Abstract
Inherited genetic variation has a critical but as yet largely uncharacterized role in human disease. Here we report a public database of common variation in the human genome: more than one million single nucleotide polymorphisms (SNPs) for which accurate and complete genotypes have been obtained in 269 DNA samples from four populations, including ten 500-kilobase regions in which essentially all information about common DNA variation has been extracted. These data document the generality of recombination hotspots, a block-like structure of linkage disequilibrium and low haplotype diversity, leading to substantial correlations of SNPs with many of their neighbours. We show how the HapMap resource can guide the design and analysis of genetic association studies, shed light on structural variation and recombination, and identify loci that may have been subject to natural selection during human evolution.
Despite the ever-accelerating pace of biomedical research, the root causes of common human diseases remain largely unknown, preventative measures are generally inadequate, and available treatments are seldom curative. Family history is one of the strongest risk factors for nearly all diseases—including cardiovascular disease, cancer, diabetes, autoimmunity, psychiatric illnesses and many others—providing the tantalizing but elusive clue that inherited genetic variation has an important role in the pathogenesis of disease. Identifying the causal genes and variants would represent an important step in the path towards improved prevention, diagnosis and treatment of disease.
More than a thousand genes for rare, highly heritable ‘mendelian’ disorders have been identified, in which variation in a single gene is both necessary and sufficient to cause disease. Common disorders, in contrast, have proven much more challenging to study, as they are thought to be due to the combined effect of many different susceptibility DNA variants interacting with environmental factors.
Studies of common diseases have fallen into two broad categories: family-based linkage studies across the entire genome, and population-based association studies of individual candidate genes. Although there have been notable successes, progress has been slow due to the inherent limitations of the methods; linkage analysis has low power except when a single locus explains a substantial fraction of disease, and association studies of one or a few candidate genes examine only a small fraction of the ‘universe’ of sequence variation in each patient.
A comprehensive search for genetic influences on disease would involve examining all genetic differences in a large number of affected individuals and controls. It may eventually become possible to accomplish this by complete genome resequencing. In the meantime, it is increasingly practical to systematically test common genetic variants for their role in disease; such variants explain much of the genetic diversity in our species, a consequence of the historically small size and shared ancestry of the human population.
Recent experience bears out the hypothesis that common variants have an important role in disease, with a partial list of validated examples including HLA (autoimmunity and infection)1, APOE4 (Alzheimer’s disease, lipids)2, Factor VLeiden (deep vein thrombosis)3, PPARG (encoding PPARγ; type 2 diabetes)4,5, KCNJ11 (type 2 diabetes)6, PTPN22 (rheumatoid arthritis and type 1 diabetes)7,8, insulin (type 1 diabetes)9, CTLA4 (autoimmune thyroid disease, type 1 diabetes)10, NOD2 (inflammatory bowel disease)11,12, complement factor H (age-related macular degeneration)13–15 and RET (Hirsch-sprung disease)16,17, among many others.
Systematic studies of common genetic variants are facilitated by the fact that individuals who carry a particular SNP allele at one site often predictably carry specific alleles at other nearby variant sites. This correlation is known as linkage disequilibrium (LD); a particular combination of alleles along a chromosome is termed a haplotype.
LD exists because of the shared ancestry of contemporary chromosomes. When a new causal variant arises through mutation—whether a single nucleotide change, insertion/deletion, or structural alteration—it is initially tethered to a unique chromosome on which it occurred, marked by a distinct combination of genetic variants. Recombination and mutation subsequently act to erode this association, but do so slowly (each occurring at an average rate of about 10−8 per base pair (bp) per generation) as compared to the number of generations (typically 104 to 105) since the mutational event.
The correlations between causal mutations and the haplotypes on which they arose have long served as a tool for human genetic research: first finding association to a haplotype, and then subsequently identifying the causal mutation(s) that it carries. This was pioneered in studies of the HLA region, extended to identify causal genes for mendelian diseases (for example, cystic fibrosis18 and diastrophic dysplasia19), and most recently for complex disorders such as age-related macular degeneration13–15.
Early information documented the existence of LD in the human genome20,21; however, these studies were limited (for technical reasons) to a small number of regions with incomplete data, and general patterns were challenging to discern. With the sequencing of the human genome and development of high-throughput genomic methods, it became clear that the human genome generally displays more LD22 than under simple population genetic models23, and that LD is more varied across regions, and more segmentally structured24–30, than had previously been supposed. These observations indicated that LD-based methods would generally have great value (because nearby SNPs were typically correlated with many of their neighbours), and also that LD relationships would need to be empirically determined across the genome by studying polymorphisms at high density in population samples.
The International HapMap Project was launched in October 2002 to create a public, genome-wide database of common human sequence variation, providing information needed as a guide to genetic studies of clinical phenotypes31. The project had become practical by the confluence of the following: (1) the availability of the human genome sequence; (2) databases of common SNPs (subsequently enriched by this project) from which genotyping assays could be designed; (3) insights into human LD; (4) development of inexpensive, accurate technologies for high-throughput SNP genotyping; (5) web-based tools for storing and sharing data; and (6) frameworks to address associated ethical and cultural issues32. The project follows the data release principles of an international community resource project (http://www.wellcome.ac.uk/doc_WTD003208.html), sharing information rapidly and without restriction on its use.
The HapMap data were generated with the primary aim of guiding the design and analysis of medical genetic studies. In addition, the advent of genome-wide variation resources such as the HapMap opens a new era in population genetics, offering an unprecedented opportunity to investigate the evolutionary forces that have shaped variation in natural populations.
The Phase I HapMap
Phase I of the HapMap Project set as a goal genotyping at least one common SNP every 5 kilobases (kb) across the genome in each of 269 DNA samples. For the sake of practicality, and motivated by the allele frequency distribution of variants in the human genome, a minor allele frequency (MAF) of 0.05 or greater was targeted for study. (For simplicity, in this paper we will use the term ‘common’ to mean a SNP with MAF ≥ 0.05.) The project has a Phase II, which is attempting genotyping of an additional 4.6 million SNPs in each of the HapMap samples.
To compare the genome-wide resource to a more complete database of common variation—one in which all common SNPs and many rarer ones have been discovered and tested—a representative collection of ten regions, each 500 kb in length, was selected from the ENCODE (Encyclopedia of DNA Elements) Project33. Each 500-kb region was sequenced in 48 individuals, and all SNPs in these regions (discovered or in dbSNP) were genotyped in the complete set of 269 DNA samples.
The specific samples examined are: (1) 90 individuals (30 parent–offspring trios) from the Yoruba in Ibadan, Nigeria (abbreviation YRI); (2) 90 individuals (30 trios) in Utah, USA, from the Centre d’Etude du Polymorphisme Humain collection (abbreviation CEU); (3) 45 Han Chinese in Beijing, China (abbreviation CHB); (4) 44 Japanese in Tokyo, Japan (abbreviation JPT).
Because none of the samples was collected to be representative of a larger population such as ‘Yoruba’, ‘Northern and Western European’, ‘Han Chinese’, or ‘Japanese’ (let alone of all populations from ‘Africa’, ‘Europe’, or ‘Asia’), we recommend using a specific local identifier (for example, ‘Yoruba in Ibadan, Nigeria’) to describe the samples initially. Because the CHB and JPT allele frequencies are generally very similar, some analyses below combine these data sets. When doing so, we refer to three ‘analysis panels’ (YRI, CEU, CHB+JPT) to avoid confusing this analytical approach with the concept of a ‘population’.
Important details about the design of the HapMap Project are presented in the Methods, including: (1) organization of the project; (2) selection of DNA samples for study; (3) increasing the number and annotation of SNPs in the public SNP map (dbSNP) from 2.6 million to 9.2 million (Fig. 1); (4) targeted sequencing of the ten ENCODE regions, including evaluations of false-positive and false-negative rates; (5) genotyping for the genome-wide map; (6) intense efforts that monitored and established the high quality of the data; and (7) data coordination and distribution through the project Data Coordination Center (DCC) (http://www.hapmap.org).
Figure 1. Number of SNPs in dbSNP over time.
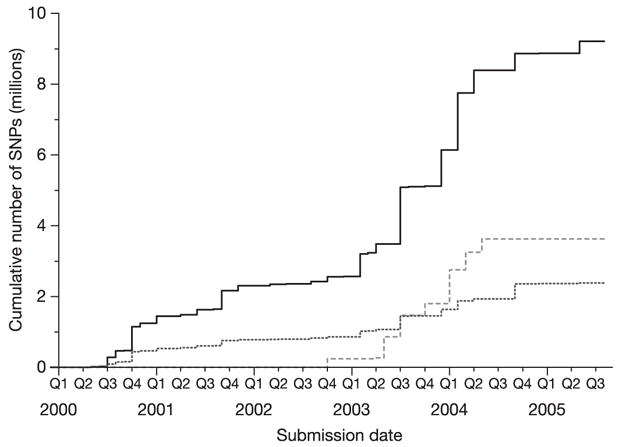
The cumulative number of non-redundant SNPs (each mapped to a single location in the genome) is shown as a solid line, as well as the number of SNPs validated by genotyping (dotted line) and double-hit status (dashed line). Years are divided into quarters (Q1–Q4).
Description of the data
The Phase I HapMap contains 1,007,329 SNPs that passed a set of quality control (QC) filters (see Methods) in each of the three analysis panels, and are polymorphic across the 269 samples. SNP genotyping was distributed across centres by chromosomal region, with several technologies employed (Table 1). Each centre followed the same standard rules for SNP selection, quality control and data release; all SNPs were genotyped in the full set of 269 samples. Some centres genotyped more SNPs than required by the rules.
Table 1.
Genotyping centres
| Centre | Chromosomes | Technology |
|---|---|---|
| RIKEN | 5, 11, 14, 15, 16, 17, 19 | Third Wave Invader |
| Wellcome Trust Sanger Institute | 1, 6, 10, 13, 20 | Illumina BeadArray |
| McGill University and Génome Québec Innovation Centre | 2, 4p | Illumina BeadArray |
| Chinese HapMap Consortium* | 3, 8p, 21 | Sequenom MassExtend, Illumina BeadArray |
| Illumina | 8q, 9, 18q, 22, X | Illumina BeadArray |
| Broad Institute of Harvard and MIT | 4q, 7q, 18p, Y, mtDNA | Sequenom MassExtend, Illumina BeadArray |
| Baylor College of Medicine with ParAllele BioScience | 12 | ParAllele MIP |
| University of California, San Francisco, with Washington University in St Louis | 7p | PerkinElmer AcycloPrime-FP |
| Perlegen Sciences | 5 Mb (ENCODE) on 2, 4, 7, 8, 9, 12, 18 in CEU | High-density oligonucleotide array |
The Chinese HapMap Consortium consists of the Beijing Genomics Institute, the Chinese National Human Genome Center at Beijing, the University of Hong Kong, the Hong Kong University of Science and Technology, the Chinese University of Hong Kong, and the Chinese National Human Genome Center at Shanghai.
Extensive, blinded quality assessment (QA) exercises documented that these data are highly accurate (99.7%) and complete (99.3%, see also Supplementary Table 1). All genotyping centres produced high-quality data (accuracy more than 99% in the blind QA exercises, Supplementary Tables 2 and 3), and missing data were not biased against heterozygotes. The Supplementary Information contains the full details of these efforts.
Although SNP selection was generally agnostic to functional annotation, 11,500 non-synonymous cSNPs (SNPs in coding regions of genes where the different SNP alleles code for different amino acids in the protein) were successfully typed in Phase I. (An effort was made to prioritize cSNPs in Phase I in choosing SNPs for each 5-kb region; all known non-synonymous cSNPs were attempted as part of Phase II.)
Across the ten ENCODE regions (Table 2), the density of SNPs was approximately tenfold higher as compared to the genome-wide map: 17,944 SNPs across the 5 megabases (Mb) (one per 279 bp).
Table 2.
ENCODE project regions and genotyping
| Available SNPs
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region name |
Chromosome band |
Genomic interval (NCBI) (base numbers)† |
Gene density (%)‡ |
Conservation score (%)§ |
Pedigree-based recombination rate (cM Mb−1)|| |
Population-based recombination rate (cM Mb−1)¶ |
G+C content# |
dbSNP⋆ | Sequence** | Total | Successfully genotyped SNPs†† |
Sequencing centre/genotyping centre(s)‡‡ |
| ENr112 | 2p16.3 | 51,633,239–52,133,238 | 0 | 3.8 | 0.8 | 0.9 | 0.35 | 1,570 | 1,762 | 3,332 | 2,275 | Broad/McGill- GQIC |
| ENr131 | 2q37.1 | 234,778,639–235,278,638 | 4.6 | 1.3 | 2.2 | 2.5 | 0.43 | 1,736 | 1,259 | 2,995 | 1,910 | Broad/McGill- GQIC |
| ENr113 | 4q26 | 118,705,475–119,205,474 | 0 | 3.9 | 0.6 | 0.9 | 0.35 | 1,444 | 2,053 | 3,497 | 2,201 | Broad/Broad |
| ENm010 | 7p15.2 | 26,699,793–27,199,792 | 5.0 | 22.0 | 0.9 | 0.9 | 0.44 | 1,220 | 1,795 | 3,015 | 1,271 | Baylor/UCSF-WU, Broad |
| ENm013* | 7q21.13 | 89,395,718–89,895,717 | 5.5 | 4.4 | 0.4 | 0.5 | 0.38 | 1,394 | 1,917 | 3,311 | 1,807 | Broad/Broad |
| ENm014* | 7q31.33 | 126,135,436–126,632,577 | 2.9 | 11.2 | 0.4 | 0.9 | 0.39 | 1,320 | 1,664 | 2,984 | 1,966 | Broad/Broad |
| ENr321 | 8q24.11 | 118,769,628–119,269,627 | 3.2 | 11.4 | 0.6 | 1.1 | 0.41 | 1,430 | 1,508 | 2,938 | 1,758 | Baylor/Illumina |
| ENr232 | 9q34.11 | 127,061,347–127,561,346 | 5.9 | 8.3 | 2.7 | 2.6 | 0.52 | 1,444 | 1,523 | 2,967 | 1,324 | Baylor/Illumina |
| ENr123 | 12q12 | 38,626,477–39,126,476 | 3.1 | 1.7 | 0.3 | 0.8 | 0.36 | 1,877 | 1,379 | 3,256 | 1,792 | Baylor/Baylor |
| ENr213 | 18q12.1 | 23,717,221–24,217,220 | 0.9 | 7.4 | 1.2 | 0.9 | 0.37 | 1,330 | 1,459 | 2,789 | 1,640 | Baylor/Illumina |
| Total | – | – | – | – | – | – | – | 14,765 | 16,319 | 31,084 | 17,944 | – |
McGill-GQIC, McGill University and Génome Québec Innovation Centre.
These regions were truncated to 500 kb for resequencing.
Sequence build 34 coordinates.
Gene density is defined as the percentage of bases covered either by Ensembl genes or human mRNA best BLAT alignments in the UCSC Genome Browser database.
Non-exonic conservation with mouse sequence was measured by taking 125 base non-overlapping sub-windows inside the 500,000 base windows. Sub-windows with less than 75% of their bases in a mouse alignment were discarded. Of the remaining sub-windows, those with at least 80% base identity were used to calculate the conservation score. The mouse alignments in regions corresponding to the following were discarded: Ensembl genes, all GenBank mRNA Blastz alignments, FGenesh++ gene predictions, Twinscan gene predictions, spliced EST alignments, and repeats.
The pedigree-based sex-averaged recombination map is from deCODE Genetics48.
Recombination rate based on estimates from LDhat46.
G + C content calculated from the sequence of the stated coordinates from sequence build 34.
SNPs in dbSNP build 121 at the time the ENCODE resequencing began and SNPs added to dbSNP in builds 122–125 independent of the resequencing.
New SNPs discovered through the resequencing reported here (not found by other means in builds 122–125).
SNPs successfully genotyped in all analysis panels (YRI, CEU, CHB + JPT).
Perlegen genotyped a subset of SNPs in the CEU samples.
More than 1.3 million SNP genotyping assays were attempted (Table 3) to generate the Phase I data on more than 1 million SNPs. The 0.3 million SNPs not part of the Phase I data set include 73,652 that passed QC filters but were monomorphic in all 269 samples. The remaining SNPs failed the QC filters in one or more analysis panels mostly because of inadequate completeness, non-mendelian inheritance, deviations from Hardy–Weinberg equilibrium, discrepant genotypes among duplicates, and data transmission discrepancies.
Table 3.
