf1pΔN209 followed by EMSA (lanes 1–4). Increasing amounts (10, 20, and 40 nM) of recombinant p110 (lanes 5–7), p64 (lanes 8–10 and 17–19), p58 (lanes 11–13), or p23 (lanes 14–16 and 20–22) were added to binding reaction mixtures, which already contained CEN-DNA and 0.5 nM Cbf1pΔN209 (lanes 5–16) or no Cbf1pΔN209 (lanes 17–22). (c) Cbf1/CEN-DNA mixtures that did not result in any detectable complex formation were incubated with increasing amounts of the indicated proteins and subjected to EMSA. Free and shifted DNA was quantitated by densitometry of shift images. The amount of shifted DNA was plotted against the protein concentration. Data points represent mean values from three independent experiments, and standard deviation was not higher than 9%. (d) The DNA-binding stimulation activities of p64 and p23 are additive. Reaction mixtures containing no (lane 1) or 0.05 nM Cbf1pΔN209 (lanes 2–10) were incubated with increasing amounts (10, 20, 40 nM) of p64 (lanes 2–4 and 8–10) or p23 (lanes 5–7 and 8–10). (e) Size-fractionation of p23. After metalchelate chromatography, the purified preparation of p23 (a, lane 4) was subjected to gel filtration chromatography on a Sephadex-G75 column. The full-length p23 (lane 1) and the truncated N-terminal peptide comprising amino acids 1–70 of p23 (p23N70, lane 2) are shown. (f) The DNA-binding stimulation activity of p23 resides in its N-terminal 70 amino acids. Reaction mixtures containing 0.5 nM of Cbf1pΔN209 were incubated with increasing amounts (40 and 80 nM) of p64 (lanes 2 and 3), column-fractionated full-length p23 (lanes 4 and 5), or p23N70 (lanes 6 and 7). CEN, free CEN3-DNA; CX, protein–DNA complex. (g) CEN-DNA was incubated without protein (lanes 1 and 4), with Cbf1p (80 nM, lanes 2 and 5), or with Cbf1p in combination with p64 (lane 3) or p23 (lane 6).
In accordance to previous findings (30), p64 and p23 were not able by themselves to bind to the CEN–DNA, excluding the possibility that the observed band shifts were attributable to CBF3–CDEIII interaction (Fig. 2b, lanes 17–22). Furthermore, heat-denatured p64 and p23 and a variety of unrelated proteins, such as GST, BSA, immunoglobulins, or a mixture of dry milk, did not increase Cbf1p binding activity (data not shown), indicating that functional and active p64 and p23 are required for the stimulation of Cbf1p binding to the CEN DNA. From a series of such experiments, we determined by densitometry of the shift complexes the concentration dependence of CBF3-mediated DNA binding stimulation (Fig. 2c). An increase of stimulation was observed between 5 and 80 nM of p64 or p23. On a molar basis, p64 seemed to be more efficient in the stimulation activity than p23 (Fig. 2c). We also tested whether p64 and p23 could act additively to stimulate Cbf1p binding. P64 and p23 were added individually or as a mixture to binding reactions containing very low amounts of Cbf1p (Fig. 2d). Whereas the addition of either p64 (lanes 2–4) or p23 (lanes 5–7) showed little or no stimulation, the presence of both proteins resulted in a DNA-binding stimulation activity of Cbf1p (lanes 8–10). This result indicates that the stimulation activity mediated by p64 and p23 was at least additive when both proteins were present in the binding reaction. When CDEII and CDEIII were deleted from the CEN–DNA, p64- and p23-mediated DNA binding stimulation was still observed, indicating that CDEII and CDEIII have no effect on this activity (data not shown).
The p23 preparation contained a discrete truncated product migrating at 18 kDa (Fig. 2a). These proteins were separated by gel chromatography (Fig. 2e), and a mass spectrometric analysis of the truncated protein revealed that this protein contained amino acids 1 to 70 of the p23 protein sequence. The protein was therefore designated as p23N70. The full-length p23 protein and the p23N70 peptide obtained after size fractionation were tested separately for their Cbf1p–DNA-binding stimulation activity (Fig. 2f). At similar concentrations, p23N70 exhibited a lower, but still significant Cbf1p–DNA-binding stimulation activity when compared with full-length p23 (Fig. 2f, compare lanes 4 and 5 with lanes 6 and 7, respectively). Thus, the DNA-binding stimulation activity of p23 can be localized to the N-terminal part of p23.
