Abstract
The RNAs of many plant viruses lack a 5′ cap and must be translated by a cap-independent mechanism. Here, we discuss the remarkably diverse cap-independent translation elements that have been identified in members of the Potyviridae, Luteoviridae, and Tombusviridae families, and genus Tobamovirus. Many other plant viruses have uncapped RNAs but their translation control elements are uncharacterized. Cap-independent translation elements of plant viruses differ strikingly from those of animal viruses: they are smaller (<200 nt), some are located in the 3′ untranslated region, some require ribosome scanning from the 5′ end of the mRNA, and the 5′ UTR elements are much less structured than those of animal viruses. We discuss how these elements may interact with host translation factors, and speculate on their mechanism of action and their roles in the virus replication cycle. Much remains to be learned about how these elements enable plant viruses to usurp the host translational machinery.
Keywords: Viral genome-linked protein, Long-distance base pairing, Translation initiation factors, Internal ribosome entry site, Potyviridae, Luteoviridae, Tombusviridae
1. Introduction
Like viruses of all organisms, plant viruses must gain access to the host translation machinery. Because recognition of the 5′ cap structure on mRNA is a key step in cellular regulation of translation, viral mRNAs that translate in the absence of a 5′ cap can bypass host regulatory mechanisms to sustain translation. Thus, plant viruses in diverse families use cap-independent translation as a gene expression strategy. By understanding the mechanisms of plant viral RNA translation, we can gain insight on how to reduce crop losses due to plant virus infection. Also, plant virus-derived sequences can be useful tools to control transgene expression in genetically engineered plants. Plant viruses serve as excellent model viral systems and shed light on basic eukaryotic translation processes, owing to their small genomes, high titer, and lack of human pathogenicity. However, as is obvious in this review, many plant virus translation mechanisms are unknown in the animal virus world. This review summarizes what (little) is known about cap-independent translation of plant viruses and discusses the role of this property in viral life cycles.
Two features are necessary for efficient translation of cellular mRNAs: the 5′ m7G(5′)ppp(5′)N cap and the 3′ poly(A) tail. On cellular mRNAs, interaction of initiation factor 4E (eIF4E) with the cap structure (von der Haar et al., 2004), and the interaction of poly(A) binding protein (PABP) with the poly(A) tail facilitate translation initiation (Kahvejian et al., 2005). Through the large adapter protein, eIF4G, which binds eIF4E and PABP simultaneously, a closed loop complex is formed to bring the mRNA ends into proximity (Hentze, 1997; Sachs et al., 1997). This closed-loop is thought to increase the affinity of translation initiation factors, the ribosome, and the mRNA for each other, to initiate scanning (Pestova et al., 2001). In plants, eIF4E has two isoforms, eIF4E and eIFiso4E. Together with the two isoforms of eIF4G and eIFiso4G, respectively, these proteins complex to form eIF4F and eIFiso4F. The plant eIF4G isoforms are less similar to each other than the different forms of eIF4G in mammals or yeast are to each other. Wheat eIF4G (180 kDa) is twice as large as eIFiso4G (86 kDa). This large difference suggests there may be significant functional differences between the eIF4F isoforms in plants (Browning, 2004).
To compete with host mRNAs for the translation apparatus and circumvent host translational regulation, many viruses contain an internal ribosome entry site (IRES) in their 5′ untranslated region (UTR). IRESes obviate the need for a 5′ cap, recruiting the ribosome directly to the vicinity of the start codon without ribosome scanning from the 5′ end. In animal virus RNAs, IRESes are usually long (200–500 nt), structured and located in the 5′ UTR. By contrast, those in plants are smaller, less structured, and sometimes located in the 3′ UTR. Here, we discuss the known plant viral sequences that facilitate cap-independent translation (Table 1 and Fig. 1): (i) elements in the 5′ end (Potyviridae and other picorna-like viruses), (ii) inter-and intragenic elements of the Tobamovirus and Polerovirus genera, respectively, and (iii) elements in the 3′ UTRs of viruses in the Tombusviridae family and Luteovirus genus. Beyond the scope of this review are translation enhancer elements in capped RNAs such as those of Potato virus X (Smirnyagina et al., 1991) and Tobacco mosaic virus (Gallie and Walbot, 1992). Nor will we discuss elements that facilitate poly(A) tail-independent translation (Leathers et al., 1993; Neeleman et al., 2001).
Table 1.
Features of cap-independent translation elements on plant viral RNAsa
| Virus | 5′ end | 3′ end | Required viral element | Complementary to 18S rRNA | 5′–3′ base pairing | References |
|---|---|---|---|---|---|---|
| 5′ Elements | ||||||
| Potyviridae | ||||||
| Tobacco etch virus | VPg | Poly(A) | CIREs: PK1 and upstream Ω-like sequence | √ | Carrington and Freed (1990), Niepel and Gallie (1999), Zeenko and Gallie (2005) | |
| Plum pox virus | VPg | Poly(A) | 5′ UTR | Simón-Buela et al. (1997) | ||
| Potato virus Y | VPg | Poly(A) | 5′ UTR | Yang et al. (1997), Levis and Astier-Manifacier (1993) | ||
| Turnip mosaic virus | VPg | Poly(A) | 5′ UTR | Basso et al. (1994) | ||
| Comoviridae | ||||||
| Cowpea mosaic virus | VPg | Poly(A) | IRES | Thomas et al. (1991), Verver et al. (1991), Belsham and Lomonossoff (1991) | ||
| Intergenic elements | ||||||
| Luteoviridae | ||||||
| Potato leafroll virus | VPg | −OH | IRES | Jaag et al. (2003) | ||
| Tobamoviridae | ||||||
| Tobacco mosaic virus U1 | m7GpppN on gRNA and CP sgRNA | TLS | on I2 sgRNA | Skulachev et al. (1999) | ||
| Crucifer-infecting tobamovirus | m7GpppN on gRNA | TLS |
on I2 sgRNA
on gRNA and I2 sgRNA |
√ | Ivanov et al. (1997), Skulachev et al. (1999) | |
| 3′ Elements | ||||||
| Luteoviridae | ||||||
| Barley yellow dwarf virus | 5′ pppN | −OH | 5′ SLD–3′ BTE | √ | √ | Wang et al. (1997), Guo et al. (2000, 2001) |
| Bean leafroll virus | 5′ pppN | −OH | 5′ UTRb–3′ BTEb | √ | √b | Domier et al. (2002) |
| Soybean dwarf virus | 5′ pppN | −OH | 5′ UTRb–3′ BTEb | √ | √b | Guo et al. (2001) |
| Tombusviridae | ||||||
| Tobacco necrosis virus A | 5′ pppN | −OH | 5′ UTR–3′ BTE | √ | √ | Meulewaeter et al. (2004) |
| Tobacco necrosis virus D | 5′ pppN | −OH | 5′ UTR–3′ BTE | √ | √ | Shen and Miller (2004) |
| Olive latent virus | 5′ pppN | −OH | 5′ UTRb–3′ BTEb | √ | √b | Shen and Miller (2004) |
| Leek white stripe virus | 5′ pppN | −OH | 5′ UTRb–3′ BTEb | √ | √b | Shen and Miller (2004) |
| Tomato bushy stunt virus | 5′ pppN | −OH | 5′ TSD–3′ CITE | √ | Wu and White (2000), Fabian and White (2004) | |
| Maize necrotic streak virus | 5′ pppN | −OH | 5′ TSD–3′ CITE | √ | P. Redinbaugh and K, Scheets, personal communication | |
| Red clover necrotic mosaic virus RNA1 | 5′ pppN | −OH | TE-DR1 (3′ BTE) | √b | Mizumoto et al. (2003), Fabian and White (2004) | |
| Turnip crinkle virus | 5′ pppN | −OH | 5′ UTR, 3′ UTR | Qu and Morris (2000) | ||
| Hibiscus chlorotic ringspot virus | 5′ pppN | −OH | IRES, 3′ UTR | √ | Koh et al. (2002, 2003) | |
| Satellite tobacco necrosis virus | 5′ pppN | −OH | 5′ leader, 3′ TED | √ | ? | Danthinne et al. (1993), Timmer et al. (1993), Meulewaeter et al. (1998a,b) |
Abbreviations: BTE: barley yellow dwarf (like) translation element; CITE: cap-independent translation enhancer; CIRE: cap-independent regulatory element; IRES: internal ribosome entry site; PK1: 45 nt RNA pseudoknot 1; TED: translation enhancer domain; TE-DR1; translation enhancer of dianthovirus RNA 1; TLS: tRNA-like structure; TSD: T-shaped domain; UTR: untranslated region; VPg: viral protein linked to the genome.
Predicted.
Fig. 1.