HapMap Phase I genotyping success measures
| Analysis panel
|
|||
|---|---|---|---|
| SNP categories | YRI | CEU | CHB + JPT |
| Assays submitted | 1,273,716 | 1,302,849 | 1,273,703 |
| Passed QC filters | 1,123,296 (88%) | 1,157,650 (89%) | 1,134,726 (89%) |
|
| |||
| Did not pass QC filters* | 150,420 (12%) | 145,199 (11%) | 138,977 (11%) |
| > 20% missing data | 98,116 (65%) | 107,626 (74%) | 93,710 (67%) |
| > 1 duplicate inconsistent | 7,575 (5%) | 6,254 (4%) | 10,725 (8%) |
| > 1 mendelian error | 22,815 (15%) | 13,600 (9%) | 0 (0%) |
| < 0.001 Hardy–Weinberg P-value | 12,052 (8%) | 9,721 (7%) | 16,176 (12%) |
| Other failures† | 23,478 (16%) | 17,692 (12%) | 23,722 (17%) |
|
| |||
| Non-redundant (unique) SNPs | 1,076,392 | 1,104,980 | 1,087,305 |
| Monomorphic | 156,290 (15%) | 234,482 (21%) | 268,325 (25%) |
| Polymorphic | 920,102 (85%) | 870,498 (79%) | 818,980 (75%) |
|
| |||
| All analysis panels | |||
|
| |||
| Unique QC-passed SNPs | 1,156,772 | ||
| Passed in one analysis panel | 52,204 (5%) | ||
| Passed in two analysis panels | 97,231 (8%) | ||
| Passed in three analysis panels | 1,007,337 (87%) | ||
|
| |||
| Monomorphic across three analysis panels | 75,997 | ||
| Polymorphic in all three analysis panels | 682,397 | ||
|
| |||
| MAF ≥ 0.05 in at least one of three analysis panels | 877,351 | ||
Out of 95 samples in CEU, YRI; 94 samples in CHB + JPT.
‘Other failures’ includes SNPs with discrepancies during the data transmission process. Some SNPs failed in more than one way, so these percentages add up to more than 100%.
SNPs on the Phase I map are evenly spaced, except on Y and mtDNA
The Phase I data include a successful, common SNP every 5 kb across most of the genome in each analysis panel (Supplementary Fig. 1): only 3.3% of inter-SNP distances are longer than 10 kb, spanning 11.9% of the genome (Fig. 2; see also Supplementary Fig. 2). One exception is the X chromosome (Supplementary Fig. 1), where a much higher proportion of attempted SNPs were rare or monomorphic, and thus the density of common SNPs is lower.
Figure 2. Distribution of inter-SNP distances.
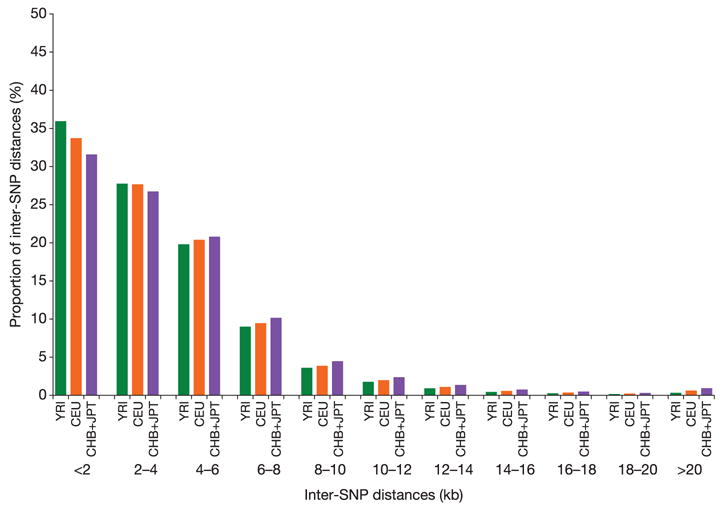
The distributions are shown for each analysis panel for the HapMappable genome (defined in the Methods), for all common SNPs (with MAF ≥ 0.05).
Two intentional exceptions to the regular spacing of SNPs on the physical map were the mitochondrial chromosome (mtDNA), which does not undergo recombination, and the non-recombining portion of chromosome Y. On the basis of the 168 successful, polymorphic SNPs, each HapMap sample fell into one of 15 (of the 18 known) mtDNA haplogroups34 (Table 4). A total of 84 SNPs that characterize the unique branches of the reference Y genealogical tree35–37 were genotyped on the HapMap samples. These SNPs assigned each Y chromosome to 8 (of the 18 major) Y haplogroups previously described (Table 4).
Table 4.
mtDNA and Y chromosome haplogroups
| DNA sample* |
||||
|---|---|---|---|---|
| MtDNA haplogroup | YRI (60) | CEU (60) | CHB (45) | JPT (44) |
| L1 | 0.22 | – | – | – |
| L2 | 0.35 | – | – | – |
| L3 | 0.43 | – | – | – |
| A | – | – | 0.13 | 0.04 |
| B | – | – | 0.33 | 0.30 |
| C | – | – | 0.09 | 0.07 |
| D | – | – | 0.22 | 0.34 |
| M/E | – | – | 0.22 | 0.25 |
| H | – | 0.45 | – | – |
| V | – | 0.07 | – | – |
| J | – | 0.08 | – | – |
| T | – | 0.12 | – | – |
| K | – | 0.03 | – | – |
| U | – | 0.23 | – | – |
| W | – | 0.02 | – | – |
|
DNA sample*
|
||||
| Y chromosome haplogroup | YRI (30) | CEU (30) | CHB (22) | JPT (22) |
|
| ||||
| E1 | 0.07 | – | – | – |
| E3a | 0.93 | – | – | – |
| F, H, K | – | 0.03 | 0.23 | 0.14 |
| I | – | 0.27 | – | – |
| R1 | – | 0.70 | – | – |
| C | – | – | 0.09 | 0.09 |
| D | – | – | – | 0.45 |
| NO | – | – | 0.68 | 0.32 |
Number of chromosomes sampled is given in parentheses.
Highly accurate phasing of long-range chromosomal haplotypes
Despite having collected data in diploid individuals, the inclusion of parent–offspring trios and the use of computational methods made it possible to determine long-range phased haplotypes of extremely high quality for each individual. These computational algorithms take advantage of the observation that because of LD, relatively few of the large number of possible haplotypes consistent with the genotype data actually occur in population samples.
The project compared a variety of algorithms for phasing haplotypes from unrelated individuals and trios38, and applied the algorithm that proved most accurate (an updated version of PHASE39) separately to each analysis panel. (Phased haplotypes are available for download at the Project website.) We estimate that ‘switch’ errors—where a segment of the maternal haplotype is incorrectly joined to the paternal—occur extraordinarily rarely in the trio samples (every 8 Mb in CEU; 3.6 Mb in YRI). The switch rate is higher in the CHB+JPT samples (one per 0.34 Mb) due to the lack of information from parent–offspring trios, but even for the unrelated individuals, statistical reconstruction of haplotypes is remarkably accurate.
Estimating properties of SNP discovery and dbSNP
Extensive sequencing and genotyping in the ENCODE regions characterized the false-positive and false-negative rates for dbSNP, as well as polymerase chain reaction (PCR)-based resequencing (see Methods). These data reveal two important conclusions: first, that PCR-based sequencing of diploid samples may be biased against very rare variants (that is, those seen only as a single heterozygote), and second, that the vast majority of common variants are either represented in dbSNP, or show tight correlation to other SNPs that are in dbSNP (Fig. 3).
Figure 3. Allele frequency and completeness of dbSNP for the ENCODE regions.
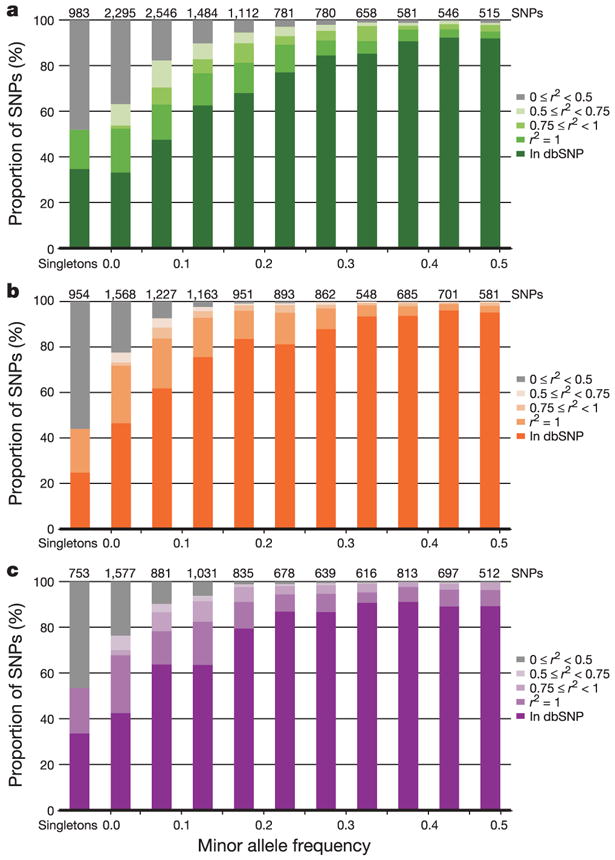
a–c, The fraction of SNPs in dbSNP, or with a proxy in dbSNP, are shown as a function of minor allele frequency for each analysis panel (a, YRI; b, CEU; c, CHB+JPT). Singletons refer to heterozygotes observed in a single individual, and are broken out from other SNPs with MAF < 0.05. Because all ENCODE SNPs have been deposited in dbSNP, for this figure we define a SNP as ‘in dbSNP’ if it would be in dbSNP build 125 independent of the HapMap ENCODE resequencing project. All remaining SNPs (not in dbSNP) were discovered only by ENCODE resequencing; they are categorized by their correlation (r2) to those in dbSNP. Note that the number of SNPs in each frequency bin differs among analysis panels, because not all SNPs are polymorphic in all analysis panels.
Allele frequency distributions within population samples
The underlying allele frequency distributions for these samples are best estimated from the ENCODE data, where deep sequencing reduces bias due to SNP ascertainment. Consistent with previous studies, most SNPs observed in the ENCODE regions are rare: 46% had MAF < 0.05, and 9% were seen in only a single individual (Fig. 4). Although most varying sites in the population are rare, most heterozygous sites within any individual are due to common SNPs. Specifically, in the ENCODE data, 90% of heterozygous sites in each individual were due to common variants (Fig. 4). With ever-deeper sequencing of DNA samples the number of rare variants will rise linearly, but the vast majority of heterozygous sites in each person will be explained by a limited set of common SNPs now contained (or captured through LD) in existing databases (Fig. 3).
Figure 4. Minor allele frequency distribution of SNPs in the ENCODE data, and their contribution to heterozygosity.
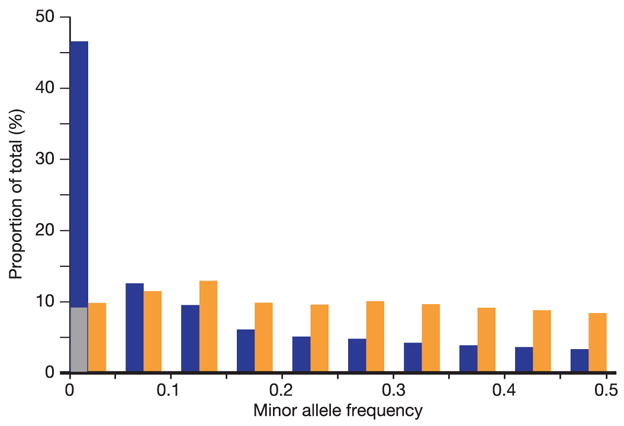
This figure shows the polymorphic SNPs from the HapMap ENCODE regions according to minor allele frequency (blue), with the lowest minor allele frequency bin (<0.05) separated into singletons (SNPs heterozygous in one individual only, shown in grey) and SNPs with more than one heterozygous individual. For this analysis, MAF is averaged across the analysis panels. The sum of the contribution of each MAF bin to the overall heterozygosity of the ENCODE regions is also shown (orange).
Consistent with previous descriptions, the CEU, CHB and JPT samples show fewer low frequency alleles when compared to the YRI samples (Fig. 5), a pattern thought to be due to bottlenecks in the history of the non-YRI populations.
Figure 5. Allele frequency distributions for autosomal SNPs.
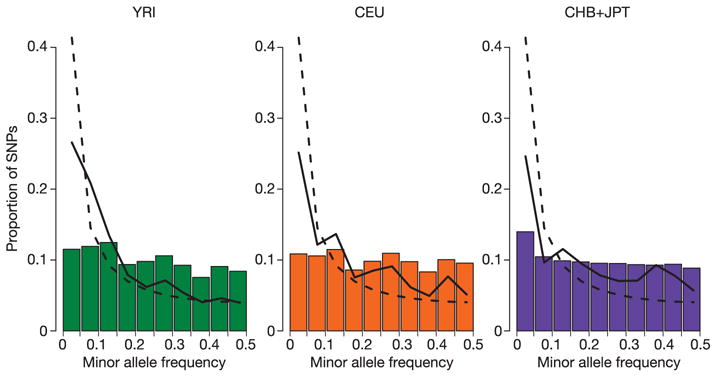
For each analysis panel we plotted (bars) the MAF distribution of all the Phase I SNPs with a frequency greater than zero. The solid line shows the MAF distribution for the ENCODE SNPs, and the dashed line shows the MAF distribution expected for the standard neutral population model with constant population size and random mating without ascertainment bias.
In contrast to the ENCODE data, the distribution of allele frequencies for the genome-wide data is flat (Fig. 5), with much more similarity in the distributions observed in the three analysis panels. These patterns are well explained by the inherent and intentional bias in the rules used for SNP selection: we prioritized using validated SNPs in order to focus resources on common (rather than rare or false positive) candidate SNPs from the public databases. For a fuller discussion of ascertainment issues, including a shift in frequencies over time and an excess of high-frequency derived alleles due to inclusion of chimpanzee data in determination of double-hit status, see the Supplementary Information (Supplementary Fig. 3).
SNP allele frequencies across population samples
Of the 1.007 million SNPs successfully genotyped and polymorphic across the three analysis panels, only a subset were polymorphic in any given panel: 85% in YRI, 79% in CEU, and 75% in CHB+JPT. The joint distribution of frequencies across populations is presented in Fig. 6 (for the ENCODE data) and Supplementary Fig. 4 (for the genome-wide map). We note the similarity of allele frequencies in the CHB and JPT samples, which motivates analysing them jointly as a single analysis panel in the remainder of this report.
Figure 6. Comparison of allele frequencies in the ENCODE data for all pairs of analysis panels and between the CHB and JPT sample sets.
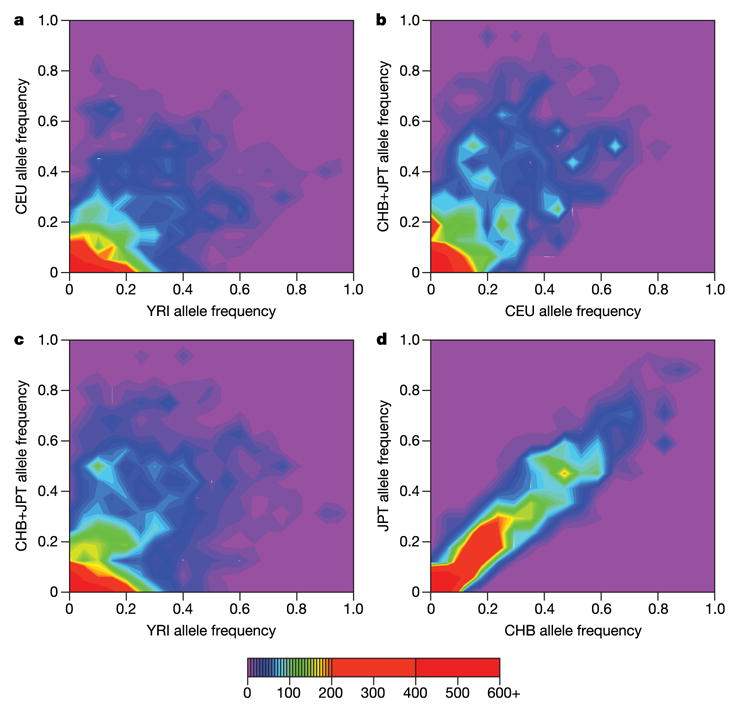
For each polymorphic SNP we identified the minor allele across all panels (a–d) and then calculated the frequency of this allele in each analysis panel/sample set. The colour in each bin represents the number of SNPs that display each given set of allele frequencies. The purple regions show that very few SNPs are common in one panel but rare in another. The red regions show that there are many SNPs that have similar low frequencies in each pair of analysis panels/sample sets.
A simple measure of population differentiation is Wright’s FST, which measures the fraction of total genetic variation due to between-population differences40. Across the autosomes, FST estimated from the full set of Phase I data is 0.12, with CEU and CHB+JPT showing the lowest level of differentiation (FST = 0.07), and YRI and CHB+JPT the highest (FST = 0.12). These values are slightly higher than previous reports41, but differences in the types of variants (SNPs versus microsatellites) and the samples studied make comparisons difficult.