Proteins containing a bHLH domain can form higher order aggregates because of homodimerization (35, 36). Multimerization of Cbf1p has previously been observed in the presence of MET16 UAS DNA (20). When high amounts of Cbf1p were incubated with CEN–DNA, multimerization of the Cbf1p–CEN complex was observed (Fig. 2g, lane 2). The addition of p64 to such reactions resulted in the appearance of additional higher molecular weight shift complexes (Fig. 2g, lane 3). However, p64 is not a part of the higher molecular weight complexes as the multimere bands in reactions with or without p64 always comigrated (Fig. 2g and data not shown). This effect was not found with p23 (Fig. 2g, compare lanes 5 and 6). Thus, we conclude that p64 but not p23 can promote the homodimerization activity of Cbf1p.
The DNA-Binding Stimulation Activity of p64 and p23 Is Specific to Centromeres.
To assess the specificity of p64 and p23 DNA-binding stimulation activity for the Cbf1p–CDEI interaction, we tested the effects of those proteins on other DNA–protein complexes. Binding of the helix-turn-helix cAMP binding protein from E. coli to its cognate site was not influenced by p64 or p23 (data not shown). We also tested the effect of p64 and p23 on the binding of the bHLH protein Pho4 to a centromere in which CDEI was replaced for the UAS of PHO5 (ref. 37, data not shown). The CEN–PHO5UAS hybrid DNA fragment was efficiently bound by Pho4p at the same concentrations that were used in the Cbf1p–CEN binding reactions, but no increase of the DNA binding activity of Pho4p upon addition of p23 and p64 was observed (data not shown). These observations indicate that p64 and p23 neither effect the activity of a DNA binding protein carrying an helix-turn-helix–DNA binding motif nor increase the activity of the bHLH domain of Pho4p, which has strong structural homology to Cbf1p. These results suggest that the p64- and p23-mediated DNA-binding stimulation activity is specific for the Cbf1p–CDEI complex.
Discussion
CFB3 Subunits Interact with Cbf1p.
GST-pull-down assays are a valuable tool to examine whether proteins can physically interact, and they have recently been used to show interactions between p58 and other components of the CBF3 complex (27). In our study, we have used this technique to identify the direct physical interaction of the kinetochore components p110, p64, p58, and Mif2p with Cbf1p in vitro. A line of evidence from previous studies supports our findings. First, double-point mutations in CDEI and CDEIII have a synergistic effect (38). Second, in analyses of genetic interactions between centromere protein genes, synthetic lethality was observed between CEP1 and the CBF3 genes encoding p110 and p64, suggesting direct or indirect molecular interactions between Cbf1p and the CBF3 subunits in vivo (8, 39). Third, according to the current model that predicts that the 125-bp CEN DNA is wrapped around a specialized nucleosome (8, 10, 21, 22, 40), CDEI and CDEIII would be brought adjacent to each other on the surface of the nucleosome, and direct interaction between Cbf1p and one or more CBF3 subunits would thus be possible (9, 38). Our in vitro data strongly support the idea of a direct interaction between Cbf1p and the CBF3 subunits p110, p64, and p58 in vivo. However, attempts to show the direct contact between Cbf1p and CBF3 components by two-hybrid experiments have proven unsuccessful so far (J.L., unpublished data), for reasons that are still unclear.
Based on the ability of Cbf1p, Mif2p, and Cse4p to interact with CBF3 in vitro and in vivo, it has been suggested that the interaction between CDEI and CDEIII is mediated by a putative three-protein complex containing Ctf19p, Mcm21p, and Okp1p (11). Our findings suggest additional direct interactions between CBF3 subunits and Cbf1p. The binding of Mif2p to Cbf1p in vitro is in accordance with previous data (9, 11). Thus, in addition to the contact of Mif2p with CBF3, the presence of Mif2p at the centromere might be facilitated by its interaction with Cbf1p.