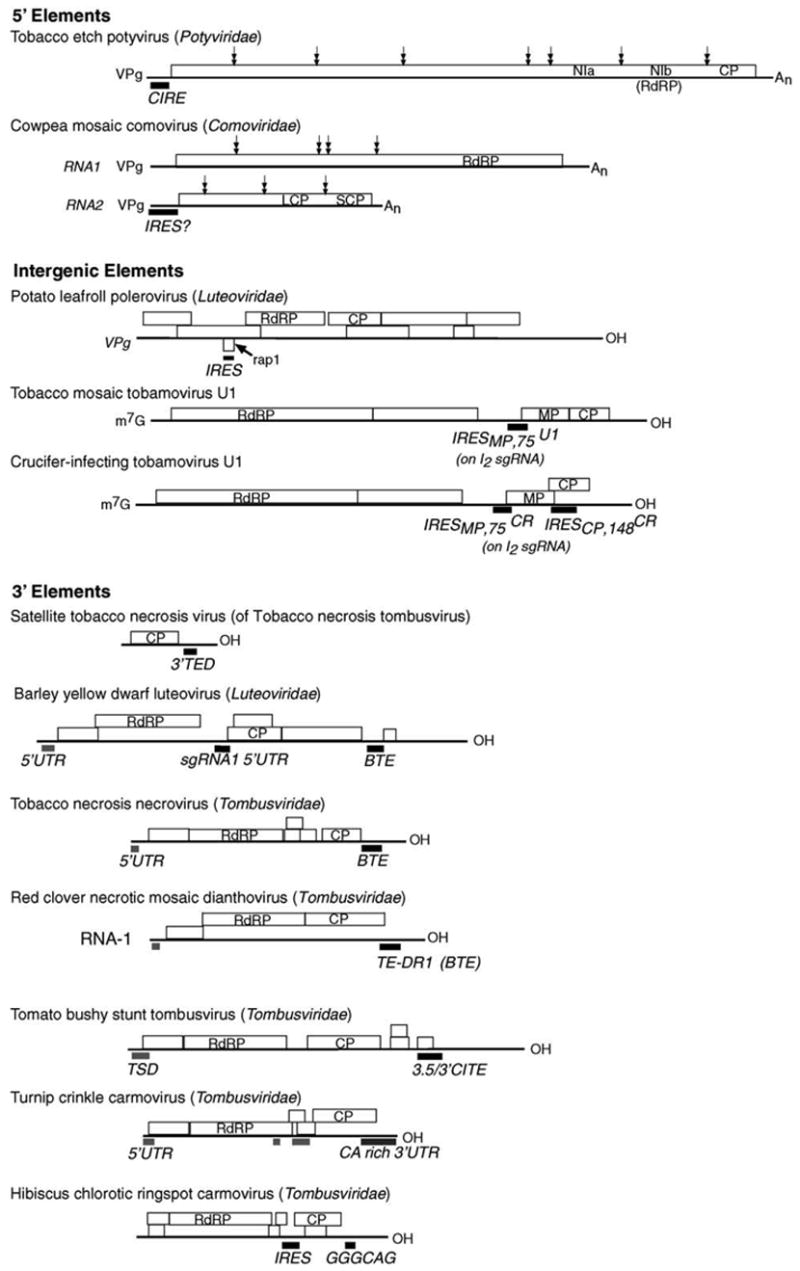
Genome organizations of selected plant viruses with cap-independent translation elements. Bold black bars beneath genomes indicate location of cap-independent translation element. Gray bars indicate upstream sites with which 3′ elements interact. Double-headed arrows indicate positions of polyprotein cleavage on TEV and CPMV. Positions of ORFs are indicated in boxes above the RNA genome. For simplicity, only replication proteins, capsid proteins, and those proteins mentioned in the text are shown. RdRP: RNA dependent RNA polymerase; CP: coat protein; LCP: large coat protein; SCP: small coat protein; MP: movement protein. MP and CP ORFs of tobamoviruses are translated from subgenomic RNAs (not shown) as discussed in text.
2. Cap-independent translation elements in the 5′ UTR
2.1. Potyviridae
The RNAs of the large Potyviridae family resemble those of the picornaviruses, in that they have a VPg (viral protein, genome-linked) at their 5′ end and a poly(A) tail. Their positive-sense RNA genomes (Fig. 1) code for a single polyprotein, which is processed by virus-encoded proteases. Potyviruses differ from picornaviruses in that their VPgs are several fold larger and the 5′ UTRs are several fold shorter (~150 nt for potyviruses versus 600–1200 nt for picornaviruses), less structured than those of picornaviruses and do not contain more than one upstream AUGs.
The 5′ leader (143 nt) of Tobacco etch potyvirus (TEV) mediates cap-independent translation (Carrington and Freed, 1990), and promotes efficient translation in conjunction with the poly(A) tail (Gallie et al., 1995; Niepel and Gallie, 1999). This is facilitated by two cap-independent regulatory elements (CIREs) that span nucleotides (nts) 28–118 in the 5′ UTR (Niepel and Gallie, 1999). Smaller segments of the TEV 5′ UTR, 1–20, 37–65, 67–113 and 110–114 nt were also reported to enhance translation (Kawarasaki et al., 2000). Zeenko and Gallie (2005) recently reported that the 5′ proximal domain (38–75 nt) can fold into a 45 nt RNA pseudoknot (PK1), that is required for full cap-independent translation (Fig. 2). However, mutations expected to disrupt and restore the pseudoknot did not behave as expected, as compensatory mutations did not restore translation activity. In addition to the pseudoknot, upstream unstructured sequences (28–37 nt), that resemble the cap-dependent TMV Ω translation enhancer sequence are clearly important. One loop (L3) of PK1 is complementary to 1117–1123 nt of 18S rRNA. Mutations within L3 that disrupt base pairing to the 18S rRNA substantially reduced translation, while mutations that retain complementarity did not affect translatability. This interaction may enhance translation by facilitating the recruitment of the 40S ribosomal subunit through direct base pairing of the viral RNA to the ribosome. When placed in the intergenic region of a dicistronic reporter construct, the TEV 5′ UTR facilitates translation of the second ORF, suggesting that it has IRES activity (Gallie, 2001). However, a stable stem loop placed upstream of the leader in its natural 5′ UTR context reduced translation as much as 10-fold, indicating that the TEV leader normally requires ribosomal scanning from the 5′ end (Niepel and Gallie, 1999).
Fig. 2.
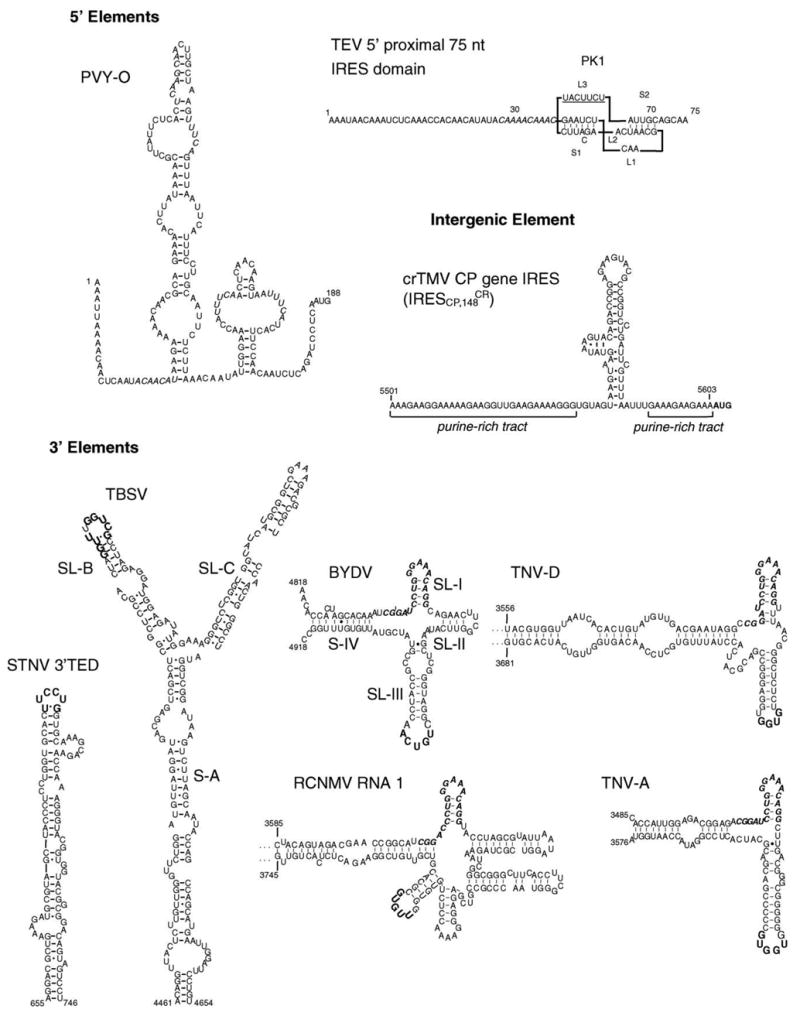
Secondary structures of plant viral translation elements. Predicted structures for 5′ and intergenic elements: PVY 5′ UTR (Yang et al., 1997). Italics in PVY indicate UUUCA, CAA, and box motifs. TEV 5′ proximal 75 nucleotide IRES domain (Zeenko and Gallie, 2005), which includes the 45 nt RNA pseudoknot PK1 with complementary sequence to the 18S rRNA in L3 (underlined), and the upstream Ω-like sequence (italics). CrTMV CP IRES ( ) (Dorokhov et al., 2002). Functionally tested structures for 3′ elements. Bold (not italic) text indicates bases known or predicted to base pair to a loop in the 5′ UTR. Bold italics indicate conserved 17 nt sequence common to all BYDV-like cap-independent translation elements.