As expected, we observed very few fixed differences (that is, cases in which alternate alleles are seen exclusively in different analysis panels). Across the 1 million SNPs genotyped, only 11 have fixed differences between CEU and YRI, 21 between CEU and CHB+JPT, and 5 between YRI and CHB+JPT, for the autosomes.
The extent of differentiation is similar across the autosomes, but higher on the X chromosome (FST = 0.21). Interestingly, 123 SNPs on the X chromosome were completely differentiated between YRI and CHB+JPT, but only two between CEU and YRI and one between CEU and CHB+JPT. This seems to be largely due to a single region near the centromere, possibly indicating a history of natural selection at this locus (see below; M. L. Freedman et al., personal communication).
Haplotype sharing across populations
We next examined the extent to which haplotypes are shared across populations. We used a hidden Markov model in which each haplotype is modelled in turn as an imperfect mosaic of other haplotypes (see Supplementary Information)42. In essence, the method infers probabilistically which other haplotype in the sample is the closest relative (nearest neighbour) at each position along the chromosome.
Unsurprisingly, the nearest neighbour most often is from the same analysis panel, but about 10% of haplotypes were found most closely to match a haplotype in another panel (Supplementary Fig. 5). All individuals have at least some segments over which the nearest neighbour is in a different analysis panel. These results indicate that although analysis panels are characterized both by different haplotype frequencies and, to some extent, different combinations of alleles, both common and rare haplotypes are often shared across populations.
Properties of LD in the human genome
Traditionally, descriptions of LD have focused on measures calculated between pairs of SNPs, averaged as a function of physical distance. Examples of such analyses for the HapMap data are presented in Supplementary Fig. 6. After adjusting for known confounders such as sample size, allele frequency distribution, marker density, and length of sampled regions, these data are highly similar to previously published surveys43.
Because LD varies markedly on scales of 1–100 kb, and is often discontinuous rather than declining smoothly with distance, averages obscure important aspects of LD structure. A fuller exploration of the fine-scale structure of LD offers both insight into the causes of LD and understanding of its application to disease research.
LD patterns are simple in the absence of recombination
The most natural path to understanding LD structure is first to consider the simplest case in which there is no recombination (or gene conversion), and then to add recombination to the model. (For simplicity we ignore genotyping error and recurrent mutation in this discussion, both of which seem to be rare in these data.)
In the absence of recombination, diversity arises solely through mutation. Because each SNP arose on a particular branch of the genealogical tree relating the chromosomes in the current populations, multiple haplotypes are observed. SNPs that arose on the same branch of the genealogy are perfectly correlated in the sample, whereas SNPs that occurred on different branches have imperfect correlations, or no correlation at all.
We illustrate these concepts using empirical genotype data from 36 adjacent SNPs in an ENCODE region (ENr131.2q37), selected because no obligate recombination events were detectable among them in CEU (Fig. 7). (We note that the lack of obligate recombination events in a small sample does not guarantee that no recombinants have occurred, but it provides a good approximation for illustration.)
Figure 7. Genealogical relationships among haplotypes and r2 values in a region without obligate recombination events.
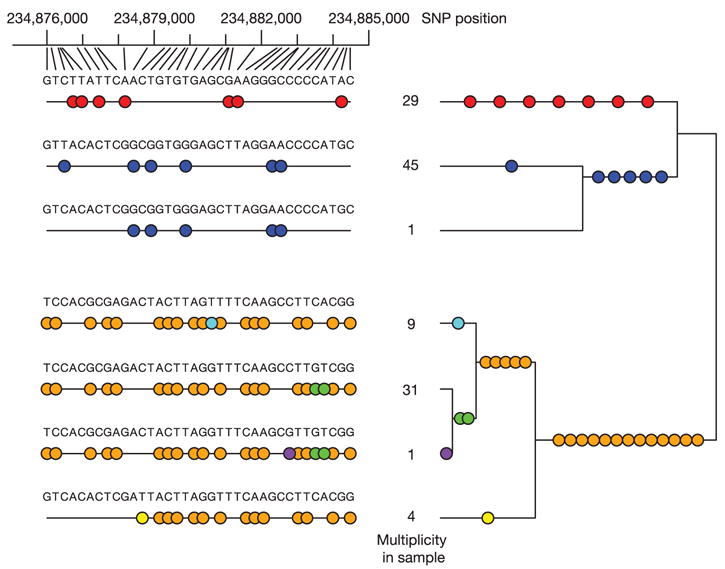
The region of chromosome 2 (234,876,004–234,884,481 bp; NCBI build 34) within ENr131.2q37 contains 36 SNPs, with zero obligate recombination events in the CEU samples. The left part of the plot shows the seven different haplotypes observed over this region (alleles are indicated only at SNPs), with their respective counts in the data. Underneath each of these haplotypes is a binary representation of the same data, with coloured circles at SNP positions where a haplotype has the less common allele at that site. Groups of SNPs all captured by a single tag SNP (with r2 ≥ 0.8) using a pairwise tagging algorithm53,54 have the same colour. Seven tag SNPs corresponding to the seven different colours capture all the SNPs in this region. On the right these SNPs are mapped to the genealogical tree relating the seven haplotypes for the data in this region.
In principle, 36 such SNPs could give rise to 236 different haplotypes. Even with no recombination, gene conversion or recurrent mutation, up to 37 different haplotypes could be formed. Despite this great potential diversity, only seven haplotypes are observed (five seen more than once) among the 120 parental CEU chromosomes studied, reflecting shared ancestry since their most recent common ancestor among apparently unrelated individuals.
In such a setting, it is easy to interpret the two most common pairwise measures of LD: D′ and r2. (See the Supplementary Information for fuller definitions of these measures.) D′ is defined to be 1 in the absence of obligate recombination, declining only due to recombination or recurrent mutation27. In contrast, r2 is simply the squared correlation coefficient between the two SNPs. Thus, r2 is 1 when two SNPs arose on the same branch of the genealogy and remain undisrupted by recombination, but has a value less than 1 when SNPs arose on different branches, or if an initially strong correlation has been disrupted by crossing over.
In this region, D′ = 1 for all marker pairs, as there is no evidence of historical recombination. In contrast, and despite great simplicity of haplotype structure, r2 values display a complex pattern, varying from 0.0003 to 1.0, with no relationship to physical distance. This makes sense, however, because without recombination, correlations among SNPs depend on the historical order in which they arose, not the physical order of SNPs on the chromosome.
Most importantly, the seeming complexity of r2 values can be deconvolved in a simple manner: only seven different SNP configurations exist in this region, with all but two chromosomes matching five common haplotypes, which can be distinguished from each other by typing a specific set of four SNPs. That is, only a small minority of sites need be examined to capture fully the information in this region.
Variation in local recombination rates is a major determinant of LD
Recombination in the ancestors of the current population has typically disrupted the simple picture presented above. In the human genome, as in yeast44, mouse45 and other genomes, recombination rates typically vary dramatically on a fine scale, with hotspots of recombination explaining much crossing over in each region28. The generality of this model has recently been demonstrated through computational methods that allow estimation of recombination rates (including hotspots and coldspots) from genotype data46,47.
The availability of nearly complete information about common DNA variation in the ENCODE regions allowed a more precise estimation of recombination rates across large regions than in any previous study. We estimated recombination rates and identified recombination hotspots in the ENCODE data, using methods previously described46 (see Supplementary Information for details). Hotspots are short regions (typically spanning about 2 kb) over which recombination rates rise dramatically over local background rates.
Whereas the average recombination rate over 500 kb across the human genome is about 0.5 cM48, the estimated recombination rate across the 500-kb ENCODE regions varied nearly tenfold, from a minimum of 0.19 cM (ENm013.7q21.13) to a maximum of 1.25 cM (ENr232.9q34.11). Even this tenfold variation obscures much more dramatic variation over a finer scale: 88 hotspots of recombination were identified (Fig. 8; see also Supplementary Fig. 7)—that is, one per 57 kb—with hotspots detected in each of the ten regions (from 4 in 12q12 to 14 in 2q37.1). Across the 5 Mb, we estimate that about 80% of all recombination has taken place in about 15% of the sequence (Fig. 9, see also refs 46, 49).
Figure 8. Comparison of linkage disequilibrium and recombination for two ENCODE regions.
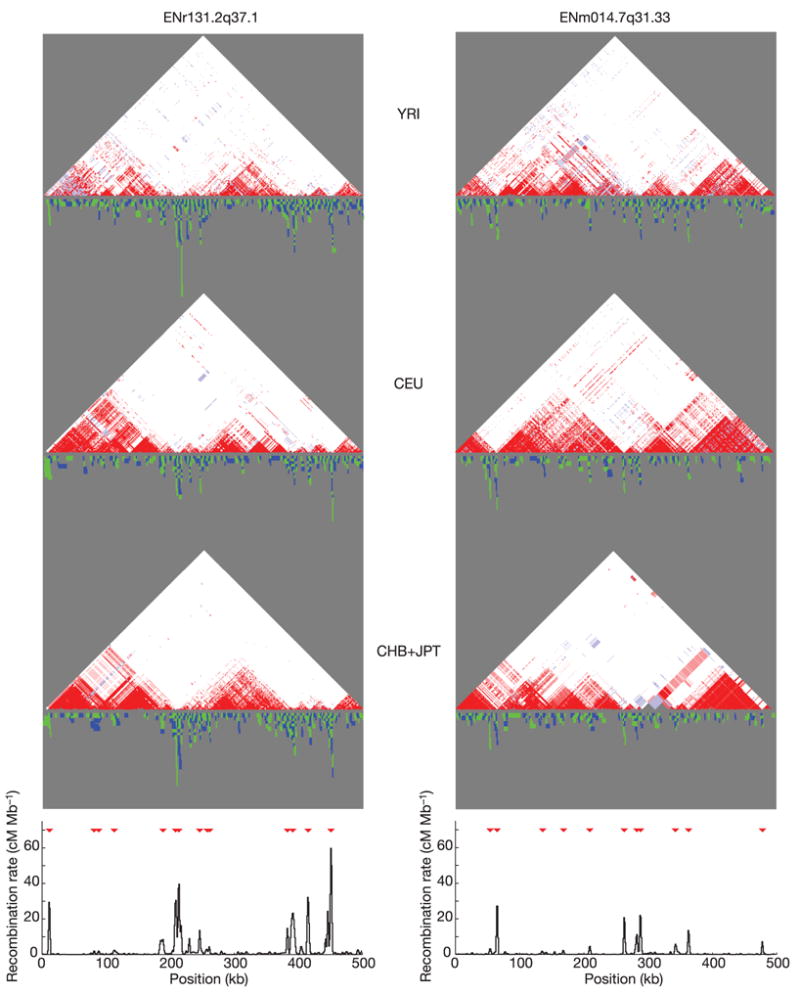
For each region (ENr131.2q37.1 and ENm014.7q31.33), D′ plots for the YRI, CEU and CHB+JPT analysis panels are shown: white, D′ < 1 and LOD < 2; blue, D′ = 1 and LOD < 2; pink, D′ < 1 and LOD ≥ 2; red, D′ = 1 and LOD ≥ 2. Below each of these plots is shown the intervals where distinct obligate recombination events must have occurred (blue and green indicate adjacent intervals). Stacked intervals represent regions where there are multiple recombination events in the sample history. The bottom plot shows estimated recombination rates, with hotspots shown as red triangles46.
Figure 9. The distribution of recombination events over the ENCODE regions.
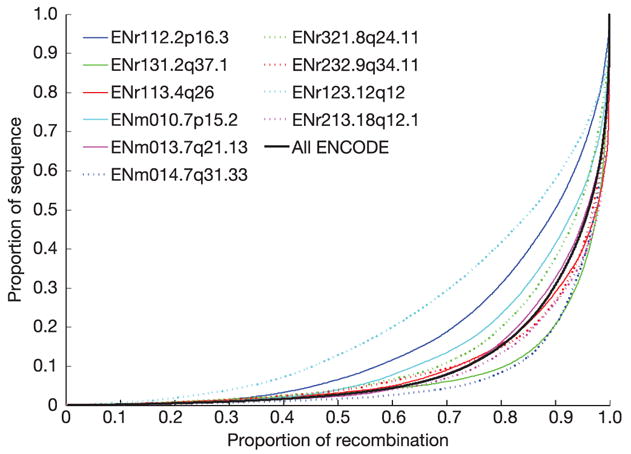
Proportion of sequence containing a given fraction of all recombination for the ten ENCODE regions (coloured lines) and combined (black line). For each line, SNP intervals are placed in decreasing order of estimated recombination rate46, combined across analysis panels, and the cumulative recombination fraction is plotted against the cumulative proportion of sequence. If recombination rates were constant, each line would lie exactly along the diagonal, and so lines further to the right reveal the fraction of regions where recombination is more strongly locally concentrated.
A block-like structure of human LD
With most human recombination occurring in recombination hotspots, the breakdown of LD is often discontinuous. A ‘block-like’ structure of LD is visually apparent in Fig. 8 and Supplementary Fig. 7: segments of consistently high D′ that break down where high recombination rates, recombination hotspots and obligate recombination events50 all cluster.
When haplotype blocks are more formally defined in the ENCODE data (using a method based on a composite of local D′ values30, or another based on the four gamete test51), most of the sequence falls into long segments of strong LD that contain many SNPs and yet display limited haplotype diversity (Table 5).
Table 5.
Haplotype blocks in ENCODE regions, according to two methods
| Parameter | YRI | CEU | CHB + JPT |
|---|---|---|---|
| Method based on a composite of local D’ values30 | |||
| Average number of SNPs per block | 30.3 | 70.1 | 54.4 |
| Average length per block (kb) | 7.3 | 16.3 | 13.2 |
| Fraction of genome spanned by blocks (%) | 67 | 87 | 81 |
| Average number of haplotypes (MAF ≥ 0.05) per block | 5.57 | 4.66 | 4.01 |
| Fraction of chromosomes due to haplotypes with MAF ≥ 0.05 (%) | 94 | 93 | 95 |
| Method based on the four gamete test51 | |||
| Average number of SNPs per block | 19.9 | 24.3 | 24.3 |
| Average length per block (kb) | 4.8 | 5.9 | 5.9 |
| Fraction of genome spanned by blocks (%) | 86 | 84 | 84 |
| Average number of haplotypes (MAF ≥ 0.05) per block | 5.12 | 3.63 | 3.63 |
| Fraction of chromosomes due to haplotypes with MAF ≥ 0.05 (%) | 91 | 95 | 95 |
Specifically, addressing concerns that blocks might be an artefact of low marker density52, in these nearly complete data most of the sequence falls into blocks of four or more SNPs (67% in YRI to 87% in CEU) and the average sizes of such blocks are similar to initial estimates30. Although the average block spans many SNPs (30–70), the average number of common haplotypes in each block ranged only from 4.0 (CHB + JPT) to 5.6 (YRI), with nearly all haplotypes in each block matching one of these few common haplotypes. These results confirm the generality of inferences drawn from disease-mapping studies27 and genomic surveys with smaller sample sizes29 and less complete data30.
Long-range haplotypes and local patterns of recombination
Although haplotypes often break at recombination hotspots (and block boundaries), this tendency is not invariant. We identified all unique haplotypes with frequency more than 0.05 across the 269 individuals in the phased data, and compared them to the fine-scale recombination map. Figure 10 shows a region of chromosome 19 over which many such haplotypes break at identified recombination hotspots, but others continue. Thus, the tendency towards co-localization of recombination sites does not imply that all haplotypes break at each recombination site.
Figure 10. The relationship among recombination rates, haplotype lengths and gene locations.
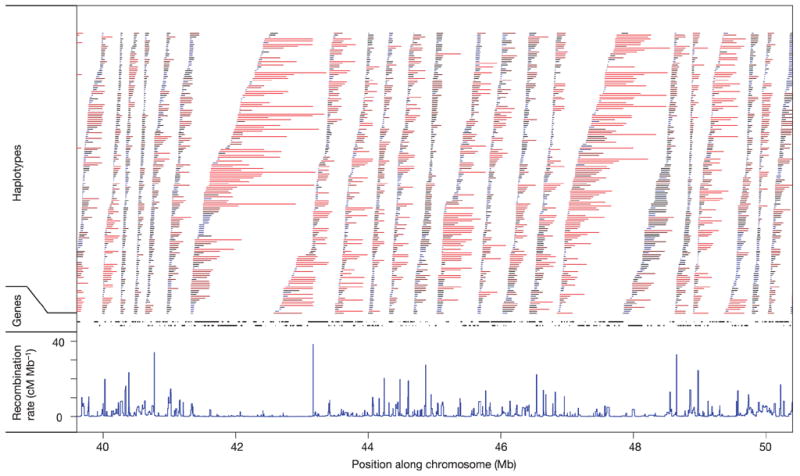
Recombination rates in cM Mb−1 (blue). Non-redundant haplotypes with frequency of at least 5% in the combined sample (bars) and genes (black segments) are shown in an example gene-dense region of chromosome 19 (19q13). Haplotypes are coloured by the number of detectable recombination events they span, with red indicating many events and blue few.