Because the TCACGTG core motif is found in several locations in the yeast genome, it had been proposed that Cbf1p is a general DNA binding protein that acts in vivo to modulate chromatin structure at multiple loci, including centromeres (refs. 14–16; for review, see ref. 41). How is Cbf1p binding to a particular binding site regulated in vivo? Possible mechanisms to discriminate between different TCACGTG sequences may be modification of the affinity or the specificity for a specific binding site by additional cofactors. This has been described for the MET16 promoter, where binding of Cbf1p is stimulated by Met28p (20). Our study shows that p64 and p23 can individually and in combination stimulate the binding of Cbf1p to the CDEI site, suggesting that those proteins are both needed for in vivo binding of Cbf1p to the CEN DNA. This finding clarifies the results of previous chromatin immunoprecipitation experiments, which show that in vivo Cbf1p cannot bind to the centromere when the CBF3 complex is absent (9), although it can bind to CDEI independently of other factors in vitro (13). Thus, it is possible that the interaction of p64 and p23 with Cbf1p provides the necessary increase in binding affinity to ensure stable binding of Cbf1p to CDEI in vivo.
P64 contains a Zn2Cys6 zinc finger region and is the only CBF3 component with a known DNA binding motif (29). It has previously been shown that p64 is active when in solution and that it can form homodimers (27). However, the mechanism by which p64 enhances the DNA binding of Cbf1p is not clear. One hint might be the ability of p64 to promote the multimerization of Cbf1p on the DNA. Multimerization most likely occurs in vitro as the result of a large excess of Cbf1p in the binding reaction, after all proteins have formed dimers (43). Cbf1p binds to DNA as a dimer, and the dimerization is a prerequisite for DNA binding (16). The essential region for dimerization and DNA binding in vitro and in vivo is located C-terminal to the bHLH motif and contains a leucine zipper (ZIP)-like element (44). It has been demonstrated that the DNA binding activity of certain bZIP proteins, such as ATFs or CREB, can be enhanced by the human T-lymphotrophic virus zinc finger protein Tax by promoting dimerization of bZIP domains (45). Likewise, it is possible that p64 stimulates DNA binding of Cbf1p by promoting the association of Cbf1p dimers via its zipper-like element. Interestingly, Tax not only stimulates dimerization of bZIPs but also alters the relative affinity of a bZIP protein for different DNA binding sites via recognition of the conserved basic region (46). We do not know, however, if p64, via similar mechanisms, is able to modify DNA site selection by Cbf1p.
The Cbf1p binding stimulation activity of p23 is a new finding that underlines the importance of p23 for the structural integrity of the CBF3 complex (30, 47). Whereas previous findings had suggested that p23 might not be directly associated with the CBF3 complex (42, 48), chromatin immunoprecipitation experiments showed the presence of p23 on the centromere in vivo (11). Our findings provide further evidence that p23 is an active structural part of the centromere and suggest that p23 can have an additional role, namely recruiting Cbf1p to the centromere.
Has the Yeast Centromere Evolved from a Promoter?
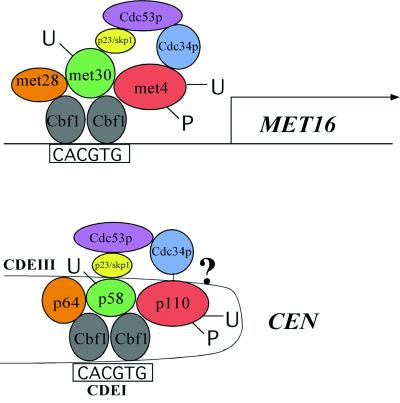
Interestingly, our findings hint toward the evolutionary similarity of budding yeast centromeres and methionine promoters. Thus, the question arises whether the yeast centromere has evolved from a promoter. All of the components involved in regulating the MET16 promoter can, by their features, be correlated to centromeric proteins (Fig. 3). The role of p64 at the centromere resembles that of Met28p in the MET16 promoter in that both proteins are able to stimulate the DNA binding affinity of Cbf1p and are unable to bind to the DNA by themselves (20, 29). It has been described earlier that the E3 ubiquitin ligase complex composed of Skp1p/Cdc53p and an F-box protein (SCF) is involved in the regulation of cell division and methionine biosynthesis in yeast (49). Ubiquitination of the target proteins is catalyzed by the E2 ubiquitin-conjugating enzyme Cdc34p, and ubiquitinated proteins are rapidly destroyed by the 28 S proteasome (49). Both, the MET promoters and the centromeres contain an F-box protein that is rapidly degraded when in solution, Met30p and p58, respectively, and for both proteins it has been speculated that ubiquitination targets the proteins for destruction (50, 51, 48). However, it is unclear how the ubiquitination takes place and whether additional proteins are involved. A novel proteolysis-independent function for Cdc34/SCF has been described recently (52). The critical target for SCFMet30 is the transcription factor Met4p (50, 52). It has been shown that ubiquitinated Met4p associates with target promoters but fails to form functional transcription complexes (52). Met4p is also regulated by phosphorylation (52). Likewise, the centromere protein p110 is regulated by phosphorylation (53, 54) and Cdc34-dependent ubiquitination (55, 56). Both, p110 and Met4p bind to Cbf1p (ref. 52 and our findings), and both are stable proteins (48, 52). It is thus tempting to speculate that p58 (the Met30p equivalent) is the F-box protein mediating the ubiquitination of p110 and that p110, in analogy to Met4p, might be regulated by ubiquitination in a reversible manner without degradation. The CSL4 gene which was found in a synthetic lethal screen with a cbf1 null mutant contains a promoter with centromere-like structure (39). However, neither deletion of the CDEI binding motif in the promoter nor deletion of the CBF1 gene has an effect on transcription of CSL4, and its regulation remains to be elucidated. Taken together, these analogies suggest that the budding yeast centromere might have evolutionary origins in primitive promoter-like structures.
