Abstract
Cell division is characterized by orchestrated events of chromosome segregation, distribution of cellular organelles, and the eventual partitioning and separation of the two daughter cells. Mitotic kinases, including polo-like kinases (Plk), influence multiple events in mitosis. In yeast two-hybrid screens using mammalian Plk C-terminal domain baits, we have identified Golgi peripheral protein GRASP65 (Golgi reassembly stacking protein of 65 kDa) as a Plk-binding protein. GRASP65 appears to function in the postmitotic reassembly of Golgi stacks. In this report we demonstrate binding between Plk and GRASP65 and provide in vitro and in vivo evidence that Plk is a GRASP65 kinase. Moreover, we show that Cdc2 can also phosphorylate GRASP65. In addition, we present data which support the observation that the conserved C terminus of Plk is important for its function. Deletion or frameshift mutations in the conserved C-terminal domain of Plk greatly diminish its ability to phosphorylate GRASP65. These and previous findings suggest that phosphorylation of Golgi components by mitotic kinases may regulate mechanisms of Golgi inheritance during cell division.
Cell-cycle progression in eukaryotic cells is regulated by multiple phosphorylation events (reviewed in refs. 1 and 2). Cyclin-dependent kinases play key roles in initiating events that characterize specific phases of the cell cycle (2). In particular, the G2-M transition is mediated by a complex consisting of cyclin B and Cdc2. Cdc2 activates many of the processes required for cell division, such as chromosome condensation, nuclear envelope breakdown, and spindle formation (2). In addition to Cdc2, other protein kinases, including members of the polo-like kinase (Plk) family, are also involved in regulating progression through mitosis. Plks have been highly conserved through evolution, and family members are present in fungi and higher organisms. Mitotic Plks influence multiple events during cell division as well, including modulation of Cdc2 activity, centrosome and spindle maturation and function, chromosome segregation, anaphase-promoting complex regulation, and execution of cytokinesis (reviewed in refs. 3 and 4). Data from yeast complementation assays suggest that the conserved C-terminal region (which consists of domains termed polo-boxes) of the mammalian mitotic polo-like kinase Plk is important for its localization and function (5–7). Based on these observations, we hypothesized that Plk localization, regulation, and function are mediated by protein–protein interactions through its C-terminal domain. To test this hypothesis, we conducted yeast two-hybrid screens of a mouse embryo cDNA library by using a variety of Plk baits. In screens for Plk-binding proteins, we isolated multiple clones encoding peripheral Golgi protein GRASP65 (Golgi reassembly stacking protein of 65 kDa).
Cell cycle-dependent mechanisms exist to ensure fidelity and reliability in partitioning single or low copy organelles, such as the Golgi, into daughter cells (reviewed in ref. 8). Studies of Golgi structure during cell division have yielded two models of Golgi inheritance (reviewed in ref. 9). The first suggests that the Golgi becomes fragmented at the onset of mitosis, distributes throughout the dividing cell via the microtubule network, and then reassembles into the familiar cisternal structures on exit from mitosis (10). The second model proposes that anterograde transport of actively cycling Golgi vesicles is halted during mitosis, and that the Golgi redistributes to the endoplasmic reticulum via retrograde transport mechanisms (11). At telophase, anterograde transport of vesicles resumes, and Golgi vesicles emerge from the endoplasmic reticulum and fuse to form stacks in each daughter cell. It is unclear whether these models represent two distinct mechanisms for Golgi inheritance, i.e., different end points in the process, or differences in the experimental systems. In both of these models, however, the changes in Golgi morphology at the onset of mitosis and the reestablishment of the cisternal structures as the cells exit mitosis are likely to involve protein complexes on the periphery of the Golgi membrane. Proteinaceous elements have been shown to bridge adjacent cisternae and may function in the maintenance of Golgi stacks (12, 13).
GRASP65 was identified in a biochemical screen for peripheral Golgi proteins involved in postmitotic Golgi reassembly in a cell-free system (14). It forms a stable complex with GM130, a cis-Golgi matrix protein, throughout the cell cycle (14, 15). Both GRASP65 and GM130 are phosphorylated under mitotic conditions, and Cdc2 appears to be a GM130 kinase (14, 16–18). Phosphorylation of GM130 by Cdc2 inhibits binding with p115, a protein involved in tethering incoming vesicles to the Golgi, and this inhibition is thought to be a mechanism for Golgi fragmentation (17). Indeed, depletion of Cdc2 from mitotic cytosol abrogates Golgi fragmentation in vitro, but it is not clear whether Cdc2 and GM130 are involved directly or indirectly in this process. Evidence suggests that p115, in conjunction with GM130 and another Golgi protein, giantin, may also facilitate the initial events of postmitotic reassembly of Golgi stacks, before GRASP65-mediated stacking, by tethering adjacent cisternae (19). The functional significance of GRASP65 phosphorylation and the identity of the GRASP65 kinase have yet to be determined.