Some regions display remarkably extended haplotype structure based on a lack of recombination (SupplementaryFig. 8a, b). Most striking, if unsurprising, are centromeric regions, which lack recombination: haplotypes defined by more than 100 SNPs span several megabases across the centromeres. The X chromosome has multiple regions with very extensive haplotypes, whereas other chromosomes typically have a few such domains.
Most global measures of LD become more consistent when measured in genetic rather than physical distance. For example, when plotted against physical distance, the extent of pairwise LD varies by chromosome; when plotted against average recombination rate on each chromosome (estimated from pedigree-based genetic maps) these differences largely disappear (Supplementary Fig. 6). Similarly, the distribution of haplotype length across chromosomes is less variable when measured in genetic rather than physical distance. For example, the median length of haplotypes is 54.4 kb on chromosome 1 compared to 34.8 kb on chromosome 21. When measured in genetic distance, however, haplotype length is much more similar: 0.104 cM on chromosome 1 compared to 0.111 cM on chromosome 21 (Supplementary Fig. 9).
The exception is again the X chromosome, which has more extensive haplotype structure after accounting for recombination rate (median haplotype length = 0.135 cM). Multiple factors could explain different patterns on the X chromosome: lower SNP density, smaller sample size, restriction of recombination to females and lower effective population size.
A view of LD focused on the putative causal SNP
Although genealogy and recombination provide insight into why nearby SNPs are often correlated, it is the redundancies among SNPs that are of central importance for the design and analysis of association studies. A truly comprehensive genetic association study must consider all putative causal alleles and test each for its potential role in disease. If a causal variant is not directly tested in the disease sample, its effect can nonetheless be indirectly tested if it is correlated with a SNP or haplotype that has been directly tested.
The typical SNP is highly correlated with many of its neighbours
The ENCODE data reveal that SNPs are typically perfectly correlated to several nearby SNPs, and partially correlated to many others.
We use the term proxy to mean a SNP that shows a strong correlation with one or more others. When two variants are perfectly correlated, testing one is exactly equivalent to testing the other; we refer to such collections of SNPs (with pairwise r2 = 1.0 in the HapMap samples) as ‘perfect proxy sets’.
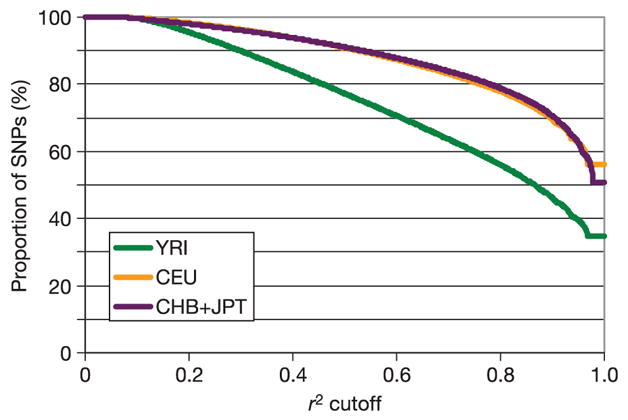
Considering only common SNPs (the target of study for the HapMap Project) in CEU in the ENCODE data, one in five SNPs has 20 or more perfect proxies, and three in five have five or more. In contrast, one in five has no perfect proxies. As expected, perfect proxy sets are smaller in YRI, with twice as many SNPs (two in five) having no perfect proxy, and a quarter as many (5%) having 20 or more (Figs 11 and 12). These patterns are largely consistent across the range of frequencies studied by the project, with a trend towards fewer proxies at MAF < 0.10 (Fig. 11). Put another way, the average common SNP in ENCODE is perfectly redundant with three other SNPs in the YRI samples, and nine to ten other SNPs in the other sample sets (Fig. 13).
Figure 11.
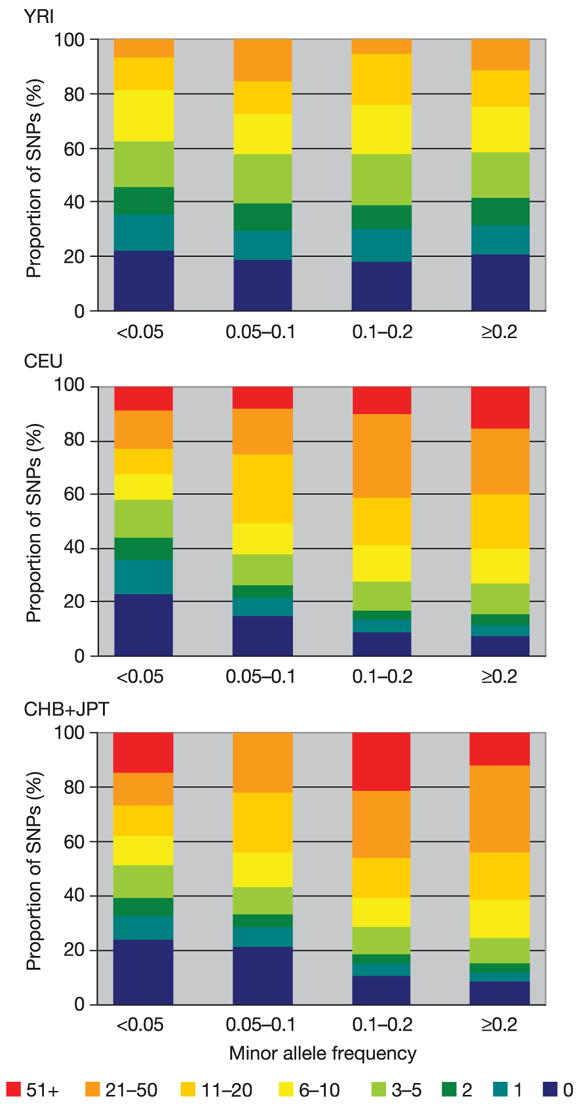
The number of proxy SNPs (r2 ≥ 0.8) as a function of MAF in the ENCODE data.
Figure 12.
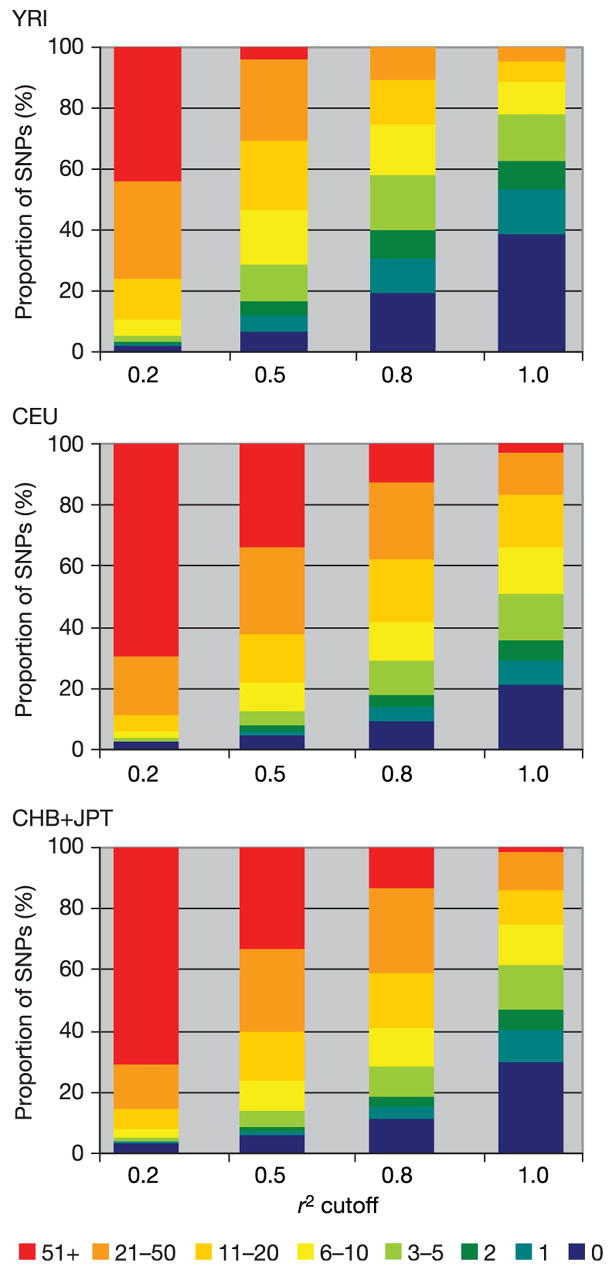
The number of proxies per SNP in the ENCODE data as a function of the threshold for correlation (r2).
Figure 13.
Relationship in the Phase I HapMap between the threshold for declaring correlation between proxies and the proportion of all SNPs captured.
Of course, to be detected through LD in an association study, correlation need not be complete between the genotyped SNP and the causal variant. For example, under a multiplicative disease model and a single-locus χ2 test, the sample size required to detect association to an allele scales as 1/r2. That is, if the causal SNP has an r2 = 0.5 to one tested in the disease study, full power can be maintained if the sample size is doubled.
The number of SNPs showing such substantial but incomplete correlation is much larger. For example, using a looser threshold for declaring correlation (r2 ≥ 0.5), the average number of proxies found for a common SNP in CHB+JPT is 43, and the average in YRI is 16 (Fig. 12). These partial correlations can be exploited through haplotype analysis to increase power to detect putative causal alleles, as discussed below.
Evaluating performance of the Phase I map
To estimate the proportion of all common SNPs captured by the Phase I map, we evaluated redundancy among SNPs on the genome-wide map, and performed simulations based on the more complete ENCODE data. The two methods give highly similar answers, and indicate that Phase I should provide excellent power for CEU, CHB and JPT, and substantial power for YRI. Phase II, moreover, will provide nearly complete power for all three analysis panels.
Redundancies among SNPs in Phase I HapMap
Redundancy offers one measure that Phase I has sampled densely in comparison to the underlying scale of correlation. Specifically, 50% (YRI) to 75% (CHB+JPT, CEU) of all SNPs on the Phase I map are highly correlated (r2 ≥ 0.8) to one or more others on the map (Fig. 13; see also Supplementary Fig. 10). Over 90% of all SNPs on the map have highly statistically significant correlation to one or more neighbours. These partial correlations can be combined to form haplotypes that are even better proxies for a SNP of interest.
Modelling Phase I HapMap from complete ENCODE data
A second approach to evaluating the completeness of the Phase I data involves thinning the more complete ENCODE data to match Phase I for allele frequency and SNP density. Simulated Phase I HapMaps were used to evaluate coverage in relation to the full set of common SNPs (Table 6), and provided nearly identical estimates to those above: 45% (YRI) to 74% (CHB+JPT, CEU) of all common SNPs are predicted to have a proxy with r2 ≥ 0.8 to a SNP included in the Phase I HapMap (Supplementary Fig. 11).
Table 6.
Coverage of simulated Phase I and Phase II HapMap to capture all common SNPs in the ten ENCODE regions
| Analysis panel | Per cent maximum r2≥ 0.8 | Mean maximum r2 |
|---|---|---|
| Phase I HapMap | ||
| YRI | 45 | 0.67 |
| CEU | 74 | 0.85 |
| CHB+JPT | 72 | 0.83 |
| Phase II HapMap | ||
| YRI | 81 | 0.90 |
| CEU | 94 | 0.97 |
| CHB+JPT | 94 | 0.97 |
Simulated Phase I HapMaps were generated from the phased ENCODE data (release 16c1) by randomly picking SNPs that appear in dbSNP build 121 (excluding ‘non-rs’ SNPs in release 16a) for every 5-kb bin until a common SNP was picked (allowing up to three attempts per bin). The Phase II HapMap was simulated by picking SNPs at random to achieve an overall density of 1 SNP per 1 kb. These numbers are averages over 20 independent iterations for all ENCODE regions in all three analysis panels.
Statistical power in association studies may be more closely approximated by the average (maximal) correlation value between a SNP and its best proxy on the map, rather than by the proportion exceeding an arbitrary (and stringent) threshold. The average values for maximal r2 to a nearby SNP range from 0.67 (YRI) to 0.85 (CEU and CHB+JPT).
Modelling Phase II HapMap from complete ENCODE data
A similar procedure was used to generate simulated Phase II HapMaps from ENCODE data (Table 6). Phase II is predicted to capture the majority of common variation in YRI: 81% of all common SNPs should have a near perfect proxy (r2 ≥ 0.8) to a SNP on the map, with the mean maximal r2 value of 0.90. Unsurprisingly, the CEU, CHB and JPT samples, already well served by Phase I, are nearly perfectly captured: 94% of all common sites have a proxy on the map with r2 ≥ 0.8, with an average maximal r2 value of 0.97.
These analyses indicate that the Phase I and Phase II HapMap resources should provide excellent coverage for common variation in these population samples.
Selection of tag SNPs for association studies
A major impetus for developing the HapMap was to guide the design and prioritization of SNP genotyping assays for disease association studies. We refer to the set of SNPs genotyped in a disease study as tags. A given set of tags can be analysed for association with a phenotype using a variety of statistical methods which we term tests, based either on the genotypes of single SNPs or combinations of multiple SNPs.
The shared goal of all tag selection methods is to exploit redundancy among SNPs, maximizing efficiency in the laboratory while minimizing loss of information24,27. This literature is extensive and varied, despite its youth. Some methods require that a single SNP serve as a proxy for other, untyped variants, whereas other methods allow combinations of alleles (haplotypes) to serve as proxies; some make explicit use of LD blocks whereas others are agnostic to such descriptions. Although it is not practical to implement all such methods at the project website, the HapMap genotypes are freely available and investigators can apply their method of choice to the data. To assist users, both a single-marker tagging method and a more efficient multimarker method have been implemented at http://www.hapmap.org.
Tagging using a simple pairwise method
To illustrate general principles of tagging, we first applied a simple and widely used pairwise algorithm53,54: SNPs are selected for genotyping until all common SNPs are highly correlated (r2 ≥ 0.8) to one or more members of the tag set.
Starting from the substantially complete ENCODE data, the density of common SNPs can be reduced by 75–90% with essentially no loss of information (Fig. 14). That is, the genotyping burden can be reduced from one common SNP every 500 bp to one SNP every 2 kb (YRI) to 5 kb (CEU and CHB+JPT). Because LD often extends for long distances, studies of short gene segments tend to underestimate the redundancy across the genome43.
Figure 14. Tag SNP information capture.
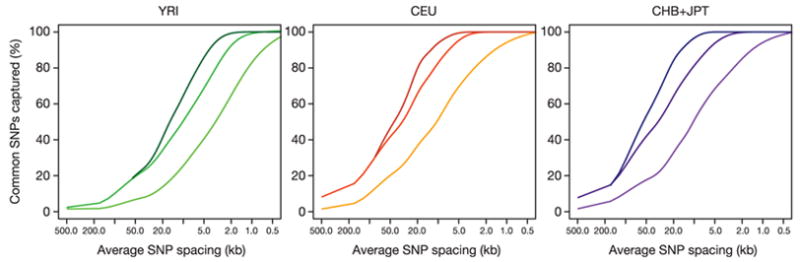
The proportion of common SNPs captured with r2 ≥ 0.8 as a function of the average tag SNP spacing is shown for the phased ENCODE data, plotted (left to right) for tag SNPs prioritized by Tagger (multimarker and pairwise) and for tag SNPs picked at random. Results were averaged over all the ENCODE regions.
Although tags selected based on LD offer the greatest improvements in efficiency and information capture, even randomly chosen subsets of SNPs offer considerable efficiencies (Fig. 14).
The data also reveal a rule of diminishing returns: a small set of highly informative tags captures a large fraction of all variation, with additional tags each capturing only one or a few proxies. For example, in CHB+JPT the most informative 1% of all SNPs (one per 50 kb) is able to proxy (at r2 ≥ 0.8) for 40% of all common SNPs, whereas a substantial proportion of SNPs have no proxies at all.
These observations are encouraging with respect to genome-wide association studies. A set of SNPs typed every 5–10 kb across the genome (within the range of current technology) can capture nearly all common variation in the genome in the CEU and CHB+JPT samples, with more SNPs required in the YRI samples.
Tagging from the genome-wide map
Whereas analysis of the complete ENCODE data set reveals the maximal efficiency likely to be possible with this tag selection strategy, analysis of the Phase I map illuminates the extent to which the current resource can be used for near-term studies. Specifically, using the same pairwise tagging approach above, 260,000 (CHB+JPT) to 474,000 (YRI) SNPs are required to capture all common SNPs in the Phase I data set (Table 7). That is, being incomplete and thus less redundant, the Phase I data are much less compressible by tag SNP selection than are the ENCODE data. Nevertheless, even at this level a half to a third of all SNPs can be selected as proxies for the remainder (and, by inference, the bulk of other common SNPs in the genome).