The TEV 5′ UTR functions in vivo in tobacco (Carrington and Freed, 1990) and carrot (Gallie et al., 1995) cells. It confers a competitive advantage against uncapped non-viral mRNAs in translation extracts depleted of eIF4F. This advantage is lost when eIF4F (but not eIFiso4F), particularly the eIF4G subunit, is added back (Gallie, 2001). Thus, the TEV 5′ UTR has either no or reduced dependence on eIF4F (or eIF4G) compared to non-viral mRNAs.
Like that of TEV, the 5′ UTRs of other potyviruses, including Potato virus Y (PVY), Plum pox virus (PPV), and Turnip mosaic virus (TuMV) have been shown to stimulate cap-independent translation in various assay systems. When placed in the inter-cistronic region of a dicistronic RNA, the 5′ UTR of PVY (Fig. 2) stimulates translation of the downstream ORF in rabbit reticulocyte lysates (Levis and Astier-Manifacier, 1993). The 3′ 55 nucleotides of the PVY 5′ UTR are the most important for the cap-independent translation in tobacco protoplasts (Yang et al., 1997). Conserved motifs that may participate in cap-independent translation include several UUUCA and (CAA)n blocks that are also found in the cap-dependent TMV Ω translation enhancer sequence (Gallie and Walbot, 1992). In contrast, the 5′ UTR of PPV, a very serious disease agent of fruit trees, does not seem to require specific sequences. Large deletions in the PPV 5′ UTR did not affect the efficient translation of uncapped mRNAs in reticulocyte lysates or in tobacco protoplasts (Simón-Buela et al., 1997). Instead, the 147 nt PPV 5′ UTR harbors an initiation codon in a poor context upstream of the primary initiation codon. Mutation of the upstream start codon to place it in a good initiation context substantially reduced expression of the viral genes. Thus, leaky scanning plays a role in cap-independent translation of PPV RNA (Simón-Buela et al., 1997). The TuMV 5′ UTR inhibits translation of capped transcripts when added in trans, suggesting that a translation factor may interact with the 5′ UTR (Basso et al., 1994).
There is little sequence conservation among the 5′ UTRs of viral RNAs in the diverse Potyviridae family. We (unpublished data) and Maiss et al. (1989) found similarities only among the first 25 nt of the leaders discussed here. In general, potyvirus 5′ leaders appear to lack extensive secondary structure (e.g. Fig. 2), and have low GC content (Simón-Buela et al., 1997). They are so simple that they may function simply by their lack of structure, which reduces factor dependence (i.e. helicase eIF4A), combined with help from their rRNA complementary. 5′ UTR elements like these may also be present in other plant virus groups such as the chrysoviruses (Covelli et al., 2004), so if motifs in the known 5′ UTR elements are delineated, broader understanding of how the RNA elements function in non-potyviruses may be gained.
2.2. Potyvirus VPg binds host translation factors
Given that potyvirus RNAs lack a 5′ cap, it is intriguing that the VPg at the 5′ end of potyviral RNA binds the host cap-binding translation factors, eIF4E and eIFiso4E. The TuMV VPg binds eIFiso4E in vitro and in the yeast two-hybrid system (Wittmann et al., 1997), and in Arabidopsis plants (Léonard et al., 2000). PABP co-purifies with the complex of VPg–Pro and eIF4E or eIFiso4E (Léonard et al., 2004). VPg–Pro has also been shown to interact with the eIFiso4F complex (Plante et al., 2004). Mutations in the VPg domain necessary for interaction with eIFiso4E reduced TuMV infectivity (Léonard et al., 2000). There is also substantial genetic evidence for a role of eIF4E in virus infection. Recessive resistance genes to potyviruses in pepper (Ruffel et al., 2002), lettuce (Duprat et al., 2002; Lellis et al., 2002; Nicaise et al., 2003), and pea (Gao et al., 2004a), and loss-of-susceptibility mutations to potyviruses in Arabidopsis (Lellis et al., 2002) all proved to be caused by mutations in eIF4E or eIFiso4E, or by reduced expression of one of these factors.
The above observations are consistent with a direct role for the VPg in recruiting the translation factors, perhaps replacing the cap structure (Thivierge et al., 2005). However, we are unaware of any direct evidence that the eIF4E/eIFiso4E interaction with VPg participates in translation. In fact, some evidence suggests that this interaction serves other function(s) in the virus life cycle. First, as discussed above, the VPg is unnecessary for efficient cap-independent translation. The 5′ UTR alone confers this function, although it has not been ruled out that the VPg may also stimulate translation (Thivierge et al., 2005), as observed for caliciviruses (Herbert et al., 1997). Secondly, the VPg competes with m7GTP for binding eIFiso4F, leading Plante et al. (2004) to propose that it inhibits host translation. Thirdly, potyviral proteins other than the VPg also interact with eIF4E. The TEV nuclear inclusion protein, Nla, binds eIF4E from tomato and tobacco in a strain specific manner (Schaad et al., 2000). The role of eIF4E in the infection cycle of Pea seedborne mosaic potyvirus (PSbMV) is postulated to be in cell-to-cell movement (Gao et al., 2004b). Mutations in eIF4E (resistance gene cum1) or eIF4G (resistance gene cum2) prevent replication of Cucumber mosaic cucumovirus (CMV), which has capped RNAs (Yoshii et al., 2004). Finally, a screen of Arabidopsis mutant lines showed that mutations of 4E (and not iso4E) are associated with susceptibility to Clover yellow vein potyvirus, while mutations in iso4E (and not 4E) are associated with susceptibility to TuMV (Sato et al., 2005). These observations suggest there are alternative roles for translation factors in the life cycle of potyviruses.
2.3. Other picorna-like plant viruses
Cowpea mosaic comovirus (CPMV, Comoviridae) is even more similar than the Potyviridae to the Picornaviridae (Argos et al., 1984). The presence of an IRES in the 5′ UTR of the middle component RNA was reported in vitro (Thomas et al., 1991; Verver et al., 1991); however internal initiation could not be demonstrated in animal cells (Belsham and Lomonossoff, 1991). Leaky scanning was also suggested as a mechanism of translation for CPMV RNA, as it has an upstream AUG in the middle component RNA (Belsham and Lomonossoff, 1991; Thomas et al., 1991; Verver et al., 1991). The picorna-like waikaviruses, in the Sequiviridae family also have uncapped RNAs and very long 5′ UTRs (454–515 nt) preceding a polyprotein ORF, and it has been suggested that three nucleotide differences between isolates in a potential IRES may explain differences in symptoms, although there is no experimental evidence for an IRES (Chaouch-Hamada et al., 2004).
3. Cap-independent translation elements within or between ORFs
3.1. Tobamoviruses
In polycistronic plant RNA viruses, 5′-distal ORFs were thought to be translated via normal 5′ end-dependent ribosome scanning on subgenomic viral mRNAs containing 5′ truncations of the viral genome (Hull, 2002). Surprisingly, some tobamoviruses have been found recently to translate ORFs on subgenomic RNAs via IRESes. Tobamoviruses, including the prototype Tobacco mosaic virus U1 (TMV U1), have four ORFs (Fig. 1). Two ORFs comprising the replicase are translated from the capped genomic (gRNA) while the downstream genes encoding the movement (MP) and coat (CP) proteins are translated from separate subgenomic RNAs (sgRNAs). Unlike the CP sgRNA, the dicistronic I2 sgRNA, with the MP ORF at its 5′ end, is uncapped (Hunter et al., 1983; Joshi et al., 1983). The 75 nt leader sequence of I2 sgRNA, called , promotes translation of the downstream ORF in a dicistronic reporter construct, even in the presence of a stable hairpin that blocks scanning at the 5′ terminus of the RNA (Skulachev et al., 1999). However, is incapable of mediating MP expression from its internal location in viral genomic RNA. The authors propose that may be inaccessible in the full-length RNA context (Skulachev et al., 1999). A functionally similar element is present in the unrelated Crucifer-infecting tobamovirus (crTMV) ( ) (Skulachev et al., 1999). The intracellular transport of a movement-deficient TMV U1 mutant lacking was restored with the insertion of , supporting the notion that confers cap-independent translation (Zvereva et al., 2004).