In this report we demonstrate binding between Plk and GRASP65 and provide evidence that GRASP65 is a Plk substrate in vitro and in vivo. In addition, we show that Cdc2 is also a GRASP65 kinase. Finally, we demonstrate that the conserved C-terminal domain of Plk is important for GRASP65 phosphorylation. The implications of these interactions for Golgi inheritance during cell division are also discussed.
Materials and Methods
Yeast Strain, Growth Conditions, Transformation, and Two-Hybrid Assays.
Saccharomyces cerevisiae strain Y153 [MATa leu2–3 leu2–112 ura3–52 trp1–901 his3-Δ200 ade2–101 gal4Δ gal80Δ URA3∷GAL-lacZ LYS2∷GAL-HIS3] was cultured in yeast extract/peptone (1% yeast extract/2% Bacto-peptone) and 2% glucose. Yeast cells were transformed with bait and prey plasmids by the lithium acetate method (20). Selection for plasmid maintenance was carried out in synthetic medium (21) supplemented with the appropriate nutrients. Interactions between bait and prey constructs were determined by growth on Trp−, Leu−, His−, and 3-amino-1,2,4-triazole selection plates, and β-galactosidase filter assays (22).
Generation of Plasmid Constructs.
Plk323–603 was generated by PCR and then subcloned into pGBT9 two-hybrid bait vector (provided by S. Fields, University of Washington, Seattle). Plk323–450, -323–499, and -323–550 bait constructs were derived from the pGBT9Plk323–603 construct by restriction digests. pGBT9Plk1–603, -1–400, and -400–603 were generated previously by Y.-L. O. Yuan (unpublished data). The GRASP65 prey construct, encoding amino acids 7–120, in the pVP16 vector was isolated from a 9.5/10.5-day-old mouse embryo cDNA two-hybrid library (provided by S. Hollenberg, Oregon Health Sciences University, Portland) in screens for Plk-binding proteins. Full-length GRASP65 was cloned by PCR using primers based on reported sequence information (14) and rat liver cDNA as template. Mutation in the myristoylation site and the C-terminal FLAG epitope tag were introduced into the forward and reverse PCR primers, respectively. PCR products were subcloned into the pBluescript II SK(−) (Stratagene) vector and the pCI-neo (Promega) mammalian expression vector. The generation of wild-type (WT), kinase-defective (KD), C-terminal deletion (ΔC), and constitutively active (T210D) Plk constructs was described previously (5). Plk polo-box 1 frameshift (FS) mutant constructs were created by using the Sculptor in vitro mutagenesis system (Amersham Pharmacia). An oligonucleotide encoding the N-terminal end of polo-box 1 was designed to shift the reading frame by removing a base pair. A one-base addition in an oligonucleotide encoding the C-terminal end of polo-box 1 restored the original reading frame. The frameshift mutation introduced multiple missense mutations within polo-box 1 (amino acids 410–439).
Protein Expression in Vitro and in Mammalian and Insect Cell Lines.
GRASP65 expression and [35S]methionine (Amersham Pharmacia) labeling were carried out in vitro by using the TNT T7 quick coupled transcription/translation system (Promega), following the manufacturer's protocol. Cos-7 cells were cultured in DMEM supplemented with 10% FBS (HyClone) and then transfected with vector, GRASP65, and Plk mammalian expression constructs by the calcium phosphate precipitation method or GenePORTER (Gene Therapy Systems, San Diego, CA) transfection reagent. Transient expression of GRASP65 and Plk was enhanced by the addition of pAdVAntage (Promega) vector to the transfection mixtures. Hi5 insect cells cultured in Ex-Cell 401 medium (JRH Bioscience, Lenexa, KS) were infected with baculovirus encoding glutathione S-transferase (GST)-Plk, GST-cyclin B, and Cdc2, and harvested 36 h postinfection. Recombinant GST-fusion proteins were purified by incubation with glutathione-agarose beads followed by elution with soluble glutathione (30 mg/ml).
Immunoprecipitation, Kinase Assays, and Western Analysis.
Transfected cells were lysed in TBSN buffer [20 mM Tris (pH 8.0)/150 mM NaCl/0.5% Nonidet P-40/5 mM EGTA/1.5 mM EDTA/0.5 mM Na3VO4/20 mM p-nitrophenyl phosphate] supplemented with protease inhibitors, and the lysate was clarified by centrifugation at 15,000 × g for 30 min. The resultant supernatant was divided into aliquots for immunoprecipitations and diluted to 1 ml with additional TBSN buffer. Coupled transcription/translation reaction mixtures containing GRASP65-FLAG were also diluted to 1 ml with TBSN buffer before immunoprecipitation. Anti-hemagglutinin epitope (HA) antibody and protein A-Sepharose beads (Zymed) or anti-FLAG antibody (Sigma) and protein G-agarose beads (Santa Cruz Biotechnology) were added to cell lysates, and immunoprecipitations were carried out overnight at 4°C. Immunocomplexes were washed four times in TBSN buffer before kinase assays or Western analysis. Cell lysates and immunoprecipitates were resolved by SDS/PAGE and transferred to Immobilon-P membranes (Millipore) for Western analysis. Recombinant proteins were detected by anti-HA and anti-FLAG primary antibodies, anti-mouse Ig horseradish peroxidase-linked secondary antibody (Amersham Pharmacia), and enhanced chemiluminescence reagents (Amersham Pharmacia). Plk and Cdc2 kinase assays were carried out in TBMD buffer [50 mM Tris (pH 7.5)/10 mM MgCl2/5 mM DTT/2 mM EGTA/0.5 mM Na3VO4/20 mM p-nitrophenyl phosphate] supplemented with 25 μM ATP and 25–50 μCi of [γ-32P]ATP. Reaction mixtures were incubated at 30°C for 30 min, and products were resolved by SDS/PAGE and subjected to autoradiography.