Figure 3.
Model illustrating the analogy between the yeast centromere and the yeast MET16 promoter. Two different sets of proteins associate with the Cbf1p binding domains in CEN-DNA (CDEI) and in the MET16 promoter. Proteins in these complexes share analogous functions and are colored identically. Note that the spatial arrangement in the drawing reflects their functional interactions and is not meant to indicate their exact structural relationship.
Acknowledgments
We thank K. Sanyal for critically reading the manuscript, P. Kaiser and S. Reed for sharing results before publication, and J. Hegemann and S. Meyers for helpful discussions. Furthermore, we thank M. Brown for plasmid pMB027 containing the MIF2 gene, T. Steitz for recombinant cAMP binding protein, D. Thomas for the generous gift of Cbf1p expression plasmids, and H. Meyer for performing mass spectrometry analyses of p23N70.
Abbreviations
- Cbf1p
centromere binding factor 1
- CEN
centromere DNA locus
- GST
glutathione S-transferase
- CDE
centromere DNA element
- EMSA
electrophoretic mobility shift assay
- UAS
upstream activating sequence
- bHLHzip
basic region helix-loop-helix zipper protein
References
- 1.Choo K H A. In: The Centromere. Choo K H A, editor. Oxford: Oxford University Press; 1997. [Google Scholar]
- 2.Lechner J, Ortiz J. FEBS Lett. 1996;389:70–74. doi: 10.1016/0014-5793(96)00563-7. [DOI] [PubMed] [Google Scholar]
- 3.Clark L. Curr Opin Genet Dev. 1998;8:212–218. doi: 10.1016/s0959-437x(98)80143-3. [DOI] [PubMed] [Google Scholar]
- 4.Hegemann J, Fleig U N. BioEssays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- 5.Bram R J, Kornberg R D. Mol Cell Biol. 1987;7:403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lechner J, Carbon J. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 7.Stoler S, Keith K C, Curnick K E, Fitzgerald-Hayes M. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 8.Meluh P B, Koshland D. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meluh P B, Koshland D. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meluh P B, Yang P, Glowczewski L, Koshland D, Smith M M. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 11.Ortiz J, Stemmann O, Rank S, Lechner J. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyland K, Kingsbury J, Koshland D, Hieter P. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker R E, Fitzgerald-Hayes M, O'Brian T C. J Biol Chem. 1989;264:10843–10850. [PubMed] [Google Scholar]
- 14.Baker R E, Masison D C. Mol Cell Biol. 1990;10:2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai M, Davies R. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 16.Mellor J, Jiang W, Funk M, Rathjen J, Barnes C A, Hinz T, Hegemann J H, Phillipsen P. EMBO J. 1990;9:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connell K F, Baker R E. Genetics. 1992;132:63–73. doi: 10.1093/genetics/132.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas D, Cherest H, Surdin-Kerjan Y. Mol Cell Biol. 1989;12:1719–1727. doi: 10.1128/mcb.12.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D. EMBO J. 1996;15:2519–2529. [PMC free article] [PubMed] [Google Scholar]
- 20.Kuras L, Barbey R, Thomas D. EMBO J. 1997;16:2441–2451. doi: 10.1093/emboj/16.9.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng R, Ness J, Carbon J. Basic Life Sci. 1986;40:479–492. doi: 10.1007/978-1-4684-5251-8_36. [DOI] [PubMed] [Google Scholar]
- 22.Sorger P K, Severin F F, Hyman A A. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy M R, Fowlkes D M, Fitzgerald-Hayes M. Chromosoma. 1991;101:189–197. doi: 10.1007/BF00355368. [DOI] [PubMed] [Google Scholar]