Unlike TMV U1, the CP gene of crTMV is translated from both the I2 sgRNA and genomic RNA by internal ribosome entry (Fig. 1) (Ivanov et al., 1997; Dorokhov et al., 2002). Translation is controlled by a 148 nt element located within the MP coding sequence that overlaps the CP ORF by 25 codons (Ivanov et al., 1997). has been reported to confer higher cap-independent translation activity than the widely used encephalomyocarditis virus IRES in plant and animal cells (Dorokhov et al., 2002). The sequence and structure analysis of revealed a bulged stem loop structure flanked by two polypurine A-rich sequences (PARS), crucial for IRES activity (Fig. 2) (Dorokhov et al., 2002; Ivanov et al., 1997). The authors showed that a simplified version of the PARS consisting only of a (GAAA)16 repeat gave very efficient cap-independent translation. The corresponding 148 nt region in the 5′ UTR of TMV U1 CP sgRNA has no homology to and does not stimulate cap-independent translation (Ivanov et al., 1997).
3.2. Potato leafroll polerovirus
Viral RNAs in the Polerovirus and Enamovirus genera (Luteoviridae) contain a VPg, so one would predict that these viruses may contain an IRES at the 5′ end of their genomic RNAs (gRNA). The three 5′-proximal ORFs (Fig. 1) are translated from gRNA, and downstream ORFs are translated from two subgenomic RNAs (Tacke et al., 1990; Ashoub et al., 1998). While a sequence element that confers cap-independent translation of the 5′ proximal ORFs has not been reported, translation can initiate at a site within ORF 1. This leads to translation of a 5 kDa protein dubbed Rap1, encoded by an ORF within ORF 1, in a different frame (Jaag et al., 2003). The function of Rap1 is unknown, but it is indispensable for viral replication. Rap1 translation initiates about 1500 nt downstream of the 5′ end of the gRNA by internal ribosome entry (Jaag et al., 2003). Translation initiation is mediated by a motif, GGAGAGAGAGG, located 22 nt downstream of the Rap1 AUG initiation start (Jaag et al., 2003). This IRES element in conjunction with the 22 nt spacer sequence is sufficient to mediate cap-independent translation in vitro but not in vivo (Jaag et al., 2003). It is noteworthy that, like the crTMV , the essential region of the PLRV IRES is a polypurine tract.
4. Cap-independent translation elements in the 3′ UTR
Viral RNAs in the Luteoviridae and Tombusviridae appear to be unmodified at the 5′ end, and they lack a poly(A) tail (Miller et al., 2002). Unlike all other known viruses of plants and animals, members of genus Luteovirus (but not the Polerovirus or Enamovirus genera) of the Luteoviridae family and most genera of the Tombusviridae contain a sequence in the 3′ UTR that facilitates cap-independent translation initiation at the 5′ end of the genome. From a 3′ position, the translation element facilitates expression of the ORFs located kilobases upstream at the 5′ end of the mRNA.
4.1. Satellite tobacco necrosis virus (STNV)
Although it is a satellite of Tobacco necrosis necrovirus (TNV), STNV RNA bears no sequence homology to its helper virus or any other member of the Tombusviridae family. Thus, perhaps it should be classified separately. STNV RNA is approximately 1200 nt long and encodes only the coat protein. STNV RNA is entirely dependent on TNV for replication but is encapsidated in its own coat protein. STNV RNA contains an efficient cap-independent translation enhancer domain (TED) located in the 5′ end of the 600 nt 3′ UTR (Fig. 1) (645–746 nt) (Danthinne et al., 1993; Timmer et al., 1993; Meulewaeter et al., 1998a). Mutations within TED that abolish translation can be fully compensated by a cap at the 5′ end of the mRNA (Meulewaeter et al., 1998b). Independent of its position within the RNA, the TED mediates cap-independent translation in vivo and in vitro of STNV and heterologous RNAs (Meulewaeter et al., 1998b). Most likely, TED adopts an extended stem loop structure (Fig. 2) for its translation activity, while an alternative pseudoknot-containing structure is unnecessary for translation and may participate in a different stage of viral infection (van Lipzig et al., 2002).
The TED translation activity correlates with its ability to bind two host factors of about 28 and 30 kDa (van Lipzig et al., 2001). Subsequently, TED was shown to bind specifically and directly to eIF4E (26 kDa) and eIFiso4E (28 kDa) (Gazo et al., 2004), which are probably the proteins detected by van Lipzig et al. TED inhibited translation of cap- or TED-containing mRNAs when added in trans, whereas no trans-inhibition was caused by mutant TED sequences that lost their ability to bind to translation factors (van Lipzig et al., 2001). Trans-inhibition by TED was reversed by the addition of eIF4F or eIFiso4F (Gazo et al., 2004). The addition of free cap analogue m7GTP to the translation extract did not inhibit TED-mediated translation, suggesting that the TED may not compete with the cap for binding of eIF4E (Gazo et al., 2004).
To achieve translation initiation at the 5′-proximal AUG, TED must interact with the STNV RNA 5′ UTR (Meulewaeter et al., 1998b). Both elements bear complementary sequences (Meulewaeter et al., 1998b), which led the authors to test the hypothesis that TED and the 5′ UTR interact by direct base pairing. However, mutations in either sequence that disrupted the predicted base pairing reduced translation only slightly. Compensatory double mutations predicted to restore base pairing did not restore cap-independent translation. Thus, any interaction between TED and the 5′ UTR may not rely on Watson–Crick base pairing. However, the results do not rule out the possibility that base pairing is required, and that the particular mutations altered the structures in unpredicted ways to prevent base pairing between UTRs, or that specific sequence as well as base pairing is required.
Both the STNV 5′ UTR and 3′ TED have sequences complementary to 18S rRNA (Meulewaeter et al., 1998a), which may facilitate recruitment of the 40S ribosomal subunit by direct base pairing (Gazo et al., 2004). The eIF4F complex recruited by the TED would then assemble the translation complex at the 5′ end of the mRNA either by interaction of both ends with the 40S subunit or by tertiary structure of the mRNA (Gazo et al., 2004).
4.2. Barley yellow dwarf virus (-like) translation element (BTE)
Barley yellow dwarf virus (BYDV, Luteoviridae), all other members of genus Luteovirus (Domier et al., 2002), and all members of the Necrovirus and Dianthovirus genera (Tombusviridae), harbor the BYDV (or BYDV-like) cap-independent translation element (BTE) in the 5′ end of the 3′ UTR. We define BTEs as cap-independent translation elements with two structural features (Fig. 2): (i) a conserved 17 nt sequence, GGAUC-CUGGGAAACAGG that includes a stem loop (SL-I) with a GNRNA loop motif (Legault et al., 1998), and (ii) a loop (not in SL-I) that can base pair to a loop in the 5′ UTR of the RNA. BTEs are functionally indistinguishable from the STNV TED, but bear no similarity in sequence or secondary structure to TED (Wang et al., 1997).
The BYDV BTE facilitates cap-independent translation of viral proteins from the genomic RNA (gRNA) and sgRNAs in vivo and in vitro (Wang et al., 1997). It folds into a roughly cruciform secondary structure of about 100 nt with three major stem loops (SL-I, SL-II, SL-III) and a terminal stem or “stalk” (S-IV) that allows it to protrude from the viral genome (Guo et al., 2000; E. Pettit Kneller, unpublished). The BTE mediates cap-independent translation when placed in the 5′ UTR, but fails to promote translation of a downstream gene when located in a dicistronic context, indicating that the BTE is not an IRES (S. Wang and E. Allen, unpublished).
As observed for TED (Gazo et al., 2004), pull-down and UV cross-linking assays revealed that eIF4F and eIFiso4F bind specifically to the BTE and not non-functional mutants (E. Allen, E. Pettit Kneller, unpublished). A wheat germ extract depleted of the cap-binding factors is unable to sustain BTE-mediated translation, while the addition of small amounts of eIF4F or eIFiso4F to the extract restores translation of BTE-containing RNA preferentially to capped mRNA (E. Pettit Kneller, unpublished). When added in trans, the BTE inhibits both cap-dependent and BTE-dependent translation (Wang et al., 1997). This inhibition is reversed by addition of eIF4F, suggesting that the BTE sequesters the translation factors. Higher concentrations of cap analogue are necessary to inhibit BTE-mediated translation than translation of capped mRNAs, indicating that the BTE may not compete for the cap-binding pocket of eIF4E (Wang and Miller, 1995). In addition to recruiting initiation factors that facilitate ribosome recruitment, the BTE may bind the 40S subunit directly. A hexamer, GAUCCU, within the 17 nt conserved sequence, is complementary to 18S rRNA at exactly the same distance from the 3′ end of 18S rRNA as is the RNA binding site (Shine–Dalgarno binding site) of prokaryotic 16S ribosomal RNA (Wang et al., 1997).