Table 7.
Number of selected tag SNPs to capture all observed common SNPs in the Phase I HapMap
| r2threshold* | YRI | CEU | CHB +JPT |
|---|---|---|---|
| r2 ≥ 0.5 | 324,865 | 178,501 | 159,029 |
| r2 ≥ 0.8 | 474,409 | 293,835 | 259,779 |
| r2 = 1.0 | 604,886 | 447,579 | 434,476 |
Tag SNPs were picked to capture common SNPs in HapMap release 16c1 using the software program Haploview.
Pairwise tagging at different r2 thresholds.
Increasing the efficiency of tag SNPs
Although the pairwise method is simple, complete and straightforward, efficiency can be improved with a number of simple changes. First, relaxing the threshold on r2 for tag SNP selection substantially reduces the number of tag SNPs selected, with only a modest decrease in the correlations among SNPs (Table 7). For example, reducing the r2 threshold from 0.8 to 0.5 decreases the number of tag SNPs selected from the HapMap by 39% in CHB+JPT (260,000 to 159,000) and 32% in YRI (474,000 to 325,000). The average r2 value between tags and other (unselected) SNPs falls much less dramatically than the number of tags selected, increasing efficiency. Whether such a loss of power is justified by the disproportionate reduction in work is a choice each investigator will need to make.
A second enhancement exploits multimarker haplotypes. Many investigators have discussed using multiple SNPs (in haplotypes and regression models) to serve as proxies for untyped sites55–58, which may reduce the number of tags required and increase the power of analyses performed. Figure 14 illustrates the point with one such method55, showing that a multimarker method allows greater coverage for a fixed set of markers (or, alternatively, fewer markers to achieve the same coverage). Although a full consideration of this issue is beyond the scope of this paper, the availability of these and other data should allow the comparison and application of such methods.
A third approach to increasing efficiency is to prioritize tags based on the number of other SNPs captured. Whereas 260,000 SNPs are required to provide r2 ≥ 0.8 for all SNPs in the Phase I HapMap (CHB+JPT), the best 10,000 such SNPs (4%) capture 22% of all common variable sites with r2 ≥ 0.8 (Table 8). Such prioritization can be applied using different weights for SNPs based on genomic annotation (for example, non-synonymous coding SNPs, SNPs in conserved non-coding sequence, and candidate genes of biological interest).
Table 8.
Proportion of common SNPs in Phase I captured by sets of tag SNPs
| Common SNPs captured (%)
|
|||
|---|---|---|---|
| Tag SNP set size | YRI | CEU | CHB +JPT |
| 10,000 | 12.3 | 20.4 | 21.9 |
| 20,000 | 19.1 | 30.9 | 33.2 |
| 50,000 | 32.7 | 50.4 | 53.6 |
| 100,000 | 47.2 | 68.5 | 72.2 |
| 250,000 | 70.1 | 94.1 | 98.5 |
As in Table 7, tag SNPs were picked to capture common SNPs in HapMap release 16c1 using Haploview, selecting SNPs in order of the fraction of sites captured. Common SNPs were captured by fixed-size sets of pairwise tags at r2 ≥ 0.8.
Tag transferability across populations
The most complete set of tags would be those based on all 269 samples; however, many studies may be performed in individuals more closely related to one particular HapMap population, and efficiency may be gained by selecting tags only from that population sample. (Selecting tags in a HapMap population sample that is known to be more distantly related than is another, for example, using CEU to pick tags for a study of Japanese, seems inefficient.)
An important question is how tags selected in one or more analysis panels will transfer to disease studies performed in these or other populations. Our data do not address this question directly, although the known similarity of allele and haplotype frequencies across populations within continents41 is encouraging. More data are clearly needed, however.
Tag selection based on initial genotyping
Whereas the discussions above assume de novo selection of SNPs, many investigators will have already performed initial studies, and wish to design follow-on experiments. The HapMap data can be used to highlight SNPs that might potentially explain a positive association signal, or those that were poorly captured (and thus still need to be tested) after a negative scan. In cases where multiple SNPs are both associated with the trait and with each other, the HapMap data can be queried to identify whether samples from any other analysis panel show a breakdown of LD in that region, and thus the possibility of narrowing the span over which the causal variant may reside.
Applications to the analysis of association data
Beyond guiding selection of tag SNPs, HapMap data can inform the subsequent analysis and interpretation in disease association studies.
Analysis of an existing genotype data set
The HapMap can be used to inform association testing, regardless of how tags were selected. Specifically, as long as the SNPs genotyped in a disease study have also been typed in the HapMap samples, it is possible to identify which SNPs are well captured by the genotyped SNPs (either singly, or in haplotype combinations), and which are not55.
This is of particular importance for genome-wide association studies performed using array-based, standardized genotyping reagents, which do not allow investigators to choose their own sets of tag SNPs. The Affymetrix 120K SNP array data included in Phase I of the HapMap provides a simple example: in CEU 48% of HapMap SNPs have substantial pairwise correlation (r2 ≥ 0.5) to one or more of the 120K SNPs on the array. An additional 13%, however, are not correlated to a single SNP, but are to a specific haplotype of two members of the 120K panel. By identifying such haplotype predictors in the HapMap, and testing them (in addition to the single SNPs) in a disease study, it is likely that power will be increased (I. Pe’er et al., manuscript in preparation).
Evaluating statistical significance and interpreting results
An important challenge in genome-wide association testing is to develop statistical procedures that minimize false positives without greatly sacrificing true positives. The challenge is amplified by the correlated nature of polymorphism data, which makes simple frequentist approaches that assume independence (such as Bonferonni correction) highly conservative. To illustrate this point, we used the ENCODE data to estimate the ‘effective number of independent tests’ (the statistical burden of testing all common (MAF ≥ 0.05) variation) across large genomic regions. Specifically, we re-sampled from the phased ENCODE chromosomes to create mock case-control panels in which all common SNPs were observed, but there was not a causal allele. The resulting χ2 distribution for association indicates that complete testing of common variation in each 500-kb region is equivalent to performing about 150 independent statistical tests (in CEU and CHB+JPT) and about 350 tests (in YRI). Although it will probably be desirable to perform such empirical estimates of significance within each disease study, these results illustrate how Bonferonni correction overestimates the statistical penalty of performing many correlated tests.
Study of less common alleles
We have focused primarily on the hypothesis that a single, common causal allele exists, and needs to be tested for association to disease. Of course, in many cases the causal allele(s) will be less common, and might be missed by such an approach.
It is possible to perform additional haplotype tests, beyond those that capture known polymorphisms, in the hope of capturing less common or unrepresented alleles56. Such haplotype analysis has a long history and proven value in mendelian genetics; the causal mutation is generally rare and unexamined during initial genotyping, but is frequently recognized by its presence on a long, unique haplotype of common alleles18,19,59–62.
Admixture mapping
Although not designed specifically to enable admixture mapping63, the HapMap has helped lay the groundwork for this approach. Admixture mapping requires a map of SNPs that are highly differentiated in frequency across population groups. By typing many SNPs in samples from multiple geographical regions, the data have helped to identify such SNPs for the design of genome-wide admixture mapping panels64,65 and can be further used to identify candidate SNPs with large allele frequency differences for follow-up of positive admixture scan results66.
Loss of heterozygosity in tumours
Loss of heterozygosity (LOH) in tumour tissue can be a powerful indicator of the location of tumour suppressor genes, and genome-wide, fine-scale LOH analysis has been empowered by genome-wide SNP arrays67. Germline DNA is not always available from the same subjects, however, and even if available, typing of germline DNA doubles project costs. In lower density scans for LOH (with markers far apart relative to the scale of LD), long runs of homozygosity in tumours are nearly always indicative of LOH. However, at higher densities runs of homozygosity can be due to haplotype homozygosity in the inherited germline DNA, rather than LOH.
The HapMap data can help minimize this difficulty; previous probabilities for homozygosity based on known frequencies of haplotypes in the HapMap data can be used to distinguish homozygosity due to haplotype sharing rather than LOH68.
Identifying structural variants in HapMap data
Structural variations—segments where DNA is deleted, duplicated, or rearranged—are common69,70 and have an important role in diseases71–73. The HapMap can provide some insight into structural variation because, in many cases, structural variants reveal themselves through signatures in SNP genotype data. In particular, polymorphic deletions are important to discover, because loss of genetic material is of obvious functional relevance, and results in aberrant patterns of SNP genotypes. These include apparent non-mendelian inheritance of SNP alleles, null genotypes and deviations from Hardy–Weinberg equilibrium. However, such SNPs are routinely discarded as technical failures of genotyping.
Thus, we scanned the unfiltered Phase I HapMap data using an approach developed and validated to identify polymorphic deletions from clusters of SNPs with aberrant genotype patterns (calibrated across the multiple centres and genotyping platforms74). In total, 541 candidate deletion polymorphisms were identified, of which 150 were common enough to be observed as homozygotes.
The properties of these candidate deletions, including experimental validation of 90 candidates, are described in ref. 74. Validated polymorphisms include 10 that remove coding exons of genes, such that in many cases individuals are homozygous null for the encoded transcript. Analysis of confirmed deletions often shows strong LD with nearby SNPs, indicating that LD-based approaches can be useful for detecting disease associations due to structural (as well as SNP) variants.
Polymorphic inversions may also be reflected in the HapMap data as long regions where multiple SNPs are perfectly correlated: because recombination between an inverted and non-inverted copy is lethal, the inverted and non-inverted copies of the region evolve independently. A striking example corresponds to the known inversion polymorphism on chromosome 17, present in 20% of the CEU chromosomes, that has been associated with fertility and total recombination rate in females among Icelanders75. Long LD may also arise, however, due to a low recombination rate or certain forms of natural selection, as discussed below.
Insights into recombination and natural selection
In addition to its intended function as a resource for disease studies, the HapMap data provide clues about the biology of recombination and history of natural selection.
A genome-wide map of recombination rates at a fine scale
On the basis of the HapMap data, we created a fine-scale genetic map spanning the human genome (Supplementary Fig. 12), including 21,617 identified recombination hotspots (one per 122 kb).
Both the number and intensity of hotspots contribute to overall variation in recombination rate. For example, we selected 25 regions of 5 Mb as having the highest (>2.75 cM Mb−1) and lowest (<0.5 cM Mb−1) rates of recombination in the deCODE (pedigree-based) genetic map48. We detected recombination hotspots in all regions, even the lowest. But in the high cM Mb−1 regions hotspots are more closely spaced (one per 84 kb) and have a higher average intensity (0.124 cM) as compared to the low cM Mb−1 regions (one every 208 kb, and 0.051 cM, respectively).
Estimates of recombination rates and identified hotspots are robust to the specific markers and samples studied. Specifically, we compared these results to a similar analysis76 of the data of ref. 77 (with about 1.6 million SNPs genotyped in 71 individuals). We find nearly complete correlation in rate estimates at a coarse scale (5 Mb) between these two surveys (r2 = 0.99) and to the pedigree map (r2 = 0.95). Very substantial correlation is found at finer scales: r2 = 0.8 at 50 kb and r2 = 0.59 at 5 kb. Moreover, of the 21,617 hotspots identified using the HapMap data, 78% (16,923) were also identified using the data of ref. 77.
The ability to detect events depends on marker density, with the larger number of SNPs studied by ref. 77 increasing power to detect hotspots, and presumably precision of rate estimates. There are, however, substantial genomic regions where the HapMap data have a higher SNP density. For example, more hotspots are detected on chromosomes 9 and 19 from the HapMap data. We expect that Phase II of HapMap will provide a genome-wide recombination map of substantially greater precision than either ref. 77, or Phase I, at fine scales.
Little is yet known about the molecular determinants of recombination hotspots. In an analysis of the data of ref. 77, another study (ref. 76) found significant evidence for an excess of the THE1A/B retrotransposon-like elements within recombination hotspots, and more strikingly for a sixfold increase of a particular motif (CCTCCCT) within copies of the element in hotspots, compared to copies of the element outside hotspots. In analysing the HapMap data, we confirmed these findings (Supplementary Fig. 13). Furthermore, THE1B elements with the motif are particularly enriched within 1.5 kb of the centre of the hotspots compared to flanking sequence (P < 10−16).
Correlations of LD with genomic features
Variation in recombination rate is important, in large part, because of its impact on LD. We thus examined genome-wide LD for correlation to recombination rates, sequence composition and gene features.
We confirmed previous observations that LD is generally low near telomeres, elevated near centromeres, and correlated with chromosome length (Fig. 15; see also Supplementary Figs 8b and 14)48,78–80. These patterns are due to recombination rate variation as discussed above. We also confirmed previously described relationships between LD and G+C content78,81,82, sequence polymorphism83 and repeat composition78,82.
Figure 15. Length of LD spans.
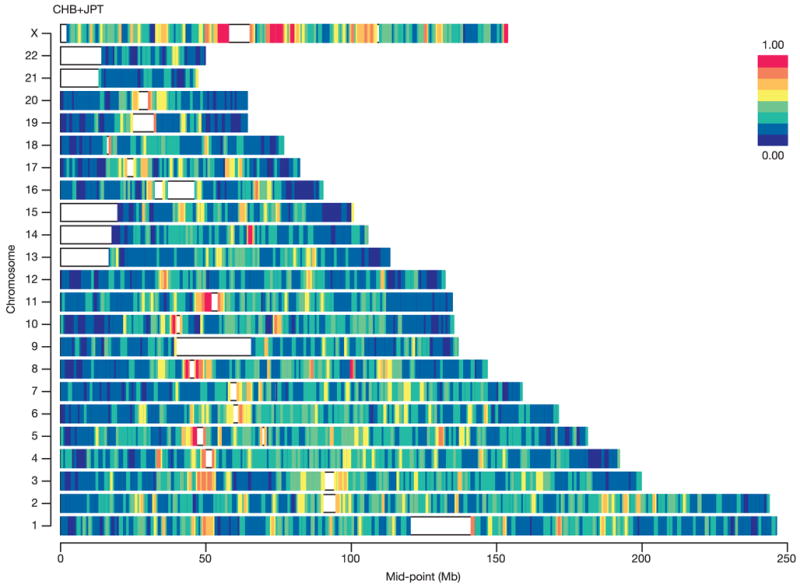
We fitted a simple model for the decay of linkage disequilibrium103 to windows of 1 million bases distributed throughout the genome. The results of model fitting are summarized for the CHB+JPT analysis panel, by plotting the fitted r2 value for SNPs separated by 30 kb. The overall pattern of variation was very similar in the other analysis panels84 (see Supplementary Information).
We observe, for the first time, that LD tracks with both the density and functional classification of genes. We examined quartiles of the genome based on extent of LD, and looked for correlations to gene density. Surprisingly, we find that both the top and bottom quartiles of the genome have greater gene density as compared to the middle quartiles (6.7 as compared to 6.1 genes per Mb), as well as percentage of bases in codons (1.24% as compared to 1.08%). We have no explanation for this observation.
Although the majority of gene classes are equally divided between these two extreme quartiles of the genome, some classes of genes show a marked skew in their distribution64,84,85. Genes involved in immune responses and neurophysiological processes are more often located in regions of low LD, whereas genes involved in DNA and RNA metabolism, response to DNA damage and the cell cycle are preferentially located in regions of strong linkage disequilibrium. It is intriguing to speculate that the extent of LD (and sequence diversity) might track with gene function due to natural selection, with increased diversity being favoured in genes involved in interface with the environment such as the immune response86, and disadvantageous for core cell biological processes such as DNA repair and packaging87,88.
Natural selection
The preceding observation highlights the hypothesis that signatures of natural selection are present in the HapMap data. The availability of genome-wide variation data makes it possible to scan the genome for such signatures to discover genes that were subject to selection during human evolution89; the HapMap data also provide a genome-wide empirical distribution against which previous claims of selection can be evaluated (rather than relying solely on theoretical computer simulations).
Natural selection influences patterns of genetic variation in various ways, such as through the removal of deleterious mutations, the fixation of advantageous variants, and the maintenance of multiple alleles through balancing selection. Each form of selection may have occurred uniformly across the world (and thus be represented in all human populations) or have been geographically localized (and thus differ among populations).
Nearly all methods for recognizing natural selection rely on the collection of complete sequence data. The HapMap Project’s focus on common variation—and the process of SNP selection that achieved a preponderance of high-frequency alleles (Fig. 5)—thus prevents their straightforward application. Adjusting for the effect of SNP choice is complex, moreover, because SNP choice varied over time as dbSNP evolved, and was implemented locally at each centre.
For these reasons, we focus here on two types of analysis. First, we examined the distributions of signatures of selection across the genome. Although the absolute value of these measures is difficult to interpret (owing to SNP ascertainment), the most extreme cases in a genome-wide distribution are important candidates to evaluate for selection. Second, we compared across functional categories, because SNP choice was largely agnostic to such features, and thus systematic differences may be a sign of selection.