Metabolic Labeling and Tryptic Mapping.
Cos-7 cells transfected with GRASP65 were arrested in mitosis by nocodazole treatment (400 ng/ml) for 16–18 h and labeled for 3 h with [32P]orthophosphate (ICN) at 1 mCi/ml in phosphate-free DMEM (GIBCO/BRL). Transfected cells were lysed, and GRASP65 was isolated by immunoprecipitation as described above. GRASP65 expressed in transcription/translation reactions was immunoprecipitated by anti-FLAG antibodies and labeled in vitro by GSTPlk and Cdc2 in kinase assay conditions. Immunoprecipitation products were resolved by SDS/PAGE and transferred to Immobilon-P membranes for autoradiography, recovery of phosphoproteins, and tryptic digestion. Eluted phosphoproteins were digested with purified sequencing grade trypsin (Promega) at 37°C. Resolution of digested peptides was performed on thin-layer cellulose plates (EM Sciences, Gibbstown, NJ) in two dimensions by electrophoresis in the first dimension [2.2% formic acid/7.8% acetic acid (pH 1.9)] and chromatography in the second dimension in an organic solvent (37.5% 1-butanol/25% pyridine/7.5% acetic acid). Phosphopeptides were visualized by autoradiography.
Results
Yeast Two-Hybrid Interactions Between GRASP65 and Plk.
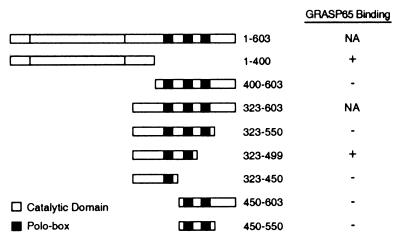
To identify proteins that interact with Plk, various two-hybrid bait constructs were generated containing the N-terminal catalytic domain or the C-terminal regions, including the three conserved polo-boxes (Fig. 1). Multiple clones encoding N-terminal fragments of GRASP65 from a 9.5/10.5-day-old mouse embryonic cDNA library were identified as Plk interactors in screens by using the Plk323–499 bait. GRASP65 was also isolated in the Plk1–400 screen as a binding protein.
Figure 1.
Yeast two-hybrid interactions between GRASP65 (amino acids 7–120) and Plk. Conserved polo-boxes (1–3, left to right) are indicated by darkly shaded boxes. Binding interaction between Plk and GRASP65 was determined by growth on selective medium and β-galactosidase activity. Data are not available for the full-length and 323–603 constructs because of background β-galactosidase activity in bait strains.
In two-hybrid interaction assays of GRASP65 (a.a. 7–120) and Plk, only Plk323–499, which lacks polo-box 3 and the extreme C terminus, and Plk1–400, which contains the catalytic domain, were able to bind GRASP65 (summarized in Fig. 1). The failure of Plk323–450 to interact with GRASP65 suggests that the binding domain is not found within the overlap between the Plk323–499 and Plk1–400 baits (a.a. 323–400), and that the interaction is likely to be mediated by chemical and structural features of both the catalytic and C-terminal domains. Furthermore, the inability of GRASP65 to bind Plk400–603 and Plk323–550 is consistent with previous observations that the C terminus of Plk includes an inhibitory domain that regulates kinase activity, and that this region may interfere with GRASP65 binding (5, 23).
Expression and Characterization of Epitope-Tagged GRASP65.
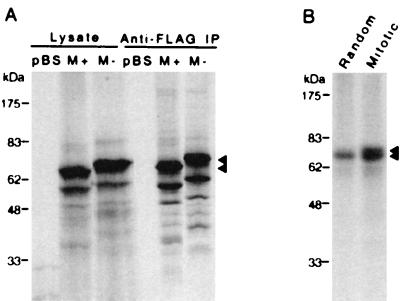
Full-length GRASP65 clones were generated from rat liver cDNA by using PCR primers based on reported sequence information (14). A FLAG epitope tag was inserted at the C terminus of each construct for Western analysis and immunoprecipitation experiments. In an in vitro expression system, GRASP65-FLAG was expressed and immunoprecipitated efficiently by anti-FLAG antibodies (Fig. 2A). Distinct populations of GRASP65 were found both in vitro (arrows in Fig. 2) and in vivo (14) and may reflect differential initiation events during translation or posttranslational modifications. Barr et al. (14) have shown that N-terminal myristoylation anchors GRASP65 to the Golgi membrane. It is not clear, however, whether the observed distinct populations of GRASP65 are the product of these modifications. Myristoylation site mutants (M−) were also generated, and the major products migrate more slowly than the WT (M+) protein. Similar expression and migration patterns were observed in mammalian cells transfected with GRASP65-FLAG constructs (Fig. 3).