In addition to recruiting ribosomes or translation factors, the 3′ BTE must deliver them to the 5′ end of the viral RNA where translation initiates. This is facilitated by kissing stem loop base pairing between the loop of SL-III in the BTE (Fig. 2) and a five base loop sequence in the 5′ UTRs of BYDV genomic RNA (Guo et al., 2001) and subgenomic RNA 1 (E. Pettit Kneller, unpublished). A single point mutation within either loop, that disrupted base pairing, abolished translation and replication. Compensatory mutations predicted to restore the kissing-stem loop base pairing also restored cap-independent translation and replication activity (Guo et al., 2001). The BTE-5′ UTR interaction may not be based solely on Watson–Crick base pairing, as many sequences predicted to retain complementarity between the BTE and 5′ UTR could not sustain translation. However, the BTE can facilitate translation of a heterologous RNA from a 3′ location using a surrogate base pairing between UTRs outside of the BTE (A. Rakotondrafara, unpublished).
BTE-mediated cap-independent translation appears to require ribosome scanning from the 5′ end, as in normal cap-dependent translation. The addition of a stable stem loop structure at the very 5′ end of the BYDV RNA blocks BTE-facilitated cap-independent translation as well as cap-dependent translation (Guo et al., 2001). When the stem loop structure was placed more distal to the 5′ terminus, the RNA translated efficiently. Furthermore, addition of an upstream AUG in a different reading frame from the main reporter gene ORF significantly reduced translation of the reporter gene (A. Rakotondrafara, unpublished). All these observations are consistent with the classical 5′ ribosome scanning mechanism (Guo et al., 2000; Kozak, 1989).
4.3. Necroviruses and dianthoviruses (Tombusviridae)
The cap-independent translation function of the BTE in two quite different necroviruses, TNV-A and TNV-D, was demonstrated by Meulewaeter et al. (2004) and Shen and Miller (2004), respectively. The necrovirus BTEs have similar secondary structures to that of BYDV except they lack SL-II (Fig. 2). Like the BYDV BTE, the TNV BTE promotes cap-independent translation from the 3′ or the 5′ UTR (Shen and Miller, 2004) and it has a stem loop that potentially base pairs to a loop in the 5′ UTR. In TNV-D RNA, this comprises only four complementary bases. Mutagenesis results are not only consistent with a requirement for base pairing between the 3′ BTE of TNV-A and TNV-D viruses and the 5′ UTRs of their genomic and subgenomic RNAs, but also indicate that the primary sequence of the paired regions is sensitive to changes (Meulewaeter et al., 2004; Shen and Miller, 2004).
Dianthoviruses, including Red clover necrotic mosaic virus (RCNMV), are the only Tombusviridae viruses with a bipartite genome. Both RNAs were reported initially to be capped (Lommel et al., 1988; Xiong and Lommel, 1989), but recent data ruled out the presence of a cap (Mizumoto et al., 2003). This finding was consistent with the infectivity of uncapped in vitro transcribed RCNMV RNA 1 in protoplasts (Mizumoto et al., 2003). Cap-independent translation is mediated by the “translation element of dianthovirus RNA 1” or TE-DR1, between bases 3596 and 3732 of the 3′ UTR. Secondary structure prediction reveals that the dianthovirus BTEs, including TE-DR1, resemble the BYDV BTE except that they contain three extra stem loops (Fig. 2). TE-DR1 was reported to mediate translation independently of the RCNMV 5′ leader, as it showed full activity with a 5′ vector-derived leader sequence (Mizumoto et al., 2003). However, the involvement of a 3′–5′ interaction could not be ruled out, because potential base pairing was predicted in silico between TE-DR1 and both the RCNMV 5′ UTR, and the vector-derived 5′ UTR that was used in the above experiment (Fabian and White, 2004).
In contrast to RCNMV RNA 1, RCNMV RNA 2 lacks both a TE-DR1-like element in its 3′ UTR and a 5′ cap (Mizumoto et al., 2003). Thus, the mechanism by which RNA 2 is translated remains unclear. We found that the same GUUGUG loop in TE-DR1 predicted by Fabian and White (2004) to base pair to the RNA 1 5′ UTR (Fig. 2), also has potential to base pair in trans to any of the three loops predicted in the 5′ UTR of RNA 2. Thus, the translation factors/ribosomes likely to be recruited to TE-DR1 could be delivered to the RNA 2 5′ UTR via this base pairing. Interactions between RCNMV RNAs 1 and 2 would be enhanced by known base pairing between internal regions of these RNAs necessary for sgRNA synthesis (Sit et al., 1998).
4.4. Other Tombusviridae
The other six genera of the Tombusviridae, while harboring some kind of cap-independent translation sequence, have no sequence resembling a BTE. Tomato bushy stunt virus (TBSV) RNA of genus Tombusvirus, lacks a BTE as evidenced by the absence of the 17 nt conserved sequence, nor is cap-independent translation clearly detectable in vitro. Instead, TBSV RNA relies on a 400 nt region composed of three contiguous domains (RIII, R3.5, RIV) referred to as the 3′ cap-independent translational enhancer or 3′ CITE, to facilitate cap-independent translation in vivo (Wu and White, 1999). The R3.5 segment represents the core translation-specific element (Oster et al., 1998; Wu and White, 1999), but requires the flanking RIII and RIV regions that are also essential for replication (Oster et al., 1998).
The R3.5 adopts a Y-shaped structure with three extended stems (S-A, SL-B, and SL-C) (Fig. 2) that is highly conserved in genus Tombusvirus (Fabian and White, 2004). Mutations on either stem loop impaired RNA translatability (Fabian and White, 2004). 3′–5′ communication is mediated by base pairing of the SL-B R3.5 to a complementary stem loop, SL3, within the 5′ T-shaped domain, TSD, essential for replication (Fabian and White, 2004; Wu et al., 2001). The interaction involves 9 nt that extend from the terminal loop to stem sequence. Sequence complementarity rather than primary sequence is important for this 3′–5′ communication (Fabian and White, 2004). Thus, like BTE-containing viruses, base pairing between loops in the 3′ and 5′ UTRs is necessary. In fact, Fabian and White found potential base pairing between 3′ and 5′ UTRs in most genera of the Tombusviridae family (Fabian and White, 2004).
Interestingly, the Maize necrotic streak virus (Tombusviridae) 3′ CITE mediates cap-independent translation in vitro from the gRNA and both sgRNAs. Rather than adopting a Y-shaped structure, it forms a long stem loop, and the loop has sequence complementary to a loop within the TSD (P. Redinbaugh and K. Scheets, personal communication).
The core translation enhancer element of Turnip crinkle carmovirus (TCV, Tombusviridae) is also located within the 5′ end of the 3′ UTR of the gRNA and sgRNA (Qu and Morris, 2000). The TCV-TE works cooperatively with the 5′ UTR in mediating cap-independent translation. This cooperation may not involve base pairing interaction as no potential complementary sequence was identified (Fabian and White, 2004; Qu and Morris, 2000).
Interestingly, the 5′ UTR of the CP sgRNA of TCV mediates translation more efficiently than the gRNA 5′ UTR. This makes sense because the level of accumulation of the CP is at least 100-fold greater than other gene products. Qu and Morris (2000) propose that the primary role of the TCV 3′ TE is to ensure efficient translation of viral RNA in competition with the host mRNA, while the 5′ UTRs modulate the level of expression to control the switch from early to late stages of viral life cycle (Qu and Morris, 2000). Yoshii et al. (2004) showed that the recessive eIF4G mutant in the cum2 gene of Arabidopsis thaliana prevents translation of TCV RNA. However, TCV translatability was not affected in the cum1 mutant line that had lost eIF4E (Yoshii et al., 2004) or with mutation within eIFiso4E gene (Lellis et al., 2002). Thus, TCV RNA translation may be eIF4G- but not eIF4E-dependent.
A 180 nt 3′ cap-independent translation enhancer was identified in another carmovirus Hibiscus chlorotic ringspot virus (HCRSV, Tombusviridae) (Koh et al., 2002). It requires a hex-anucleotide, GGGCAG (Koh et al., 2002), and acts synergistically with an IRES element located 124 bases upstream (2394–2566 bp) of the CP gene (Koh et al., 2003). The authors propose that both elements function by direct base pairing to the ribosomal RNA (Koh et al., 2002, 2003).