- 24.Bechert T, Heck S, Fleig U, Diekmann S, Hegemann J H. Nucleic Acids Res. 1999;27:1444–1449. doi: 10.1093/nar/27.6.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niedenthal R K, Sen-Gupta M, Wilmen A, Hegemann J. Nucleic Acids Res. 1993;21:4726–4733. doi: 10.1093/nar/21.20.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pietrasanta L I, Thrower D, Hsieh W, Rao S, Stemmann O, Lechner J, Carbon J, Hansma H. Proc Natl Acad Sci USA. 1999;96:3757–3762. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russel I D, Grancell A S, Sorger P K. J Cell Biol. 1999;145:933–950. doi: 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilmen A, Pick H, Niedenthal R K, Sen-Gupta M, Hegemann J. Nucleic Acids Res. 1994;22:2791–2800. doi: 10.1093/nar/22.14.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lechner J. EMBO J. 1994;13:5203–5211. doi: 10.1002/j.1460-2075.1994.tb06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stemmann O, Lechner J. EMBO J. 1996;15:3611–3620. [PMC free article] [PubMed] [Google Scholar]
- 31.Brown M T, Goetsch L, Hartwell L H. J Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 33.Vossen K M, Stickle D F, Fried M G. J Mol Biol. 1996;255:44–54. doi: 10.1006/jmbi.1996.0005. [DOI] [PubMed] [Google Scholar]
- 34.Hershko A. J Biol Chem. 1988;263:15237–15240. [PubMed] [Google Scholar]
- 35.Ferre-D'Amare A R, Pognonec P, Roeder R G, Burley S K. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anthony-Cahill S J, Benfield P A, Fairman R, Wasserman Z R, Brenner S L, Stafford W F, III, Altenbach C, Hubbell W L, DeGrado W F. Science. 1992;255:979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niedenthal R, Stoll R, Hegemann J. Mol Cell Biol. 1991;11:3545–3553. doi: 10.1128/mcb.11.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker R E, Harris K, Zhang K. Genetics. 1998;149:73–85. doi: 10.1093/genetics/149.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saunders M J, Yeh E, Grunstein M, Bloom K. Mol Cell Biol. 1990;10:5721–5727. doi: 10.1128/mcb.10.11.5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson K A, Lopes J M. Nucleic Acids Res. 2000;28:1499–1505. doi: 10.1093/nar/28.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Espelin C W, Kaplan K B, Sorger P K. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roman C, Matera A G, Cooper C, Artandi S, Blain S, Ward D C, Calame K. Mol Cell Biol. 1992;2:817–827. doi: 10.1128/mcb.12.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dowell S J, Tsang J S H, Mellor J. Nucleic Acids Res. 1992;20:4229–4236. doi: 10.1093/nar/20.16.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner S, Green R G. Science. 1993;62:395. doi: 10.1126/science.8211160. [DOI] [PubMed] [Google Scholar]
- 46.Perini G, Wagner S, Green M R. Nature (London) 1995;376:602. doi: 10.1038/376602a0. [DOI] [PubMed] [Google Scholar]
- 47.Connelly C, Hieter P. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaplan K, Hyman A A, Sorger P K. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 49.Patton E E, Willems A R, Sa D, Kauras L, Thomas D, Craig K L, Tyers M. Genes & Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas D, Kuras L, Barbey R, Cherest H, Blaiseau P-L, Surdin-Kerjan Y. Mol Cell Biol. 1995;15:6526–6534. doi: 10.1128/mcb.15.12.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rouillon A, Barbey R, Patton E E, Tyers M, Thomas D. EMBO J. 2000;19:282–294. doi: 10.1093/emboj/19.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaiser P, Flick K, Wittenberg C, Reed S. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 53.Biggins S, Severin F F, Bhalla N, Sassoon I, Hyman A A, Murray A W. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sassoon I, Severin F F, Andrews P D, Taba M-R, Kaplan K B, Ashford A J, Stark M J R, Sorger P K, Hyman A A. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon H-J, Carbon J. Mol Cell Biol. 1995;5:4835–4842. doi: 10.1128/mcb.15.9.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kopski K M, Huffaker T C. Genetics. 1997;147:409–420. doi: 10.1093/genetics/147.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]