Figure 2.
Expression and characterization of epitope-tagged GRASP65. (A) Full-length GRASP65 was cloned from rat liver cDNA and a C-terminal FLAG epitope tag was added. A 1-μg sample of vector (pBS), WT (M+), or myristoylation site mutant (M−) constructs was added to each in vitro expression reaction mixture. Products were labeled with Tran35S-label (ICN) for visualization and immunoprecipitated (IP) with anti-FLAG antibodies to determine the integrity of the epitope tag. GRASP65-FLAG is indicated by arrows. (B) Cos-7 cells transfected with GRASP65-FLAG were arrested in mitosis or allowed to divide randomly and labeled with [32P]orthophosphate for 3 h. Phosphoproteins were immunoprecipitated, resolved by SDS/PAGE, and visualized by autoradiography. Multiple phosphorylated bands are indicated by arrows.
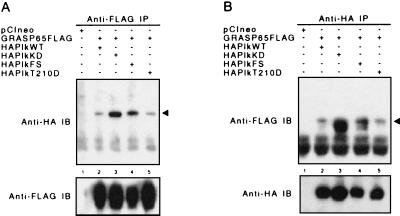
Figure 3.
GRASP65 coimmunoprecipitates with Plk. (A) Anti-FLAG immunoprecipitations (IP). HA-Plk is indicated by arrow. (B) Anti-HA immunoprecipitations. GRASP65-FLAG is indicated by arrow. Fusion proteins were detected by immunoblotting (IB) with their respective epitope antibodies. Cos-7 cells were transfected with vector (pCIneo) only (lane 1) or cotransfected with GRASP65-FLAG and HA-Plk WT (lane 2), KD (lane 3), polo-box 1 (a.a. 411–439) FS (lane 4), or T210D (lane 5) constructs at a 1:3 ratio.
GRASP65 has previously been shown to be phosphorylated in a cell cycle-dependent manner (14). To characterize the interaction between GRASP65-FLAG and endogenous proteins, Cos-7 cells were transfected with mammalian expression constructs and arrested in mitosis by nocodazole treatment. Metabolic labeling of phosphoproteins was then carried out by incubation in phosphate-free medium containing [32P]orthophosphate. Randomly growing transfected cells were also included in the in vivo labeling experiments as controls. Anti-FLAG antibodies precipitated highly phosphorylated 65-kDa proteins (at least two distinct bands) from mitotic cells (Fig. 2B). In contrast, these bands were weakly phosphorylated in a growing untreated cell population. These findings suggest that the recombinant protein is modified specifically by mitotic kinases and may function in a manner similar to native GRASP65. The availability of these constructs allowed further analysis of the Plk–GRASP65 interaction; only WT GRASP65 constructs were used in subsequent binding and phosphorylation experiments.
Interaction Between Plk and GRASP65 in Mammalian Cells.
Data from yeast two-hybrid assays indicate binding between Plk and GRASP65 fragments. To study the interaction between full-length proteins in vivo, Cos-7 cells were transfected with WT and mutant HA-Plk and GRASP65-FLAG. Fusion proteins were immunoprecipitated with their respective epitope antibodies, and the presence of binding proteins was determined by Western analysis. In cells transfected with Plk and GRASP65, the two proteins coimmunoprecipitated in experiments using either the anti-HA (Fig. 3A) or anti-FLAG (Fig. 3B) antibodies. WT and active Plk (T210D; Fig. 3 A and B, lanes 2 and 5, Upper) formed less stable complexes with GRASP65 than KD Plk (Fig. 3 A and B, lane 3, Upper). This binding profile is suggestive of an enzyme–substrate relationship between Plk and GRASP65. Similar to the KD enzyme, Plk with a FS mutation in the first polo-box also exhibited a greater affinity for GRASP65 than WT or constitutively active Plk (Fig. 3 A and B, lane 4, Upper). Comparable levels of GRASP65 (Fig. 3A, Lower) and Plk (Fig. 3B, Lower) were immunoprecipitated from cells cotransfected with GRASP65 and the various Plk constructs.
Phosphorylation of GRASP65 by Plk and Cdc2.