5. Discussion
5.1. Plant virus translation elements differ from animal virus translation elements
Even though plant viruses have similar genome organizations and expression strategies to animal viruses, their cap-independent translation signals are strikingly different from those in animal viruses. The plant elements are less complex, and some are not IRESes. These differences may reflect fundamental differences between the plant and animal viral translation apparatus. For example, in response to stress including certain viral infections, animal hosts shut down cap-dependent translation via dephosphorylation of eIF4E-binding protein (4EBP) which, in turn binds eIF4E and blocks binding of eIF4G (Richter and Sonenberg, 2005). No 4EBP homologs have been identified in plants (Browning, 2004), so plants may lack this translation system defense mechanism, and plant viruses hence do not need to bypass such regulation.
5.2. Why cap-independent translation?
Being able to translate cap-independently allows viruses to avoid the above eIF4E-mediated cellular translational control. It also permits animal viruses to actively shut off host cap-dependent translation, which prevents defense gene expression and frees ribosomes for the viral RNA. The cost to animal viruses is the requirement for large, complex IRESes to functionally replace the host factors in recruiting ribosomes. In contrast, plant viruses are not known to globally shut down host translation. They may require more of the host translational machinery and hence do not need large IRESes. Some small plant virus translation elements also can function in artificial mammalian systems using reporter genes (Dorokhov et al., 2002), but more complex IRESes are needed in infectious animal viruses, perhaps to override antiviral translation defense systems that are absent in plants.
Another advantage of cap-independent translation is simply that this capability avoids the need to encode capping enzymes. Most positive strand RNA viruses replicate in the cytoplasm, but the cellular capping machinery resides in the nucleus. Thus, capped, positive strand plant viruses encode their own capping enzymes (Ahola and Ahlquist, 1999). Considering that most plant viruses encode only four or five genes, it is a major burden to devote a gene to capping.
5.3. Why a 3′ translation element?
The presence of the translation element in the 3′ UTR of the viral RNA: (i) prevents translation initiation on 3′-truncated or degraded RNAs, (ii) ensures the presence of the translation element on all 3′ co-terminal subgenomic mRNAs, and (iii) may facilitate the switch from translation to replication (Barry and Miller, 2002). Translation and RNA replication cannot take place simultaneously on the same RNA (Gamarnik and Andino, 1998). To avoid non-productive collisions between the replicase moving from the 3′ end toward the 5′ end and ribosomes moving 5′ to 3′, while copying the 3′ UTR the replicase may disrupt the 3′ cap-independent translation element structure, hence turning off translation at the 5′ end of the genome. The resulting ribosome-free genomic RNA could then be replicated (Barry and Miller, 2002).
5.4. How does cap-independent translation relate to the closed loop model?
Base pairing between the 3′ cap-independent translation elements and the 5′ UTR circularizes the mRNA and thus may obviate the need for translation factor-mediated circularization as occurs on normal capped, polyadenylated mRNAs. However, cap-binding factors are still needed, and also sequence downstream of the BTE is required in the BYDV RNA 3′ UTR for full translation in vivo. Translation of mRNAs lacking the downstream sequence can be restored by addition of a poly(A) tail but not by adding a cap (Guo et al., 2000). These observations indicate that eIF4F and a “poly(A) mimic” sequence are necessary for aspects of translation initiation other than circularization. How plant viral RNAs that harbor the cap-independent translation element in the 5′ UTR or in intergenic regions are circularized, if at all, is unknown or has not been demonstrated. Thivierge et al. (2005) propose that the potyviral RNAs circularized by a VPg–eIF4E–eIF4G–PABP–poly(A) tail or a VPg–PABP–poly(A) tail interaction.
6. Future prospects
We have barely begun to understand how plant viral translation elements interact with the host translation machinery. Screens to identify protein/viral translation element interactions and protein/protein interactions would shed light on cap-independent translation mechanisms. So far, eIF4E has been found to bind some 3′ UTR cap-independent translation elements (Gazo et al., 2004; E. Pettit Kneller and E. Allen, unpublished). The roles of the different isoforms of eIF4E and eIF4G in viral translation are unknown. One functional difference between the isoforms is their ability to support translation on a mRNA containing a structured 5′ UTR leader (eIF4F is more efficient than eIFiso4F) (Gallie and Browning, 2001).
The roles of translation factors recruited by viruses must be elucidated. Translation factors may perform functions other than translation such as cell-to-cell movement or RNA replication. Plant viruses modify the connections between cells (plasmodesmata) to move between cells (Oparka, 2004). Given that eIFiso4G binds microtubules (Bokros et al., 1995), by binding viral RNAs it could facilitate movement by localizing viral RNA to the ER, which spans the plasmodesmatal connections. Zucchini yellow mosaic potyvirus RdRP interacts with PABP in Y2H (Wang et al., 2000); the function of this interaction is unknown.
On the other hand, translation factors are often commandeered by the viral replicase for RNA replication (Blumenthal and Carmichael, 1979; Quadt et al., 1993). The ability of potyvirus VPg–Pro to bind eIF4E may recruit translation factors to the replicase complex rather than facilitate translation. Perhaps the eIF4E mutants that confer recessive resistance (discussed earlier) prevent assembly of a functional viral replicase.
The phosphorylation status of plant initiation factors affects mRNA cap-binding (Khan and Goss, 2004), but little is known about the effect of plant virus infection on factor phosphorylation. TMV, like influenza virus, appears to modulate phosphorylation of translation initiation factor eIF2α (Bilgin et al., 2003), indicating that similarities between plant and animal translational control may exist, at least for a capped RNA virus.
Given that many plant viral elements have oligomers that are complementary to 18S rRNA (Table 1), approaches to understand the biological significance of this complementarity (Akbergenov et al., 2004) should also be investigated. In summary, we have only begun to scratch the surface with our understanding of cap-independent translation mechanisms in plants.
Acknowledgments
We thank Kay Scheets for sharing unpublished data. This work was supported by NIH grant R01-GM067104, and by a scholarship from Pioneer Hi-Bred to A.M.R.
References
- Ahola T, Ahlquist P. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J Virol. 1999;73:10061–10069. doi: 10.1128/jvi.73.12.10061-10069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbergenov R, Zhanybekova S, Kryldakov RV, Zhigailov A, Polimbetova NS, Hohn T, Iskakov BK. ARC-1, a sequence element complementary to an internal 18S rRNA segment, enhances translation efficiency in plants when present in the leader or intercistronic region of mRNAs. Nucleic Acids Res. 2004;32:239–247. doi: 10.1093/nar/gkh176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P, Kamer M, Nicklin J, Wimmer E. Similarity in gene organization and homology between proteins of animal picornaviruses and a plant comovirus suggest common ancestry of these virus families. Nucleic Acids Res. 1984;12:7251–7267. doi: 10.1093/nar/12.18.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashoub A, Rohde W, Prufer D. In planta transcription of a second subgenomic RNA increases the complexity of the subgroup 2 luteovirus genome. Nucleic Acids Res. 1998;26:420–426. doi: 10.1093/nar/26.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JK, Miller WA. A programmed -1 ribosomal frameshift that requires base-pairing across four kilobases suggests a novel mechanism for controlling ribosome and replicase traffic on a viral RNA. Proc Natl Acad Sci USA. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso J, Dallaire P, Charest PJ, Devantier Y, Laliberte JF. Evidence for an internal ribosome entry site within the 5′ non-translated region of turnip mosaic potyvirus RNA. J Gen Virol. 1994;75:3157–3165. doi: 10.1099/0022-1317-75-11-3157. [DOI] [PubMed] [Google Scholar]
- Belsham G, Lomonossoff G. The mechanism of translation of cowpea mosaic virus middle component RNA: no evidence for internal initiation from experiments in an animal cell transient expression system. J Gen Virol. 1991;72:3109–3113. doi: 10.1099/0022-1317-72-12-3109. [DOI] [PubMed] [Google Scholar]