The outcomes of these analyses confirm a number of previous hypotheses about selection and identify new loci as candidates for selection.
Evidence for selective sweeps in particular genomic regions
First we consider population differentiation, generally accepted as a clue to past selection in one of the populations. The HapMap data reveal 926 SNPs with allele frequencies that differ across the analysis panels in a manner as extreme as the well-accepted example of selection at the Duffy (FY) locus (Supplementary Fig. 8c). Of these 926 SNPs, 32 are non-synonymous coding SNPs and many others occur in transcribed regions, making them strong candidates for functional polymorphisms that have experienced geographically restricted selection pressures (see Table 9 and Supplementary Information for details). In particular, the ALMS1 gene on chromosome 2 has six amino acid polymorphisms that show very strong population differentiation.
Table 9.
High-differentiation non-synonymous SNPs
| Chromosome | Position (base number) | Gene* | SNP |
|---|---|---|---|
| 1 | 54,772,383 | THEA | rs1702003 |
| 1 | 156,000,000 | FY | rs12075 |
| 1 | 244,000,000 | Q8NGY8_human† | rs7555046 |
| 2 | 3,184,917 | COLEC11 | rs7567833 |
| 2 | 73,563,622 | ALMS1 | rs3813227 |
| 2 | 73,589,553 | ALMS1 | rs6546837 |
| 2 | 73,591,645 | ALMS1 | rs6724782 |
| 2 | 73,592,163 | ALMS1 | rs6546839 |
| 2 | 73,629,222 | ALMS1 | rs2056486 |
| 2 | 73,629,311 | ALMS1 | rs10193972 |
| 2 | 109,000,000 | EDAR | rs3827760 |
| 3 | 182,000,000 | FXR1 | rs11499 |
| 3 | 185,000,000 | MCF2L2 | rs7639705 |
| 4 | 41,844,599 | SLC30A9 | rs1047626 |
| 4 | 46,567,077 | ENSG00000172895.1 | rs5825 |
| 4 | 101,000,000 | ADH1B | rs1229984 |
| 8 | 10,517,787 | RP1L1 | rs6601495 |
| 8 | 146,000,000 | SLC39A4 | rs1871534 |
| 10 | 50,402,145 | ERCC6 | rs4253047 |
| 10 | 71,002,210 | NEUROG3 | rs4536103 |
| 11 | 46,701,579 | F2 | rs5896 |
| 15 | 46,213,776 | SLC24A5 | rs1426654 |
| 15 | 61,724,262 | HERC1 | rs7162473 |
| 16 | 30,996,126 | ZNF646 | rs749670 |
| 16 | 46,815,699 | ABCC11 | rs17822931 |
| 17 | 26,322,430 | RNF135 | rs7225888 |
| 17 | 26,399,303 | ENSG00000184253.2 | rs6505228 |
| 18 | 66,022,323 | RTTN | rs3911730 |
| 19 | 5,782,891 | FUT6 | rs364637 |
| 19 | 47,723,209 | CEACAM1 | rs8110904 |
| 22 | 18,164,095 | GNB1L | rs2073770 |
| X | 65,608,007 | EDA2R | rs1385699 |
Where no standard gene abbreviation exists, the ENSEMBL gene ID has been given.
It is unclear from current annotations whether this is a pseudogene.
Another signature of an allele having risen to fixation through selection is that all other diversity in the region is eliminated (known as a selective sweep). We identified extreme outliers in the joint distribution of heterozygosity (as assessed from shotgun sequencing SNP discovery projects) and either population differentiation or skewing of allele frequency towards rare alleles in each analysis panel (Supplementary Fig. 15). We identified 19 such genomic regions (13 on autosomes, 6 on the X chromosome) as candidates for future study (Supplementary Table 4); these include candidates for population-specific sweeps and sweeps in the ancestral population. Encouragingly, this analysis includes among its top-scoring results the LCT gene, which influences the ability to digest dairy products90 and has been shown to be subject to past natural selection91.
Long haplotypes as candidates for natural selection
Selective sweeps that fail to fix in the population, as well as balancing selection, lead to haplotypes that are relatively high in frequency and long in duration. In the HLA region (which is widely believed to have been influenced by balancing selection) multiple haplotypes of 500 SNPs that extend more than 1 cM in length are observed with a frequency in the HapMap samples of more than 1%. We identified other such occurrences of long haplotypes across the genome (Supplementary Fig. 8 and Supplementary Tables 5 and 6).
An approach to long haplotypes designed specifically to identify regions having undergone partial selective sweeps is the long range haplotype (LRH) test91,92, which compares the length of each haplo-type to that of others at the locus, matched across the genome based on frequency. Previously identified outliers to the genome-wide distribution for the LRH test (Fig. 16) that have been identified as candidates for selection include the LCT gene in the CEU sample (empirical P-value = 1.3 × 10−9), which was an outlier for the heterozygosity/allele frequency test above, and the HBB gene (empirical P-value = 1.39 × 10−5) in the YRI sample. However, most of the strongest signals in the LRH test (Table 10) were not previously hypothesized as undergoing selection.
Figure 16. The distribution of the long range haplotype (LRH92) test statistic for natural selection.
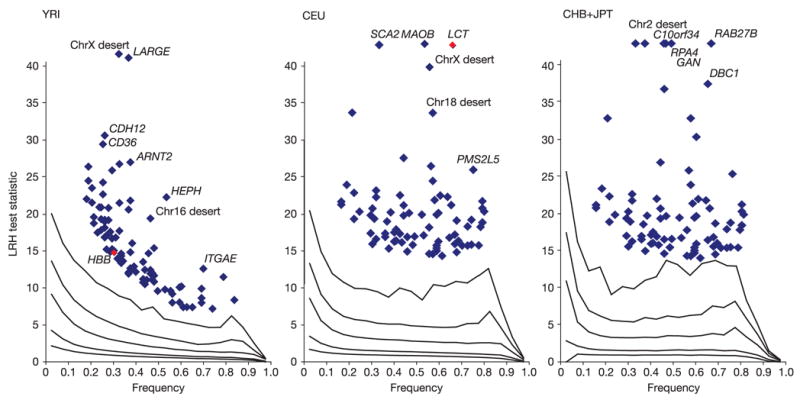
In the YRI analysis panel, diversity around the HBB gene is highlighted by the red point. In the CEU analysis panel, diversity within the LCT gene region is similarly highlighted.
Table 10.
Candidate loci in which selection occurred
| Chromosome | Position (base number) at centre | Genes in region | Population | Haplotype frequency | Empirical P-value |
|---|---|---|---|---|---|
| 2 | 137,224,699 | LCT | CEU | 0.65 | 1.25 × 10−9 |
| 5 | 22,296,347 | CDH12, PMCHL1 | YRI | 0.25 | 5.77 × 10−8 |
| 7 | 79,904,387 | CD36 | YRI | 0.24 | 2.72 × 10−6 |
| 7 | 73,747,934 | PMS2L5, WBSCR16 | CEU | 0.76 | 3.37 × 10−6 |
| 12 | 109,892,896 | CUTL2 | CEU | 0.36 | 7.95 × 10−9 |
| 15 | 78,558,508 | ARNT2 | YRI | 0.32 | 6.92 × 10−7 |
| 16 | 75,661,011 | Desert | YRI | 0.46 | 5.01 × 10−7 |
| 17 | 3,945,580 | ITGAE, GSG2, HSA277841, CAMKK1, P2RX1 | YRI | 0.70 | 9.26 × 10−7 |
| 18 | 24,502,756 | Desert | CEU | 0.57 | 2.23 × 10−7 |
| 22 | 32,459,471 | LARGE | YRI | 0.36 | 7.82 × 10−9 |
| X | 20,171,291 | Desert | YRI | 0.33 | 5.02 × 10−9 |
| X | 64,323,320 | HEPH | YRI | 0.55 | 3.02 × 10−8 |
| X | 42,763,073 | MAOB | CEU | 0.53 | 4.21 × 10−9 |
| X | 34,399,948 | Desert | CEU | 0.57 | 8.85 × 10−8 |
These four tests overlap only partially in the hypotheses they address—heterozygosity, for example, is sensitive to older sweeps, whereas the haplotype tests are most powerful for partial sweeps—but encouragingly some candidate regions are found by more than one test. In particular, six regions are identified both by long haplotypes and by low heterozygosity, and three regions (LCT on chromosome 2, and two regions on the X chromosome at 20 and 65 Mb) are identified by three different tests.
Confirming purifying selection at conserved non-coding elements
Finally, we used the HapMap data to test an important hypothesis from comparative genomics. Genomic sequencing has shown that about 5% of the human sequence is highly conserved across species, yet less than half of this sequence spans known functional elements such as exons45. It is widely assumed that conserved non-genic sequences lack diversity because of selective constraint (that is, purifying selection), but such regions may simply be coldspots for mutation, and thus be of little value as candidates for functional study.
Analysis of allele frequencies helps to resolve this uncertainty. Functional constraint, but not a low mutation rate, results in a downward skew in allele frequencies for conserved sequences as compared to neutral sequences93,94. We find that conserved non-genic sequences display a greater skew towards rare alleles than do intergenic regions, as predicted under purifying selection. This skew is less extreme than that observed for exons (Supplementary Fig. 16), reflecting either stronger purifying selection or the prioritization of coding SNPs for genotyping by the HapMap centres regardless of validation status. This novel evidence for ongoing constraint shows that conserved non-genic sequences are not mutational coldspots, and thus remain of high interest for functional study.
Conclusions
The International HapMap Project set out to create a resource that would accelerate the identification of genetic factors that influence medical traits. Analyses reported here confirm the generality of hotspots of recombination, long segments of strong LD, and limited haplotype diversity. Most important is the extensive redundancy among nearby SNPs, providing (1) the potential to extract extensive information about genomic variation without complete resequencing, and (2) efficiencies through selection of tag SNPs and optimized association analyses. Beyond the biomedical context, these data have made it possible to identify deletion variants in the genome, explore the nature of fine-scale recombination and identify regions that may have been subject to natural selection.
The HapMap Project (along with a previous genome-wide assessment of LD77) is a natural extension of the Human Genome Project. Where the reference sequence constructed by the Human Genome Project is informative about the vast majority of bases that are invariant across individuals, the HapMap focuses on DNA sequence differences among individuals. Our understanding of SNP variation and LD around common variants in the sampled populations is reasonably complete; the current picture is unlikely to change with additional data. In other aspects—such as the fine details of local correlation among SNPs, rarer alleles, structural variants, and inter-population differences—these resources are only a first step on the path towards a complete characterization of genetic variation of the human population. Planned extensions of the Phase I map include Phase II of HapMap, with genotyping of another 4.6 million SNPs attempted in the HapMap samples, and detailed genotyping of the HapMap ENCODE regions in additional members of each HapMap population sampled, as well as in samples from additional populations. These results should guide understanding of the robustness and transferability of LD inferences and tag SNPs selected from the current set of HapMap samples.
An important application of the HapMap data is to help make possible comprehensive, genome-wide association studies. There are now laboratory tools that make it practical to undertake such studies, and initial results are encouraging13. Given the low prior probability of causality for each SNP in the genome, however, rigorous standards of statistical significance will be needed to avoid a flood of false-positive results. Multiple replications in large samples provide the most straightforward path to identifying robust and broadly relevant associations. Given the potential for confusion if associations of uncertain validity are widely reported (and a persistent tendency towards genetic determinism in public discourse), we urge conservatism and restraint in the public dissemination and interpretation of such studies, especially if non-medical phenotypes are explored. It is time to create mechanisms by which all results of association studies, positive and negative, are reported and discussed without bias.
The success of the HapMap will be measured in terms of the genetic discoveries enabled, and improved knowledge of disease aetiology. Specifically, identifying which genes and pathways are causal in humans has the potential to provide a new and solid foundation for biomedical research. This is equally true whether the variants that lead to the discovery of those genes are themselves rare or common, or of large or small effect. The impact on diagnostics and targeted prevention, however, will depend on how predictive each given allele may be. Where genetic mechanisms underlie treatment responses, both more efficient trials and individualized preventive and treatment strategies may become practical95.
Success identifying alleles conferring susceptibility or resistance to common diseases will also provide a deeper understanding of the architecture of disease: how many genes are involved in each case, whether and how alleles interact with one another96 and with environmental exposures to shape clinical phenotypes. In this regard, it will be important to invest heavily in the discovery and characterization of relevant lifestyle factors, environmental exposures, detailed characterization of clinical phenotypes, and the ability to obtain such information in longitudinal studies of adequate size. Where environmental and behavioural factors vary across studies, replication will be hard to come by (as will clinical utility) unless we can learn to capture these variables with the same precision and completeness as genotypic variation. Technological innovation and international collaboration in these realms will probably be required (as they have been in the Human Genome Project and the HapMap) to advance the shared goal of understanding, and ultimately preventing, common human diseases.
METHODS
The project was undertaken by investigators from Japan, the United Kingdom, Canada, China, Nigeria and the United States, and from multiple disciplines: sample collection, sequencing and genotyping, bioinformatics, population genetics, statistics, and the ethical, legal, and social implications of genetic research. The Supplementary Information contains information about project participants and organization.
Choice of DNA samples
Any choice of DNA samples represents a compromise: a single population offers simplicity, but cannot be representative, whereas grid-sampling is representative of the current worldwide population, but is neither practical nor captures historical genetic diversity. The project chose to include DNA samples based on well-known patterns of allele frequencies across populations41, reflecting historical genetic diversity31,32.
For practical reasons, the project focused on SNPs present at a minor allele frequency (MAF) ≥ 0.05 in each analysis panel, and thus studied a sufficient number of individuals to provide good power for this frequency range31. Cell lines and DNA are available at the Coriell Institute for Medical Research (http://locus.umdnj.edu/nigms/products/hapmap.html).
Community engagement was employed to explain the project, and to learn how the project was viewed, in the communities where samples were collected31,32. Papers describing the community engagement processes are being prepared.
One JPT sample was replaced for technical reasons, but not in time for inclusion in this report. We surveyed cryptic relatedness among the study participants, and identified a small number of pairs with unexpectedly high allele sharing (Supplementary Information). As the total level of sharing is not great, and as a subset of analyses performed without these individuals were unchanged, we include these individuals in the data and analyses presented here.
Genome-wide SNP discovery
The project required a dense map of SNPs, ideally containing information about validation and frequency of each candidate SNP. When the project started, the public SNP database (dbSNP) contained 2.6 million candidate SNPs, few of which were annotated with the required information.
To generate more SNPs and obtain validation information, shotgun sequencing of DNA from whole-genome libraries and flow-sorted chromosomes was performed31, augmented by analysis of sequence traces produced by Applied Biosystems97,98, and information on 1.6 million SNPs genotyped by Perlegen Sciences77, including 425,000 not in dbSNP when released (Supplementary Table 7). The HapMap Project contributed about 6 million new SNPs to dbSNP.
At the time of writing (October 2005) dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) contains 9.2 million candidate human SNPs, of which 3.6 million have been validated by both alleles having been seen two or more times during discovery (‘double-hit’ SNPs), and 2.4 million have genotype data (Fig. 1).
Comprehensive study of common variation across 5 Mb of DNA
To study patterns of genetic variation as comprehensively as possible, we selected ten 500-kb regions from the ENCODE Project33. These ten regions were chosen in aggregate to approximate the genome-wide average for G+C content, recombination rate, percentage of sequence conserved relative to mouse sequence, and gene density (Table 2).
In each such region additional sequencing and genotyping were performed to obtain a much more complete inventory of common variation. Specifically, bidirectional PCR-based sequencing was performed across each 500-kb region in 48 individuals (16 YRI, 16 CEU, 8 CHB, 8 JPT). Although the intent was for these same DNA samples to be included in Phase I, eight Yoruba and one Han Chinese sample used in sequencing were not among the 269 samples genotyped. (The nine samples are available from Coriell.)
All variants found by sequencing, and any others in dbSNP (build 121) not found by sequencing, were genotyped in all 269 HapMap samples. If the first attempt at genotyping was unsuccessful, a second platform was tried for each SNP.
False-positive and false-negative rates in PCR-based SNP discovery
The false-positive rate of SNP discovery by PCR-based resequencing was estimated at 7–11% (for the two sequencing centres), based on genotyping of each candidate SNP in the same samples used for discovery.
The false-negative rate of SNP discovery by PCR resequencing was estimated at 6%, using as the denominator a set of SNPs previously in dbSNP and confirmed by genotyping in the specific individuals sequenced. The false-negative rate was considerably higher, however, for singletons (SNPs seen only as a single heterozygote): 15% of singletons covered by high-quality sequence data were not detected by the trace analysis, and another 25% were missed due to a failure to obtain a high-quality sequence over the relevant base in the one heterozygous individual (D. J. Richter et al., personal communication).