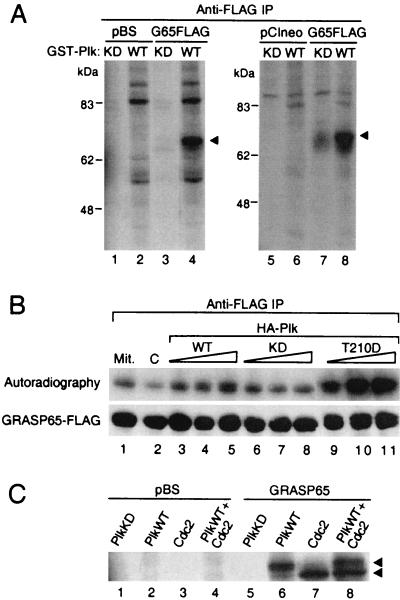
Because GRASP65 becomes phosphorylated in M phase (14), the binding data from yeast two-hybrid assays and coimmunoprecipitation experiments suggest that Plk may be a GRASP65 kinase. To determine the ability of Plk to phosphorylate GRASP65, purified GST-Plk (WT or KD) was incubated in kinase reaction buffer with GRASP65-FLAG immunoprecipitated from in vitro expression reactions or from transfected Cos-7 cells. A 65-kDa phosphoprotein was generated by WT Plk (Fig. 4A, lanes 4 and 8). Vector-only controls (Fig. 4A, lanes 2 and 6) indicate that the phosphoprotein is GRASP65. The absence or the diminished amount of phosphorylated GRASP65 in reactions with KD Plk (Fig. 4A, lanes 3 and 7) suggests that the kinase activity is attributable to Plk. The low level of phosphorylation of GRASP65 from transfected Cos-7 cells when incubated with KD Plk (Fig. 4A, lane 7) is likely due to endogenous kinases that coimmunoprecipitated with GRASP65 (see Fig. 6B, lane 2).
Figure 4.
Plk and Cdc2 are GRASP65 kinases. (A) Plk is a GRASP65 kinase. GRASP65-FLAG immunoprecipitated from in vitro expression reactions (lanes 1–4) or from transfected Cos-7 cells (lanes 5–8) was incubated with soluble GST-Plk (KD or WT) in kinase reaction buffer. A 0.5 μg sample of either vector (pBS) or GRASP65-FLAG constructs was used in each expression reaction. Equal aliquots of immunoprecipitates from transfected cells were used in each kinase reaction. (B) Expression of Plk enhances in vivo phosphorylation of GRASP65. Cos-7 cells were cotransfected with pCIneo vector (lanes 1 and 2) or HA-Plk WT (lanes 3–5), KD (lanes 6–8) or T210D (lanes 9–11), and GRASP65-FLAG. In each cotransfection experiment, 2.5, 5, or 7.5 μg of Plk constructs was used. Vector-only controls (lanes 1 and 2) were either treated with nocodazole (Mit.) or left untreated (C) to determine basal levels of GRASP65 phosphorylation. Transfected cells were incubated with [32P]orthophosphate for 3 h in phosphate-free medium, and GRASP65 was isolated by anti-FLAG immunoprecipitations (IP) and resolved by SDS/PAGE. Phosphorylated GRASP65 was detected by autoradiography (Upper), and the amount of GRASP65 in the immunocomplex was determined by Western analysis with anti-FLAG antibody (Lower). (C) GRASP65 is also phosphorylated by Cdc2. GRASP65-FLAG immunoprecipitated from in vitro expression reactions was incubated with soluble GST-Plk (KD or WT) or GST-cyclin B-Cdc2 in kinase reaction buffer.
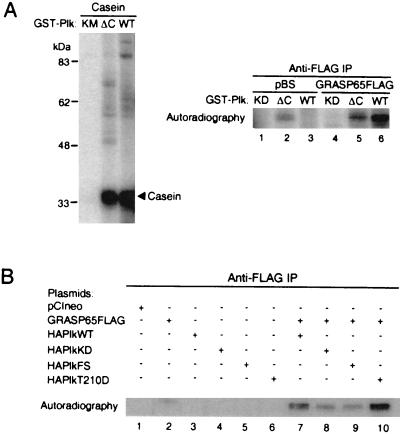
Figure 6.
The conserved C-terminal domain of Plk is important for GRASP65 phosphorylation. (A) Deletion of conserved Plk C terminus (amino acids 356–603 deleted) diminishes GRASP65 phosphorylation (lane 5). Comparable casein kinase activity was present in the WT or ΔC reactions (Left). GRASP65-FLAG expressed in vitro was immunoprecipitated and incubated in kinase reaction buffer with purified GST-PlkKD, WT, or ΔC. (B) HA-Plk polo-box 1 FS mutant coimmunoprecipitates with GRASP65-FLAG but does not efficiently phosphorylate it in an immunocomplex kinase assay (lane 9). The immunocomplexes were denatured after the kinase reaction was terminated, and GRASP65-FLAG was immunoprecipitated with anti-FLAG antibodies and subjected to autoradiography. The background kinase activity observed in KD (lane 8) and FS immunocomplexes is partly due to coimmunoprecipitating endogenous kinases (see GRASP65-FLAG-only control, lane 2).
To characterize GRASP65 phosphorylation by Plk in vivo, Cos-7 cells were cotransfected with WT, KD, or T210D Plk constructs and GRASP65, and labeled with [32P]orthophosphate in phosphate-free medium. Cells cotransfected with GRASP65 and the expression vector were either treated with nocodazole (Fig. 4B, lane 1) or left untreated (Fig. 4B, lane 2) to determine basal levels of phosphorylation in mitotic and unsynchronized cells.