- Bilgin DD, Liu Y, Schiff M, Dinesh-Kumar SP. P58(IPK), a plant ortholog of double-stranded RNA-dependent protein kinase PKR inhibitor, functions in viral pathogenesis. Dev Cell. 2003;4:651–661. doi: 10.1016/s1534-5807(03)00125-4. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Carmichael GG. RNA replication: function and structure of Qβ replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- Bokros CL, Hugdahl JD, Kim HH, Hanesworth VR, van Heerden A, Browning KS, Morejohn LC. Function of the p86 subunit of eukaryotic initiation factor (iso)4F as a microtubule-associated protein in plant cells. Proc Natl Acad Sci USA. 1995;92:7120–7124. doi: 10.1073/pnas.92.15.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning KS. Plant translation initiation factors: it is not easy to be green. Biochem Soc Trans. 2004;32:589–591. doi: 10.1042/BST0320589. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Freed DD. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch-Hamada R, Redinbaugh MG, Gingery RE, Willie K, Hogenhout SA. Accumulation of maize chlorotic dwarf virus proteins in its plant host and leafhopper vector. Virology. 2004;325:379–388. doi: 10.1016/j.virol.2004.04.039. [DOI] [PubMed] [Google Scholar]
- Covelli L, Coutts RH, Di Serio F, Citir A, Acikgoz S, Hernandez C, Ragozzino A, Flores R. Cherry chlorotic rusty spot and Amasya cherry diseases are associated with a complex pattern of mycoviral-like double-stranded RNAs. I Characterization of a new species in the genus Chrysovirus. J Gen Virol. 2004;85:3389–3397. doi: 10.1099/vir.0.80181-0. [DOI] [PubMed] [Google Scholar]
- Danthinne X, Seurinck J, Meulewaeter F, Van Montagu M, Cornelissen M. The 3′ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol Cell Biol. 1993;13:3340–3349. doi: 10.1128/mcb.13.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier LL, McCoppin NK, Larsen RC, D’Arcy CJ. Nucleotide sequence shows that Bean leafroll virus has a Luteovirus-like genome organization. J Gen Virol. 2002;83:1791–1798. doi: 10.1099/0022-1317-83-7-1791. [DOI] [PubMed] [Google Scholar]
- Dorokhov YL, Skulachev MV, Ivanov PA, Zvereva SD, Tjulkina LG, Merits A, Gleba YY, Hohn TJ, Atabekov JG. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc Natl Acad Sci USA. 2002;99:5301–5306. doi: 10.1073/pnas.082107599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 2002;32:927–934. doi: 10.1046/j.1365-313x.2002.01481.x. [DOI] [PubMed] [Google Scholar]
- Fabian MR, White KA. 5′–3′ RNA–RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA: a potential common mechanism fortombusviridae. J Biol Chem. 2004;279:28862–28872. doi: 10.1074/jbc.M401272200. [DOI] [PubMed] [Google Scholar]
- Gallie DR. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J Virol. 2001;75:12141–12152. doi: 10.1128/JVI.75.24.12141-12152.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Browning KS. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem. 2001;276:36951–36960. doi: 10.1074/jbc.M103869200. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Tanguay RL, Leathers V. The tobacco etch viral 5′ leader and poly(A) tail are functionally synergistic regulators of translation. Gene. 1995;165:233–238. doi: 10.1016/0378-1119(95)00521-7. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Walbot V. Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res. 1992;20:4631–4638. doi: 10.1093/nar/20.17.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Eyers S, Thomas C, Ellis N, Maule A. Identification of markers tightly linked to sbm recessive genes for resistance to Pea seed-borne mosaic virus. Theor Appl Genet. 2004a;109:488–494. doi: 10.1007/s00122-004-1652-6. [DOI] [PubMed] [Google Scholar]
- Gao ZH, Johansen E, Eyers S, Thomas CL, Noel Ellis TH, Maule AJ. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 2004b;40:376–385. doi: 10.1111/j.1365-313X.2004.02215.x. [DOI] [PubMed] [Google Scholar]
- Gazo BM, Murphy P, Gatchel JR, Browning KS. A novel interaction of cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J Biol Chem. 2004;279:13584–13592. doi: 10.1074/jbc.M311361200. [DOI] [PubMed] [Google Scholar]
- Guo L, Allen E, Miller WA. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA. 2000;6:1808–1820. doi: 10.1017/s1355838200001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Allen E, Miller WA. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- Hentze MW. eIF4G: a multipurpose ribosome adapter? Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. (published erratum appears in Science 1997 March 14;275(5306):1553). [DOI] [PubMed] [Google Scholar]
- Herbert TP, Brierley I, Brown TD. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J Gen Virol. 1997;78:1033–1040. doi: 10.1099/0022-1317-78-5-1033. [DOI] [PubMed] [Google Scholar]
- Hull R. Matthews’ Plant Virology. 4. Academic Press; London: 2002. [Google Scholar]
- Hunter T, Jackson R, Zimmern D. Multiple proteins and subgenomic mRNAs may be derived from a single open reading frame on tobacco mosaic virus RNA. Nucleic Acids Res. 1983;11:801–821. doi: 10.1093/nar/11.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PA, Karpova OV, Skulachev MV, Tomashevskaya OL, Rodionova NP, Dorokhov YL, Atabekov JG. A tobamovirus genome that contains an internal ribosome entry site functional in vitro. Virology. 1997;232:32–43. doi: 10.1006/viro.1997.8525. [DOI] [PubMed] [Google Scholar]
- Jaag HM, Kawchuk L, Rohde W, Fischer R, Emans N, Prufer D. An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc Natl Acad Sci USA. 2003;100:8939–8944. doi: 10.1073/pnas.1332697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Pleij CW, Haenni AL, Chapeville F, Bosch L. Properties of the tobacco mosaic virus intermediate length RNA-2 and its translation. Virology. 1983;127:100–111. doi: 10.1016/0042-6822(83)90375-6. [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawarasaki Y, Kasahara S, Kodera N, Shinbata T, Sekiguchi S, Nakano H, Yamane T. A trimmed viral cap-independent translation enhancing sequence for rapid in vitro gene expression. Biotechnol Prog. 2000;16:517–521. doi: 10.1021/bp000021x. [DOI] [PubMed] [Google Scholar]
- Khan MA, Goss DJ. Phosphorylation states of translational initiation factors affect mRNA cap binding in wheat. Biochemistry. 2004;43:9092–9097. doi: 10.1021/bi049602b. [DOI] [PubMed] [Google Scholar]
- Koh DC, Wong SM, Liu DX. Synergism of the 3′-untranslated region and an internal ribosome entry site differentially enhances the translation of a plant virus coat protein. J Biol Chem. 2003;278:20565–20573. doi: 10.1074/jbc.M210212200. [DOI] [PubMed] [Google Scholar]
- Koh DCY, Liu DX, Wong SM. A six-nucleotide segment within the 3′ untranslated region of Hibiscus chlorotic ringspot virus plays an essential role in translatoinal enhancement. J Virol. 2002;76:1144–1153. doi: 10.1128/JVI.76.3.1144-1153.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leathers V, Tanguay R, Kobayashi M, Gallie DR. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol Cell Biol. 1993;13:5331–5347. doi: 10.1128/mcb.13.9.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- Lellis AD, Kasschau KD, Whitham SA, Carrington JC. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol. 2002;12:1046–1051. doi: 10.1016/s0960-9822(02)00898-9. [DOI] [PubMed] [Google Scholar]
- Levis C, Astier-Manifacier S. The 5′ untranslated region of PVY RNA, even located in an internal position, enables initiation of translation. Virus Genes. 1993;7:367–379. doi: 10.1007/BF01703392. [DOI] [PubMed] [Google Scholar]
- Lommel SA, Weston-Fina M, Xiong Z, Lomonossoff GP. The nucleotide sequence and gene organization of red clover necrotic mosaic virus RNA-2. Nucleic Acids Res. 1988;16:8587–8602. doi: 10.1093/nar/16.17.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léonard S, Plante D, Wittmann S, Daigneault N, Fortin MG, Laliberte JF. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J Virol. 2000;74:7730–7737. doi: 10.1128/jvi.74.17.7730-7737.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léonard S, Viel C, Beauchemin C, Daigneault N, Fortin MG, Laliberte JF. Interaction of VPg–Pro of turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J Gen Virol. 2004;85:1055–1063. doi: 10.1099/vir.0.19706-0. [DOI] [PubMed] [Google Scholar]
- Maiss E, Timpe U, Brisske A, Jelkmann W, Casper R, Himmler G, Mattanovich D, Katinger HW. The complete nucleotide sequence of plum pox virus RNA. J Gen Virol. 1989;70:513–524. doi: 10.1099/0022-1317-70-3-513. [DOI] [PubMed] [Google Scholar]