False-positive and false-negative rates in dbSNP
The false positive rate (candidate SNPs that cannot be confirmed as variable sites) estimated for dbSNP was 17%. This represents an upper bound, because dbSNP entries that are monomorphic in the 269 HapMap samples could be rare variants, or polymorphic in other samples. We note that as the catalogue of dbSNP gets deeper, the rate at which candidate SNPs are monomorphic in any given sample is observed to rise (Supplementary Table 8). This is expected because the number of rare SNPs and false positives scales with depth of sequencing, whereas the number of true common variants will plateau.
SNP genotyping for the genome-wide map
Genotyping assays were designed from dbSNP, with priority given to SNPs validated by previous genotyping data or both alleles having been seen more than once in discovery. Data from the Chimpanzee Genome Sequencing Project99 were used in SNP validation if they confirmed the ancestral status of a human allele seen only once in discovery (Supplementary Information). Non-synonymous coding SNPs were also prioritized for genotyping. Two whole-genome, array-based genotyping reagents were used efficiently to increase SNP density: 40,000 SNPs from Illumina, and 120,000 SNPs from Affymetrix100.
To monitor progress, the genome was partitioned into 5-kb bins, with genotyping continuing through iterative rounds until a set of predetermined ‘stopping rules’ was satisfied in each analysis panel. (1) Minor allele frequency: in each analysis panel a common SNP (MAF ≥ 0.05) was obtained in each 5-kb bin. (2) Spacing: the distance between adjacent SNPs was 2–8 kb, with at least 9 SNPs across 50 kb. (3) ‘HapMappable’ genome: with available technologies it is challenging to study centromeres, telomeres, gaps in genome sequence, and segmental duplications. The project identified such regions101 (Supplementary Table 9), spanning 4.4% of the finished human genome sequence, in which only a single attempt to develop a genotyping assay was required. (4) Three strikes, you’re out: if the above rules were not satisfied after three attempts to develop an assay in a given 5-kb region, or if all available SNPs in dbSNP had been tried, genotyping was considered complete for Phase I. Two attempts were considered sufficient if one attempt was of a SNP previously shown to have MAF ≥ 0.05 in the appropriate population sample in a previous genome-wide survey77. (5) Quality control: ongoing and standardized quality control (QC) filters and three rounds of quality assessment (QA) were used to ensure and document the high quality of the genotype data.
QC filters were systematically performed, with each SNP tested for completeness (>80%), consistency across five duplicate genotypes (≤1 discrepancy), mendelian inheritance in 60 trios (≤1 discrepancy in each of YRI and CEU), and Hardy–Weinberg equilibrium (P > 0.001102). SNPs in the Phase I data set passed all the QC filters in all the analysis panels and were polymorphic in the HapMap samples. Failing SNPs were released (with a special flag), as they can help to identify polymorphisms under primers, insertions/deletions, paralogous loci and natural selection.
Three QA exercises were carried out. First, a calibration exercise to ‘benchmark’ each platform and laboratory protocol. Second, a mid-project evaluation of each genotyping centre. Third, a blind analysis of a random sample of the complete Phase I data set. A number of SNPs were genotyped more than once during the project, or by other investigators, providing additional information about data quality. See the Supplementary Information for full information about the QA exercises.
An exhaustive approach was taken to mtDNA. Alignment of more than 1,000 publicly available mtDNA sequences of African (n = 87), European (n = 928) and Asian (n = 238) geographical origin34 was used to identify 210 common variants (MAF ≥ 0.05 in at least one continental region) that were attempted in the samples.
Data release
Data deposited at the Data Coordination Center and released at http://www.hapmap.org, a Japanese mirror site http://hapmap.jst.go.jp/ and dbSNP include ascertainment status of each SNP at the time of selection, primer sequences, protocols for genotyping, genotypes for each sample, allele frequencies, and, for SNPs that failed QC filters, a code indicating the mode(s) of failure.
Initially, because of concern that third parties might seek patents on HapMap data, users were required to agree to a web-based ‘click-wrap license’, assenting that they would not prevent others from using the data (http://www.hapma-p.org/cgi-perl/registration). In December 2004 this license was dropped, and all data were released without restriction into the public domain.
Acknowledgments
We thank many people who contributed to this project: J. Beck, C. Beiswanger, D. Coppock, A. Leach, J. Mintzer and L. Toji (Coriell Institute for Medical Research) for transforming the Yoruba, Japanese and Han Chinese samples, distributing the DNA and cell lines, storing the samples for use in future research, and producing the community newsletters and reports; J. Greenberg and R. Anderson (NIH National Institute of General Medical Sciences) for providing funding and support for cell line transformation and storage in the NIGMS Human Genetic Cell Repository at the Coriell Institute; T. Dibling, T. Ishikura, S. Kanazawa, S. Mizusawa and S. Saito (SNP Research Center, RIKEN) for help with genotyping; C. Hind and A. Moghadam for technical support in genotyping and all members of the subcloning and sequencing teams at the Wellcome Trust Sanger Institute; X. Ke (Wellcome Trust Centre for Human Genetics at the University of Oxford) for help with data analysis; Oxford E-Science Centre for provision of high-performance computing resources; H. Chen, W. Chen, L. Deng, Y. Dong, C. Fu, L. Gao, H. Geng, J. Geng, M. He, H. Li, H. Li, S. Li, X. Li, B. Liu, Z. Liu, F. Lu, F. Lu, G. Lu, C. Luo, X. Wang, Z. Wang, C. Ye and X. Yu (Beijing Genomics Institute) for help with genotyping and sample collection; X. Feng, Y. Li, J. Ren and X. Zhou (Beijing Normal University) for help with sample collection; J. Fan, W. Gu, W. Guan, S. Hu, H. Jiang, R. Lei, Y. Lin, Z. Niu, B. Wang, L. Yang, W. Yang, Y. Wang, Z. Wang, S. Xu, W. Yan, H. Yang, W. Yuan, C. Zhang, J. Zhang, K. Zhang and G. Zhao (Chinese National Human Genome Center at Shanghai) for help with genotyping; P. Fong, C. Lai, C. Lau, T. Leung, L. Luk and W. Tong (University of Hong Kong, Genome Research Centre) for help with genotyping; C. Pang (Chinese University of Hong Kong) for help with genotyping; K. Ding, B. Qiang, J. Zhang, X. Zhang and K. Zhou (Chinese National Human Genome Center at Beijing) for help with genotyping; Q. Fu, S. Ghose, X. Lu, D. Nelson, A. Perez, S. Poole, R. Vega and H. Yonath (Baylor College of Medicine); C. Bruckner, T. Brundage, S. Chow, O. Iartchouk, M. Jain, M. Moorhead and K. Tran (ParAllele Bioscience Inc.); N. Addleman, J. Atilano, T. Chan, C. Chu, C. Ha, T. Nguyen, M. Minton and A. Phong (UCSF) for help with genotyping, and D. Lind (UCSF) for help with quality control and experimental design; R. Donaldson and S. Duan (Washington University) for help with genotyping, and J. Rice and N. Saccone (Washington University) for help with experimental design; J. Wigginton (University of Michigan) for help with implementing and testing QA/QC software; A. Clark, B. Keats, R. Myers, D. Nickerson and A. Williamson for providing advice to NIH; J. Melone, M. Weiss and E. DeHaut-Combs (NHGRI) for help with project management; M. Gray for organizing phone calls and meetings; D. Leja for help with figures; the Yoruba people of Ibadan, Nigeria, the people of Tokyo, Japan, and the community at Beijing Normal University, who participated in public consultations and community engagements; the people in these communities who were generous in donating their blood samples; and the people in the Utah CEPH community who allowed the samples they donated earlier to be used for the Project. We also thank A. Clark, E. Lander, C. Langley and R. Lifton for comments on earlier drafts of the manuscript. This work was supported by the Japanese Ministry of Education, Culture, Sports, Science, and Technology, the Wellcome Trust, Nuffield Trust, Wolfson Foundation, UK EPSRC, Genome Canada, Génome Québec, the Chinese Academy of Sciences, the Ministry of Science and Technology of the People’s Republic of China, the National Natural Science Foundation of China, the Hong Kong Innovation and Technology Commission, the University Grants Committee of Hong Kong, the SNP Consortium, the US National Institutes of Health (FIC, NCI, NCRR, NEI, NHGRI, NIA, NIAAA, NIAID, NIAMS, NIBIB, NIDA, NIDCD, NIDCR, NIDDK, NIEHS, NIGMS, NIMH, NINDS, NLM, OD), the W.M. Keck Foundation, and the Delores Dore Eccles Foundation.
The International HapMap Consortium
(Participants are arranged by institution, listed alphabetically, and then alphabetically within institutions except for Principal Investigators and Project Leaders, as indicated.)
Genotyping centres
Baylor College of Medicine and ParAllele BioScience Richard A. Gibbs (Principal Investigator)1, John W. Belmont1, Andrew Boudreau2, Suzanne M. Leal1, Paul Hardenbol2, Shiran Pasternak1, David A. Wheeler1, Thomas D. Willis2, Fuli Yu1; Beijing Genomics Institute Huanming Yang (Principal Investigator)3, Changqing Zeng (Principal Investigator)3, Yang Gao3, Haoran Hu3, Weitao Hu3, Chaohua Li3, Wei Lin3, Siqi Liu3, Hao Pan3, Xiaoli Tang3, Jian Wang3, Wei Wang3, Jun Yu3, Bo Zhang3, Qingrun Zhang3, Hongbin Zhao3, Hui Zhao3, Jun Zhou3; Broad Institute of Harvard and Massachusetts Institute of Technology Stacey B. Gabriel (Project Leader)4, Rachel Barry4, Brendan Blumenstiel4, Amy Camargo4, Matthew Defelice4, Maura Faggart4, Mary Goyette4, Supriya Gupta4, Jamie Moore4, Huy Nguyen4, Robert C. Onofrio4, Melissa Parkin4, Jessica Roy4, Erich Stahl4, Ellen Winchester4, Liuda Ziaugra4, David Altshuler (Principal Investigator)4,5; Chinese National Human Genome Center at Beijing Yan Shen (Principal Investigator)6, Zhijian Yao6; Chinese National Human Genome Center at Shanghai Wei Huang (Principal Investigator)7, Xun Chu7, Yungang He7, Li Jin7, Yangfan Liu7, Yayun Shen7, Weiwei Sun7, Haifeng Wang7, Yi Wang7, Ying Wang7, Ying Wang7, Xiaoyan Xiong7, Liang Xu7; Chinese University of Hong Kong Mary M. Y. Waye (Principal Investigator)8, Stephen K. W. Tsui8; Hong Kong University of Science and Technology Hong Xue (Principal Investigator)9, J. Tze-Fei Wong9; Illumina Launa M. Galver (Project Leader)10, Jian-Bing Fan10, Sarah S. Murray10, Arnold R. Oliphant11, Mark S. Chee (Principal Investigator)12; McGill University and Génome Québec Innovation Centre Alexandre Montpetit (Project Leader)13, Fanny Chagnon13, Vincent Ferretti13, Martin Leboeuf13, Jean-François Olivier13, Michael S. Phillips13, Stéphanie Roumy13, Clémentine Sallée14, Andrei Verner13, Thomas J. Hudson (Principal Investigator)13; Perlegen Sciences Kelly A. Frazer (Principal Investigator)15, Dennis G. Ballinger15, David R. Cox15, David A. Hinds15, Laura L. Stuve15; University of California at San Francisco and Washington University Pui-Yan Kwok (Principal Investigator)16, Dongmei Cai16, Daniel C. Koboldt17, Raymond D. Miller17, Ludmila Pawlikowska16, Patricia Taillon-Miller17, Ming Xiao16; University of Hong Kong Lap-Chee Tsui (Principal Investigator)18, William Mak18, Pak C. Sham18, You Qiang Song18, Paul K. H. Tam18; University of Tokyo and RIKEN Yusuke Nakamura (Principal Investigator)19,20, Takahisa Kawaguchi20, Takuya Kitamoto20, Takashi Morizono20, Atsushi Nagashima20, Yozo Ohnishi20, Akihiro Sekine20, Toshihiro Tanaka20, Tatsuhiko Tsunoda20; Wellcome Trust Sanger Institute Panos Deloukas (Project Leader)21, Christine P. Bird21, Marcos Delgado21, Emmanouil T. Dermitzakis21, Rhian Gwilliam21, Sarah Hunt21, Jonathan Morrison21, Don Powell21, Barbara E. Stranger21, Pamela Whittaker21, David R. Bentley (Principal Investigator)22
Analysis groups
Broad Institute of Harvard and Massachusetts Institute of Technology Mark J. Daly (Project Leader)4,5, Paul I. W. de Bakker4,5, Jeff Barrett4,5, Ben Fry4, Julian Maller4,5, Steve McCarroll4,5, Nick Patterson4, Itsik Pe’er4,5, Shaun Purcell5, Daniel J. Richter4, Pardis Sabeti4, Richa Saxena4,5, Stephen F. Schaffner4, Patrick Varilly4, David Altshuler (Principal Investigator)4,5; Cold Spring Harbor Laboratory Lincoln D. Stein (Principal Investigator)23, Lalitha Krishnan23, Albert Vernon Smith23, Gudmundur A. Thorisson23; Johns Hopkins University School of Medicine Aravinda Chakravarti (Principal Investigator)24, Peter E. Chen24, David J. Cutler24, Carl S. Kashuk24, Shin Lin24; University of Michigan Gonçalo R. Abecasis (Principal Investigator)25, Weihua Guan25, Heather M. Munro25, Zhaohui Steve Qin25, Daryl J. Thomas26; University of Oxford Gilean McVean (Project Leader)27, Leonardo Bottolo27, Susana Eyheramendy27, Colin Freeman27, Jonathan Marchini27, Simon Myers27, Chris Spencer27, Matthew Stephens28, Peter Donnelly (Principal Investigator)27; University of Oxford, Wellcome Trust Centre for Human Genetics Lon R. Cardon (Principal Investigator)29, Geraldine Clarke29, David M. Evans29, Andrew P. Morris29, Bruce S. Weir30; RIKEN Tatsuhiko Tsunoda20; US National Institutes of Health James C. Mullikin31, Stephen T. Sherry32, Michael Feolo32
Community engagement/public consultation and sample collection groups
Beijing Normal University and Beijing Genomics Institute Houcan Zhang33, Changqing Zeng3, Hui Zhao3; Health Sciences University of Hokkaido, Eubios Ethics Institute, and Shinshu University Ichiro Matsuda (Principal Investigator)34, Yoshimitsu Fukushima35, Darryl R. Macer36, Eiko Suda37; Howard University and University of Ibadan Charles N. Rotimi (Principal Investigator)38, Clement A. Adebamowo39, Ike Ajayi39, Toyin Aniagwu39, Patricia A. Marshall40, Chibuzor Nkwodimmah39, Charmaine D. M. Royal38; University of Utah Mark F. Leppert (Principal Investigator)41, Missy Dixon41, Andy Peiffer41
Ethical, legal and social issues
Chinese Academy of Social Sciences Renzong Qiu42; Genetic Interest Group Alastair Kent43; Kyoto University Kazuto Kato44; Nagasaki University Norio Niikawa45; University of Ibadan School of Medicine Isaac F. Adewole39; University of Montréal Bartha M. Knoppers14; University of Oklahoma Morris W. Foster46; Vanderbilt University Ellen Wright Clayton47; Wellcome Trust Jessica Watkin48
SNP discovery
Baylor College of Medicine Richard A. Gibbs (Principal Investigator)1, John W. Belmont1, Donna Muzny1, Lynne Nazareth1, Erica Sodergren1, George M. Weinstock1, David A. Wheeler1, Imtiaz Yakub1; Broad Institute of Harvard and Massachusetts Institute of Technology Stacey B. Gabriel (Project Leader)4, Robert C. Onofrio4, Daniel J. Richter4, Liuda Ziaugra4, Bruce W. Birren4, Mark J. Daly4,5, David Altshuler (Principal Investigator)4,5; Washington University Richard K. Wilson (Principal Investigator)49, Lucinda L. Fulton49; Wellcome Trust Sanger Institute Jane Rogers (Principal Investigator)21, John Burton21, Nigel P. Carter21, Christopher M. Clee21, Mark Griffiths21, Matthew C. Jones21, Kirsten McLay21, Robert W. Plumb21, Mark T. Ross21, Sarah K. Sims21, David L. Willey21
Scientific management
Chinese Academy of Sciences Zhu Chen50, Hua Han50, Le Kang50; Genome Canada Martin Godbout51, John C. Wallenburg52; Génome Québec Paul L’ Archevêque53, Guy Bellemare53; Japanese Ministry of Education, Culture, Sports, Science and Technology Koji Saeki54; Ministry of Science and Technology of the People’s Republic of China Hongguang Wang55, Daochang An55, Hongbo Fu55, Qing Li55, Zhen Wang55; The Human Genetic Resource Administration of China Renwu Wang56; The SNP Consortium Arthur L. Holden57; US National Institutes of Health Lisa D. Brooks58, Jean E. McEwen58, Christianne R. Bird58, Mark S. Guyer58, Patrick J. Nailer58, Vivian Ota Wang58, Jane L. Peterson58, Michael Shi59, Jack Spiegel60, Lawrence M. Sung61, Jonathan Witonsky62, Lynn F. Zacharia58, Francis S. Collins63; Wellcome Trust Karen Kennedy48, Ruth Jamieson48 & John Stewart48
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions David Altshuler, Lisa D. Brooks, Aravinda Chakravarti, Francis S. Collins, Mark J. Daly and Peter Donnelly are members of the writing group responsible for this manuscript.
Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare competing financial interests: details accompany the paper at www.nature.com/nature. Correspondence and requests for materials should be addressed to D.A. (altshuler@molbio.mgh.harvard.edu) or P.D. (donnelly@stats.ox.ac.uk).
Baylor College of Medicine, Human Genome Sequencing Center, Department of Molecular and Human Genetics, 1 Baylor Plaza, Houston, Texas 77030, USA.
ParAllele Bioscience, Inc., 7300 Shoreline Court, South San Francisco, California 94080, USA.
Beijing Genomics Institute, Chinese Academy of Sciences, Beijing 100300, China.
The Broad Institute of Harvard and Massachusetts Institute of Technology, 1 Kendall Square, Cambridge, Massachusetts 02139, USA.
Massachusetts General Hospital and Harvard Medical School, Simches Research Center, 185 Cambridge Street, Boston, Massachusetts 02114, USA.
Chinese National Human Genome Center at Beijing, 3-707 N. Yongchang Road, Beijing Economic-Technological Development Area, 100176, China.
Chinese National Human Genome Center at Shanghai, 250 Bi Bo Road, Shanghai 201203, China.
The Chinese University of Hong Kong, Department of Biochemistry, The Croucher Laboratory for Human Genomics, 6/F Mong Man Wai Building, Shatin, Hong Kong.
Hong Kong University of Science and Technology, Department of Biochemistry, Clear Water Bay, Kowloon, Hong Kong.
Illumina, 9885 Towne Centre Drive, San Diego, California 92121, USA.
15518 Markar Road, Poway, California 92064, USA.
Prognosys Biosciences, Inc., 4215 Sorrento Valley Boulevard, Suite 105, San Diego, California 92121, USA.
McGill University and Génome Québec Innovation Centre, 740 Drive Penfield Avnue, Montréal, Québec H3A 1A4, Canada.
University of Montréal, The Public Law Research Centre (CRDP), P.O. Box 6128, Downtown Station, Montréal, Québec H3C 3J7, Canada.
Perlegen Sciences, Inc., 2021 Stierlin Court, Mountain View, California 94043, USA.
University of California, San Francisco, Cardiovascular Research Institute, 513 Parnassus Avenue, Box 0793, San Francisco, California 94143, USA.
Washington University School of Medicine, Department of Genetics, 660 S. Euclid Avenue, Box 8232, St Louis, Missouri 63110, USA.
University of Hong Kong, Genome Research Centre, 6/F, Laboratory Block, 21 Sassoon Road, Pokfulam, Hong Kong.
University of Tokyo, Institute of Medical Science, 4-6-1 Sirokanedai, Minato-ku, Tokyo 108-8639, Japan.
RIKEN SNP Research Center, 1-7-22 Suehiro-cho, Tsurumi-ku Yokohama, Kanagawa 230-0045, Japan.
Wellcome Trust Sanger Institute, The Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK.
Solexa Ltd, Chesterford Research Park, Little Chesterford, Nr Saffron Walden, Essex CB10 1XL, UK.
Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor, New York 11724, USA.
Johns Hopkins University School of Medicine, McKusick-Nathans Institute of Genetic Medicine, Broadway Research Building, Suite 579, 733 N. Broadway, Baltimore, Maryland 21205, USA.
University of Michigan, Center for Statistical Genetics, Department of Biostatistics, 1420 Washington Heights, Ann Arbor, Michigan 48109, USA.
Center for Biomolecular Science and Engineering, Engineering 2, Suite 501, Mail Stop CBSE/ITI, UC Santa Cruz, Santa Cruz, California 95064, USA.
University of Oxford, Department of Statistics, 1 South Parks Road, Oxford OX1 3TG, UK.
University of Washington, Department of Statistics, Box 354322, Seattle, Washington 98195, USA.
University of Oxford/Wellcome Trust Centre for Human Genetics, Roosevelt Drive, Oxford OX3 7BN, UK.
North Carolina State University, Bioinformatics Research Center, Campus Box 7566, Raleigh, North Carolina 27695, USA.
US National Institutes of Health, National Human Genome Research Institute, 50 South Drive, Bethesda, Maryland 20892, USA.
US National Institutes of Health, National Library of Medicine, National Center for Biotechnology Information, 8600 Rockville Pike, Bethesda, Maryland 20894, USA.
Beijing Normal University, 19 Xinjiekouwai Street, Beijing 100875, China.
Health Sciences University of Hokkaido, Ishikari Tobetsu Machi 1757, Hokkaido 061-0293, Japan.
Shinshu University School of Medicine, Department of Medical Genetics, Matsumoto 390-8621, Japan.
United Nations Educational, Scientific and Cultural Organization (UNESCO Bangkok), 920 Sukhumwit Road, Prakanong, Bangkok 10110, Thailand.
University of Tsukuba, Eubios Ethics Institute, P.O. Box 125, Tsukuba Science City 305-8691, Japan.
Howard University, National Human Genome Center, 2216 6th Street, NW, Washington DC 20059, USA.
University of Ibadan College of Medicine, Ibadan, Oyo State, Nigeria.
Case Western Reserve University School of Medicine, Department of Bioethics, 10900 Euclid Avenue, Cleveland, Ohio 44106, USA.
University of Utah, Eccles Institute of Human Genetics, Department of Human Genetics, 15 North 2030 East, Salt Lake City, Utah 84112, USA.
Chinese Academy of Social Sciences, Center for Applied Ethics, 2121, Building 9, Caoqiao Xinyuan 3 Qu, Beijing 100054, China.
Genetic Interest Group, 4D Leroy House, 436 Essex Road, London N1 3QP, UK.
Kyoto University, Institute for Research in Humanities and Graduate School of Biostudies, Ushinomiya-cho, Sakyo-ku, Kyoto 606-8501, Japan.
Nagasaki University Graduate School of Biomedical Sciences, Department of Human Genetics, Sakamoto 1-12-4, Nagasaki 852-8523, Japan.
University of Oklahoma, Department of Anthropology, 455 W. Lindsey Street, Norman, Oklahoma 73019, USA.
Vanderbilt University, Center for Genetics and Health Policy, 507 Light Hall, Nashville, Tennessee 37232-0165, USA.
Wellcome Trust, 215 Euston Road, London NW1 2BE, UK.
Washington University School of Medicine, Genome Sequencing Center, Box 8501, 4444 Forest Park Avenue, St Louis, Missouri 63108, USA.
Chinese Academy of Sciences, 52 Sanlihe Road, Beijing 100864, China.
Genome Canada, 150 Metcalfe Street, Suite 2100, Ottawa, Ontario K2P 1P1, Canada.
McGill University, Office of Technology Transfer, 3550 University Street, Montréal, Québec H3A 2A7, Canada.
Génome Québec, 630, boulevard René-Lévesque Ouest, Montréal, Québec H3B 1S6, Canada.
Ministry of Education, Culture, Sports, Science, and Technology, 3-2-2 Kasumigaseki, Chiyodaku, Tokyo 100-8959, Japan.
Ministry of Science and Technology of the People’s Republic of China, 15 B. Fuxing Road, Beijing 100862, China.
The Human Genetic Resource Administration of China, b7, Zaojunmiao, Haidian District, Beijing 100081, China.
The SNP Consortium, 3 Parkway North, Deerfield, Illinois 60015, USA.
US National Institutes of Health, National Human Genome Research Institute, 5635 Fishers Lane, Suite 4076, Bethesda, Maryland 20892, USA.
Novartis Pharmaceuticals Corporation, Biomarker Development, One Health Plaza, East Hanover, New Jersey 07936, USA.
US National Institutes of Health, Office of Technology Transfer, 6011 Executive Boulevard, Rockville, Maryland 20852, USA.
University of Maryland School of Law, 500 W. Baltimore Street, Baltimore, Maryland 21201, USA.
Frost & Sullivan, 2400 Geng Road, Suite 201, Palo Alto, California 94303, USA.
US National Institutes of Health, National Human Genome Research Institute, 31 Center Drive, Bethesda, Maryland 20892, USA.
References
- 1.Lechler R, Warrens A. HLA in Health and Disease. 2. Academic Press; San Diego, California: 2005. [Google Scholar]
- 2.Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer’s disease. Annu Rev Neurosci. 1996;19:53–77. doi: 10.1146/annurev.ne.19.030196.000413. [DOI] [PubMed] [Google Scholar]
- 3.Dahlbäck B. Resistance to activated protein C caused by the factor V R506Q mutation is a common risk factor for venous thrombosis. Thromb Haemost. 1997;78:483–488. [PubMed] [Google Scholar]
- 4.Altshuler D, et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nature Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 5.Deeb SS, et al. A Pro12Ala substitution in PPARγ2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nature Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 6.Florez JC, Hirschhorn J, Altshuler D. The inherited basis of diabetes mellitus: implications for the genetic analysis of complex traits. Annu Rev Genomics Hum Genet. 2003;4:257–291. doi: 10.1146/annurev.genom.4.070802.110436. [DOI] [PubMed] [Google Scholar]
- 7.Begovich AB, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottini N, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nature Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 9.Bell GI, Horita S, Karam JH. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes. 1984;33:176–183. doi: 10.2337/diab.33.2.176. [DOI] [PubMed] [Google Scholar]
- 10.Ueda H, et al. Assocation of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 11.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 12.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 13.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 15.Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 16.Puffenberger EG, et al. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung’s disease. Cell. 1994;79:1257–1266. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 17.Emison ES, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 18.Kerem B, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 19.Hästbacka J, et al. Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nature Genet. 1992;2:204–211. doi: 10.1038/ng1192-204. [DOI] [PubMed] [Google Scholar]
- 20.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jorde LB. Linkage disequilibrium and the search for complex disease genes. Genome Res. 2000;10:1435–1444. doi: 10.1101/gr.144500. [DOI] [PubMed] [Google Scholar]
- 22.Reich DE, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 23.Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- 24.Johnson GC, et al. Haplotype tagging for the identification of common disease genes. Nature Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 25.Nickerson DA, et al. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nature Genet. 1998;19:233–240. doi: 10.1038/907. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X, et al. Localization of a small genomic region associated with elevated ACE. Am J Hum Genet. 2000;67:1144–1153. doi: 10.1016/s0002-9297(07)62945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nature Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 28.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 29.Patil N, et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294:1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 30.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 31.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789, 796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 32.The International HapMap Consortium. Integrating ethics and science in the International HapMap Project. Nature Rev Genet. 2004;5:467–475. doi: 10.1038/nrg1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 34.Herrnstadt C, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nature Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 36.The Y Chromosome Consortium. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Underhill PA, et al. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- 38.Marchini J, et al. A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet. doi: 10.1086/500808. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright S. Evolution and the Genetics of Populations Volume 2: the Theory of Gene Frequencies. Univ of Chicago Press; Chicago: 1969. pp. 294–295. [Google Scholar]
- 41.Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 42.Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pe’er I, et al. Reconciling estimates of linkage disequilibrium in the human genome. Genome Res. (submitted) [Google Scholar]
- 44.Lichten M, Goldman AS. Meiotic recombination hotspots. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 45.Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 46.McVean GA, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 47.Crawford DC, et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nature Genet. 2004;36:700–706. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- 48.Kong A, et al. A high-resolution recombination map of the human genome. Nature Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 49.Winckler W, et al. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- 50.Myers SR, Griffiths RC. Bounds on the minimum number of recombination events in a sample history. Genetics. 2003;163:375–394. doi: 10.1093/genetics/163.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phillips MS, et al. Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nature Genet. 2003;33:382–387. doi: 10.1038/ng1100. [DOI] [PubMed] [Google Scholar]
- 53.Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- 54.Carlson CS, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Bakker PIW, et al. Efficiency and power in genetic association studies. Nature Genet. doi: 10.1038/ng1669. Advance online publication, 23 October 2005. [DOI] [PubMed] [Google Scholar]
- 56.Lin S, Chakravarti A, Cutler DJ. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nature Genet. 2004;36:1181–1188. doi: 10.1038/ng1457. [DOI] [PubMed] [Google Scholar]
- 57.Weale ME, et al. Selection and evaluation of tagging SNPs in the neuronal-sodium-channel gene SCN1A: implications for linkage-disequilibrium gene mapping. Am J Hum Genet. 2003;73:551–565. doi: 10.1086/378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stram DO, et al. Choosing haplotype-tagging SNPs based on unphased genotype data using a preliminary sample of unrelated subjects with an example from the Multiethnic Cohort Study. Hum Hered. 2003;55:27–36. doi: 10.1159/000071807. [DOI] [PubMed] [Google Scholar]
- 59.de la Chapelle A, Wright FA. Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc Natl Acad Sci USA. 1998;95:12416–12423. doi: 10.1073/pnas.95.21.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mootha VK, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Engert JC, et al. ARSACS, a spastic ataxia common in northeastern Québec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nature Genet. 2000;24:120–125. doi: 10.1038/72769. [DOI] [PubMed] [Google Scholar]
- 62.Richter A, et al. Location score and haplotype analyses of the locus for autosomal recessive spastic ataxia of Charlevoix-Saguenay, in chromosome region 13q11. Am J Hum Genet. 1999;64:768–775. doi: 10.1086/302274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chakraborty R, Weiss KM. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci USA. 1988;85:9119–9123. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith MW, O’Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nature Rev Genet. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 65.Smith MW, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu X, et al. Admixture mapping for hypertension loci with genome-scan markers. Nature Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 67.Zhao X, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 68.Huang J, et al. Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum Genomics. 2004;1:287–299. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nature Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 70.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 71.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 72.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 73.Singleton AB, et al. α-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 74.McCarroll S, et al. Common deletion variants in the human genome. Nature Genet. (in the press) [Google Scholar]
- 75.Stefansson H, et al. A common inversion under selection in Europeans. Nature Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 76.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and recombination hotspots in the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 77.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 78.Yu A, et al. Comparison of human genetic and sequence-based physical maps. Nature. 2001;409:951–953. doi: 10.1038/35057185. [DOI] [PubMed] [Google Scholar]
- 79.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weissenbach J, et al. A second-generation linkage map of the human genome. Nature. 1992;359:794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- 81.Fullerton SM, Bernardo Carvalho A, Clark AG. Local rates of recombination are positively correlated with GC content in the human genome. Mol Biol Evol. 2001;18:1139–1142. doi: 10.1093/oxfordjournals.molbev.a003886. [DOI] [PubMed] [Google Scholar]
- 82.Dawson E, et al. A first-generation linkage disequilibrium map of human chromosome 22. Nature. 2002;418:544–548. doi: 10.1038/nature00864. [DOI] [PubMed] [Google Scholar]
- 83.Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 84.Smith AV, Thomas DJ, Munro HM, Abecasis GR. Sequence features in regions of weak and strong linkage disequilibrium. Genome Res. 2005;15:1519–1534. doi: 10.1101/gr.4421405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trachtenberg E, et al. Advantage of rare HLA supertype in HIV disease progression. Nature Med. 2003;9:928–935. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- 87.Pehrson JR, Fuji RN. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 1998;26:2837–2842. doi: 10.1093/nar/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 89.Nielsen R. Human genomics: disclosure of variation. Nature. 2005;434:288–289. doi: 10.1038/434288a. [DOI] [PubMed] [Google Scholar]
- 90.Enattah NS, et al. Identification of a variant associated with adult-type hypolactasia. Nature Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 91.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 93.Dermitzakis ET, et al. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature. 2002;420:578–582. doi: 10.1038/nature01251. [DOI] [PubMed] [Google Scholar]
- 94.Margulies EH, Blanchette M, Haussler D, Green ED. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Need AC, Motulsky AG, Goldstein DB. Priorities and standards in pharmacogenetic research. Nature Genet. 2005;37:671–681. doi: 10.1038/ng1593. [DOI] [PubMed] [Google Scholar]
- 96.Brem RB, Storey JD, Whittle J, Kruglyak L. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Istrail S, et al. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc Natl Acad Sci USA. 2004;101:1916–1921. doi: 10.1073/pnas.0307971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 99.The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 100.Matsuzaki H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nature Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 101.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 102.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy–Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hill WG, Weir BS. Maximum-likelihood estimation of gene location by linkage disequilibrium. Am J Hum Genet. 1994;54:705–714. [PMC free article] [PubMed] [Google Scholar]