Cotransfections with WT (Fig. 4B, lanes 3–5) and T210D (Fig. 4B, lanes 9–11) Plk enhanced GRASP65 phosphorylation in a dosage-dependent manner to levels equal to or greater than GRASP65 from mitotic cells. Consistent with previous observations that the T210D mutation constitutively increases kinase activity, GRASP65 phosphorylation increased severalfold in cells expressing T210D as compared with WT Plk (5). KD (Fig. 4B, lanes 6–8) Plk expression also enhanced GRASP65 phosphorylation to levels similar to mitotic GRASP65, although not in a dosage-dependent manner as seen with the WT and T210D constructs. Based on in vitro kinase assays (Figs. 4A) and previous observations by Lane and Nigg (24), it is likely that the increase in GRASP65 phosphorylation in cells expressing KD Plk is not due to direct interaction with the recombinant Plk but rather reflects defects or delays in progression through mitosis and subsequent accumulation of endogenous mitotic kinases that phosphorylate GRASP65. The failure to further enhance GRASP65 phosphorylation by increasing the amount of KD constructs in the transfections may be due to the efficacy of the minimum amount (2.5 μg) of constructs used in the experiment to sufficiently induce mitotic delay or arrest.
In previous studies, Cdc2 has been shown to be a GM130 kinase. Under the same conditions as the Plk assays, Cdc2 also phosphorylates GRASP65 (Fig. 4C, lane 7). However, Cdc2 appears to generate a more rapidly migrating phosphoprotein. Kinase reactions using both Plk and Cdc2 indicate that these kinases target distinct populations of GRASP65 (Fig. 4C, lane 8). It should be noted that two forms of GRASP65 have previously been detected in immunoprecipitation experiments (14). The origin and significance of these distinct populations of GRASP65 are not known.
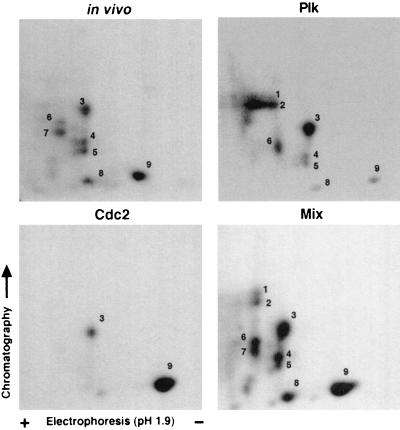
To compare GRASP65 phosphorylation by Plk and Cdc2 to in vivo phosphorylation by endogenous mitotic kinases, phosphoproteins were collected from Plk and Cdc2 kinase reactions or from metabolically labeled transfected Cos-7 cells arrested in mitosis by nocodazole, resolved by SDS/PAGE, and subjected to tryptic digestion. Recombinant GRASP65 expressed in Cos-7 cells was phosphorylated by endogenous kinases in a cell cycle-dependent manner (Figs. 2B and 3B) and multiple bands, representing distinct populations of phosphorylated GRASP65, as observed previously in this and other studies (14), were isolated from the samples labeled in vivo. Digestion products were separated in two dimensions by electrophoresis and chromatography and detected by autoradiography. Nine major phosphopeptides were present in a mixture of in vitro- and in vivo-labeled peptides (Fig. 5, Lower Right, peptides 1–9). Plk appears to specifically phosphorylate peptides 1–6, 8, and 9, whereas Cdc2 targets mainly peptide 9 and, to a lesser extent, peptide 3. Peptides 1 and 2 in Plk-phosphorylated GRASP65 have been observed in vivo, although not consistently. They may represent unstable partial digestion products or residues that are phosphorylated by Plk at a different time point in mitosis from the prometaphase arrest induced by nocodazole treatment. These findings provide evidence that Plk and Cdc2 phosphorylate GRASP65 on specific residues and on sites that are phosphorylated in vivo. The phosphopeptide (peptide 7) unaccounted for by Plk and Cdc2 suggests the existence of at least one additional GRASP65 kinase.
Figure 5.
Tryptic mapping of phosphorylated GRASP65. GRASP65-FLAG immunoprecipitated from in vitro expression reactions was labeled by purified GST-PlkWT (Upper Right), GST-cyclin B-Cdc2 (Lower Left), and radiolabeled by endogenous kinases (Upper Left) in transfected cells arrested in mitosis by nocodazole treatment and was recovered by immunoprecipitation. Phosphoproteins were eluted from the gel after SDS/PAGE and subjected to tryptic digest. Digestion products were separated in two dimensions by electrophoresis (pH 1.9) and chromatography and detected by autoradiography. Nine major phosphopeptides (labeled 1–9) were detected in a mixture (Lower Right) of in vitro and in vivo products. Plk and Cdc2 phosphorylate specific sites within GRASP65 that are relevant in vivo. The additional phosphopeptide (peptide 7) in the GRASP65 labeled in vivo suggests the existence of at least one more GRASP65 kinase other than Plk and Cdc2.
The Effects of Mutations of the C Terminus of Plk on GRASP65 Phosphorylation.