- Meulewaeter F, Van Lipzig R, Gultyaev AP, Pleij CW, Van Damme D, Cornelissen M, Van Eldik G. Conservation of RNA structures enables TNV and BYDV 5′ and 3′ elements to cooperate synergistically in cap-independent translation. Nucleic Acids Res. 2004;32:1721–1730. doi: 10.1093/nar/gkh338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulewaeter F, Van Montagu M, Cornelissen M. Features of the autonomous function of the translational enhancer domain of satellite tobacco necrosis virus. RNA. 1998a;4:1347–1356. doi: 10.1017/s135583829898092x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulewaeter F, Danthinne X, Van Montagu M, Cornelissen M. 5′- and 3′-sequences of satellite tobacco necrosis virus RNA promoting translation in tobacco. Plant J. 1998b;14:169–176. doi: 10.1046/j.1365-313x.1998.00104.x. [DOI] [PubMed] [Google Scholar]
- Miller WA, Liu S, Beckett R. Barley yellow dwarf virus: Luteoviridae or Tombusvihdae? Mol Plant Pathol. 2002;3:177–183. doi: 10.1046/j.1364-3703.2002.00112.x. [DOI] [PubMed] [Google Scholar]
- Mizumoto H, Tatsuta M, Kaido M, Mise K, Okuno T. Cap-independent translational enhancement by the 3′ untranslated region of red clover necrotic mosaic virus RNA1. J Virol. 2003;77:12113–12121. doi: 10.1128/JVI.77.22.12113-12121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeleman L, Olsthoorn RC, Linthorst HJ, Bol JF. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc Natl Acad Sci USA. 2001;98:14286–14291. doi: 10.1073/pnas.251542798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V, German-Retana S, Sanjuan R, Dubrana MP, Mazier M, Maisonneuve B, Candresse T, Caranta C, LeGall O. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus lettuce mosaic virus. Plant Physiol. 2003;132:1272–1282. doi: 10.1104/pp.102.017855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M, Gallie DR. Identification and characterization of the functional elements within the tobacco etch virus 5′ leader required for cap-independent translation. J Virol. 1999;73:9080–9088. doi: 10.1128/jvi.73.11.9080-9088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka KJ. Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci. 2004;9:33–41. doi: 10.1016/j.tplants.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Oster SK, Wu B, White KA. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J Virol. 1998;72:5845–5851. doi: 10.1128/jvi.72.7.5845-5851.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante D, Viel C, Leonard S, Tampo H, Laliberte JF, Fortin MG. Tumip mosaic virus VPg does not disrupt the translation initiation complex but interferes with cap binding. Physiol Mol Plant Pathol. 2004;64:219–226. [Google Scholar]
- Qu F, Morris TJ. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J Virol. 2000;74:1085–1093. doi: 10.1128/jvi.74.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadt R, Kao CC, Browning KS, Hershberger RP, Ahlquist P. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1993;90:1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Ruffel S, Dussault MH, Palloix A, Moury B, Bendahmane A, Robaglia C, Caranta C. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E) Plant J. 2002;32:1067–1075. doi: 10.1046/j.1365-313x.2002.01499.x. [DOI] [PubMed] [Google Scholar]
- Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- Sato M, Nakahara K, Yoshii M, Ishikawa M, Uyeda I. Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 2005;579:1167–1171. doi: 10.1016/j.febslet.2004.12.086. [DOI] [PubMed] [Google Scholar]
- Schaad MC, Anderberg RJ, Carrington JC. Strain-specific interaction of the tobacco etch virus Nla protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology. 2000;273:300–306. doi: 10.1006/viro.2000.0416. [DOI] [PubMed] [Google Scholar]
- Shen R, Miller WA. The 3 untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J Virol. 2004;78:100–111. doi: 10.1128/JVI.78.9.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Buela L, Guo HS, Garcia JA. Cap-independent leaky scanning as the mechanism of translation initiation of a plant viral genomic RNA. J Gen Virol. 1997;78:2691–2699. doi: 10.1099/0022-1317-78-10-2691. [DOI] [PubMed] [Google Scholar]
- Sit TL, Vaewhongs AA, Lommel SA. RNA-mediated transactivation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- Skulachev MV, Ivanov PA, Karpova OV, Korpela T, Rodionova NP, Dorokhov YL, Atabekov JG. Internal initiation of translation directed by the 5′-untranslated region of the tobamovirus subgenomic RNA I(2) Virology. 1999;263:139–154. doi: 10.1006/viro.1999.9928. [DOI] [PubMed] [Google Scholar]
- Smirnyagina EV, Morozov SY, Rodionova NP, Miroshnichenko NA, Solovev AG, Fedorkin ON, Atabekov JG. Translational efficiency and competitive ability of mRNAs with 5′-untranslated alpha beta-leader of potato virus X RNA. Biochimie. 1991;73:587–598. doi: 10.1016/0300-9084(91)90027-x. [DOI] [PubMed] [Google Scholar]
- Tacke E, Prufer D, Salamini F, Rohde W. Characterization of a potato leafroll luteovirus subgenomic RNA: differential expression by internal translation initiation and UAG suppression. J Gen Virol. 1990;10:2265–2272. doi: 10.1099/0022-1317-71-10-2265. [DOI] [PubMed] [Google Scholar]
- Thivierge K, Nicaise V, Dufresne PJ, Cotton S, Laliberte JF, LeGall O, Fortin MG. Plant virus RNAs coordinated recruitment of conserved host functions by (+) ssRNA viruses during early infection events . Plant Physiol. 2005;138:1822–1827. doi: 10.1104/pp.105.064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, ter Haar E, Wellink J, Voorma H. Cowpea mosaic virus middle component RNA contains a sequence that allows internal binding of ribosomes and that requires eukaryotic initiation factor 4F for optimal translation. J Virol. 1991;65:2953–2959. doi: 10.1128/jvi.65.6.2953-2959.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer RT, Benkowski LA, Schodin D, Lax SR, Metz AM, Ravel JM, Browning KS. The 5′ and 3′ untranslated regions of satellite tobacco necrosis virus RNA affect translational efficiency and dependence on a 5′ cap structure. J Biol Chem. 1993;268:9504–9510. [PubMed] [Google Scholar]
- van Lipzig R, Gultyaev AP, Pleij CW, van Montagu M, Cornelissen M, Meulewaeter F. The 5′ and 3′ extremities of the satellite tobacco necrosis virus translational enhancer domain contribute differentially to stimulation of translation. RNA. 2002;8:229–236. doi: 10.1017/s1355838202018071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lipzig R, Van Montagu M, Cornelissen M, Meulewaeter F. Functionality of the STNV translational enhancer domain correlates with affinity for two wheat germ factors. Nucleic Acids Res. 2001;29:1080–1086. doi: 10.1093/nar/29.5.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verver J, Le Gall O, van Kammen A, Wellink J. The sequence between nucleotides 161 and 512 of cowpea mosaic virus M RNA is able to support internal initiation of translation into vitro. J Gen Virol. 1991;72:2339–2345. doi: 10.1099/0022-1317-72-10-2339. [DOI] [PubMed] [Google Scholar]
- von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- Wang S, Browning KS, Miller WA. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Miller WA. A sequence located 4.5 to 5 kilobases from the 5′ end of the barley yellow dwarf virus (PAV) genome strongly stimulates translation of uncapped mRNA. J Biol Chem. 1995;270:13446–13452. doi: 10.1074/jbc.270.22.13446. [DOI] [PubMed] [Google Scholar]
- Wang X, Ullah Z, Grumet R. Interaction between zucchini yellow mosaic potyvirus RNA-dependent RNA polymerase and host poly-(A) binding protein. Virology. 2000;275:433–443. doi: 10.1006/viro.2000.0509. [DOI] [PubMed] [Google Scholar]
- Wittmann S, Chatel H, Fortin MG, Laliberte JF. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology. 1997;234:84–92. doi: 10.1006/viro.1997.8634. [DOI] [PubMed] [Google Scholar]
- Wu B, Vanti WB, White KA. An RNA domain within the 5′ untranslated region of the tomato bushy stunt virus genome modulates viral RNA replication. J Mol Biol. 2001;305:741–756. doi: 10.1006/jmbi.2000.4298. [DOI] [PubMed] [Google Scholar]
- Wu B, White KA. A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J Virol. 1999;73:8982–8988. doi: 10.1128/jvi.73.11.8982-8988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Lommel SA. The complete nucleotide sequence and genome organization of red clover necrotic mosaic virus RNA-1. Virology. 1989;171:543–554. doi: 10.1016/0042-6822(89)90624-7. [DOI] [PubMed] [Google Scholar]
- Yang LJ, Hidaka M, Sonoda J, Masaki H, Uozumi T. Mutational analysis of the potato virus Y5′ untranslated region for alteration in translational enhancement in tobacco protoplasts. Biosci Biotechnol Biochem. 1997;61:2131–2133. doi: 10.1271/bbb.61.2131. [DOI] [PubMed] [Google Scholar]
- Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol. 2004;78:6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeenko V, Gallie DR. Cap-independent translation of tobacco etch virus is conferred by an RNA pseudoknot in the 5′-leader. J Biol Chem. 2005;280:26813–26824. doi: 10.1074/jbc.M503576200. [DOI] [PubMed] [Google Scholar]
- Zvereva SD, Ivanov PA, Skulachev MV, Klyushin AG, Dorokhov YL, Atabekov JG. Evidence for contribution of an internal ribosome entry site to intercellular transport of a tobamovirus. J Gen Virol. 2004;85:1739–1744. doi: 10.1099/vir.0.79792-0. [DOI] [PubMed] [Google Scholar]