Previous studies have found that the conserved C-terminal region of Plk is important for its localization and function (5–7). Truncation of the C terminus or a point mutation in polo-box 1 of Plk abrogates its ability to complement a mutation in cdc5, the gene that encodes the polo-like kinase in yeast. To ascertain the function of the conserved Plk C terminus in GRASP65 phosphorylation, GST-PlkΔC (amino acids 358–603 deleted) and HA-PlkFS (frameshift mutation in polo-box 1) mutants were assayed for GRASP65 kinase activity.
ΔC, WT, and KD enzymes were incubated with GRASP65-FLAG immunoprecipitated from in vitro expression reactions in kinase reaction buffer. The amount of ΔC and WT Plk used in the assays had comparable levels of casein kinase activity (Fig. 6A, Left). However, the ΔC mutant exhibited reduced GRASP65 kinase activity when compared with WT Plk (Fig. 6A, Right, lanes 5 and 6). In fact, the difference between the ΔC and WT GRASP65 kinase activities is greater than what is apparent from Fig. 6A, Right, lanes 5 and 6, because some of the phosphoprotein generated in the ΔC kinase assay is attributable to autophosphorylated GST-PlkΔC (Fig. 6A, Right, lane 2), which migrates at the same molecular weight as GRASP65-FLAG. These findings indicate that the conserved C terminus of Plk is important for GRASP65 phosphorylation.
In our studies of the interaction between Plk and GRASP65, Plk with a frameshift mutation in the first polo-box exhibited a greater affinity for GRASP65 than WT or constitutively active Plk (Fig. 3). To determine the role of polo-box 1 in GRASP65 phosphorylation, Plk-GRASP65 coimmunoprecipitation experiments were carried out as described above, and the immunocomplexes were subjected to kinase assays. Western analysis of anti-FLAG immunoprecipitates indicates that the binding profile of various Plk constructs and GRASP65 is similar to that seen in Fig. 3A, with KD and frameshift mutants exhibiting greater affinity for GRASP65, and comparable levels of GRASP65 are in the immunocomplex kinase assays (data not shown). WT and T210D Plk phosphorylated GRASP65 (Fig. 6B, lanes 7 and 10), with an expected 2- to 4-fold increase in GRASP65 kinase activity in reactions with T210D (5). FS mutants, on the other hand, had similar activity to the KD construct (Fig. 6B, lanes 8 and 9), and both constructs phosphorylated GRASP65 to a lesser extent than the WT or T210D enzymes. These observations suggest that the integrity of polo-box 1 is important for productive interactions with GRASP65. The observed GRASP65 phosphorylation in reactions with KD and FS mutants is partly due to coprecipitating endogenous kinases as evidenced by the activity in the GRASP65-only control (Fig. 6B, lane 2).
Discussion
Mitotic polo-like kinases have been shown to function in multiple processes during cell division. In this report, we provide evidence that mammalian Plk may also regulate Golgi inheritance in dividing cells. Peripheral Golgi protein GRASP65 was identified as a Plk-binding protein in yeast two-hybrid screens and interacted with catalytic and conserved C-terminal domains of Plk. Coimmunoprecipitation experiments demonstrated binding between full-length proteins in vivo, and the binding profile was suggestive of an enzyme–substrate relationship. GRASP65 was phosphorylated by purified recombinant Plk and by coprecipitating Plk in anti-GRASP65 immunocomplexes. Cdc2 was also found to be a GRASP65 kinase. Tryptic mapping of GRASP65 phosphorylated by recombinant Plk, Cdc2, or endogenous mitotic kinases indicated that Plk and Cdc2 phosphorylate GRASP65 at the same sites as mitotic kinases in vivo. Moreover, mutations in the C terminus of Plk greatly diminished its ability to phosphorylate GRASP65, consistent with previous observations that the conserved C-terminal domain is important for Plk function. Finally, we have shown that ectopic expression of Plk enhances GRASP65 phosphorylation in vivo. Taken together, these findings point to GRASP65 as a target of both Plk and Cdc2 and implicate these kinases in the regulation of Golgi structure during mitosis.
Reversible protein phosphorylation appears to be an important mechanism in regulating Golgi function and structure (reviewed in ref. 25). In addition to Cdc2, mitogen-activated protein kinase kinase Mek1 and protein kinase D (PKD) have been shown to function in mitotic and drug-induced Golgi fragmentation (26, 27). Direct Mek1 and PKD substrates involved in regulating Golgi structure are presently unknown. However, in this report, we show that Plk and Cdc2 have the capacity to directly influence Golgi function and structure by phosphorylating GRASP65 G130. Whether Plk functions downstream of Cdc2, Mek1, or PKD in mediating Golgi fragmentation remains to be determined. In this regard, it should be noted that okadaic acid treatment induces Golgi fragmentation and activates both Mek1 and mitotic polo-like kinases. However, Cdc2 is not activated under these conditions (28, 29).
GRASP65 phosphorylation may contribute to events that initiate and maintain the unstacking of Golgi cisternae in mitosis, and dephosphorylation of GRASP65 on exit from mitosis enables the reassembly of cisternal structures. We have shown that Cdc2 and Plk target different populations of GRASP65 and phosphorylate distinct residues. However, the significance of these differences is not known. Additional biochemical and cell biological studies are required to elucidate the influence of Plk and Cdc2 on GRASP65 function. These findings should shed light on the relationship between Cdc2 and Plk in regulating Golgi inheritance in mitosis.
Acknowledgments
We are grateful to the late Arthur M.-E. Lee for his guidance during the early part of this project. We thank E. Erikson, J. Blenis, D. Pellman, and D. Wolf for helpful discussions and critical reading of this manuscript; K. S. Lee, Y.-L. O. Yuan, S. Fields, and S. Hollenberg for providing key reagents; and R. Hellmiss-Peralta for artistic and technical advice on the figures. C.-Y.L. is in the Biological Sciences in Public Health doctoral program. This work was supported by National Institutes of Health Grant CA42580 (R.L.E.). R.L.E. is the John F. Drum American Cancer Society Research Professor.
Abbreviations
- GRASP65
Golgi reassembly stacking protein of 65 kDa
- Plk
polo-like kinase
- GST
glutathione S-transferase
- HA
hemagglutinin epitope
- WT
wild-type
- KD
kinase-defective
- ΔC
C-terminal deletion
- FS
frameshift
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220423497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220423497
References
- 1.Nasmyth K. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- 2.Nigg E A. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 3.Nigg E A. Curr Opin Cell Biol. 1998;10:776–783. doi: 10.1016/s0955-0674(98)80121-x. [DOI] [PubMed] [Google Scholar]
- 4.Glover D M, Hagan I M, Tavares A A. Genes Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- 5.Lee K S, Erikson R L. Mol Cell Biol. 1997;17:3408–3417. doi: 10.1128/mcb.17.6.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee K S, Grenfell T Z, Yarm F R, Erikson R L. Proc Natl Acad Sci USA. 1998;95:9301–9306. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee K S, Song S, Erikson R L. Proc Natl Acad Sci USA. 1999;96:14360–14365. doi: 10.1073/pnas.96.25.14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren G, Wickner W. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- 9.Roth M G. Cell. 1999;99:559–562. doi: 10.1016/s0092-8674(00)81544-5. [DOI] [PubMed] [Google Scholar]
- 10.Lowe M, Nakamura N, Warren G. Trends Cell Biol. 1998;8:40–44. doi: 10.1016/s0962-8924(97)01189-6. [DOI] [PubMed] [Google Scholar]
- 11.Zaal K J, Smith C L, Polishchuk R S, Altan N, Cole N B, Ellenberg J, Hirschberg K, Presley J F, Roberts T H, Siggia E, et al. Cell. 1999;99:589–601. doi: 10.1016/s0092-8674(00)81548-2. [DOI] [PubMed] [Google Scholar]
- 12.Cluett E B, Brown W J. J Cell Sci. 1992;103:773–784. doi: 10.1242/jcs.103.3.773. [DOI] [PubMed] [Google Scholar]
- 13.Slusarewicz P, Nilsson T, Hui N, Watson R, Warren G. J Cell Biol. 1994;124:405–413. doi: 10.1083/jcb.124.4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barr F A, Puype M, Vandekerckhove J, Warren G. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis T E, Warren G. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura N, Lowe M, Levine T P, Rabouille C, Warren G. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- 17.Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin D J, Warren G. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- 18.Lowe M, Gonatas N K, Warren G. J Cell Biol. 2000;149:341–356. doi: 10.1083/jcb.149.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shorter J, Warren G. J Cell Biol. 1999;146:57–70. doi: 10.1083/jcb.146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman F G, Fink G R, Hicks J B. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. p. 164. [Google Scholar]
- 22.Breeden L, Nasmyth K. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 23.Mundt K E, Golsteyn R M, Lane H A, Nigg E A. Biochem Biophys Res Commun. 1997;239:377–385. doi: 10.1006/bbrc.1997.7378. [DOI] [PubMed] [Google Scholar]
- 24.Lane H A, Nigg E A. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson W J. J Cell Biol. 2000;149:243–248. doi: 10.1083/jcb.149.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acharya U, Mallabiabarrena A, Acharya J K, Malhotra V. Cell. 1998;92:183–192. doi: 10.1016/s0092-8674(00)80913-7. [DOI] [PubMed] [Google Scholar]
- 27.Jamora C, Yamanouye N, Van Lint J, Laudenslager J, Vandenheede J R, Faulkner D J, Malhotra V. Cell. 1999;98:59–68. doi: 10.1016/S0092-8674(00)80606-6. [DOI] [PubMed] [Google Scholar]
- 28.Gomez N, Cohen P. Nature (London) 1991;353:170–173. doi: 10.1038/353170a0. [DOI] [PubMed] [Google Scholar]
- 29.Qian Y W, Erikson E, Maller J L. Science. 1998;282:1701–1704. doi: 10.1126/science.282.5394.1701. [DOI] [PubMed] [Google Scholar]