Abstract
After surgical removal of two-thirds of the liver, remaining hepatocytes replicate and restore hepatic mass within 2 weeks. This process must be initiated by signals extrinsic to the hepatocyte, but it remains unclear whether subsequent events leading to DNA synthesis (S phase) are regulated by circulating or locally produced growth factors (a noncell autonomous response), or by a program intrinsic to the hepatocyte itself (a cell autonomous response). To identify the type of mechanism regulating passage to S, we exploited the difference between rat and mouse hepatocytes in the timing of DNA synthesis after partial hepatectomy, which peaks 12–16 h earlier posthepatectomy in rat compared with mouse. Four groups of animals received two-thirds partial hepatectomies: rats, mice, mice with chimeric livers composed of both transplanted rat hepatocytes and endogenous mouse hepatocytes, and mice with chimeric livers composed of both transplanted and endogenous mouse hepatocytes. Following two-thirds partial hepatectomy, both donor and endogenous hepatocytes in mouse/mouse chimeric livers displayed kinetics of DNA synthesis characteristic of the mouse, indicating that transplantation per se did not affect the response to subsequent partial hepatectomy. In contrast, rat hepatocytes in chimeric mouse livers displayed rat kinetics despite their presence in a mouse host. Thus, factors intrinsic to the hepatocyte must regulate the timing of entry into DNA synthesis. This result defines the process as cell autonomous and suggests that locally or distantly produced cytokines or growth factors may have a permissive but not an instructive role in progression to S.
After surgical removal of two-thirds of the liver, hepatocytes exit a mitotically inactive resting state (G0 phase) and traverse G1 phase, DNA synthesis (S phase), mitosis, and cytokinesis, resulting in replacement of lost cells and restoration of hepatic mass within 1–2 weeks after surgery (1, 2). This precisely regulated response to partial hepatectomy provides one of the most striking examples of the body's ability to recognize and repair tissue damage. Because the timing of entry of hepatocytes into DNA synthesis after hepatectomy is highly synchronous, this process has been studied extensively to provide insight into how the body regulates cell replication. Two important questions have been raised about the liver's early response to hepatectomy. First, what signal(s) initiate the process of restoration of liver mass? Second, once initiated, how is the process regulated?
Clues to the initiating events were provided by genetically modified mice. Mice lacking either the tumor necrosis factor α receptor I (TNFR-I) gene (3, 4) or the interleukin-6 (IL-6) gene (5) exhibited a severely blunted response to partial hepatectomy. Hepatocyte entry into S phase was blocked or delayed, and a large fraction of hepatectomized mice failed to survive the procedure. In both groups, a normal response could be restored by a single injection of IL-6 1 h before surgery. Detailed studies of these experimental systems identified the transcription factors NF-κB (nuclear factor κB) and STAT 3 (signal transducer and activator of transcription) as important targets of TNFR-I/IL-6 signaling. The molecular signals described above are thought to be completed within the first several hours posthepatectomy, and to bring about the hepatocyte transition from G0 to G1 (2).
The second question concerns the nature of the transition through G1 to S phase. Numerous reports have detailed the identity and pattern of expression of genes during G1 progression (reviewed in ref. 6). However, the signals that orchestrate this complex pattern of gene expression have not been determined. Conceptually, there are two alternative mechanisms for control of G1 to S progression (7). The first is noncell autonomous regulation by circulating or locally produced cytokines or growth factors. Candidates include hepatocyte growth factor (HGF), transforming growth factor α (TGFα), and epidermal growth factor (EGF) (reviewed in refs. 1 and 2). In this view, the timing of production or release of growth factors would have an instructive role in hepatocyte progression through G1 to S. In the second mechanism, passage through G1 is cell autonomous: the sequence and timing of molecular events is determined by a program within the hepatocyte itself that is activated on entry into G1. In this view, locally or distantly produced cytokines or growth factors may have a permissive role in the process, but would not override the internal hepatocyte “G1 clock.”
To identify the type of mechanism controlling hepatocyte passage through G1 to S, we have exploited the difference between rat and mouse hepatocytes in the timing of entry into S phase after partial hepatectomy. In rats, DNA synthesis initiates approximately 20 h posthepatectomy, whereas in mice this process initiates at 32–36 h posthepatectomy. By using chimeric livers composed of both rat and mouse hepatocytes in a mouse host, we find that hepatocytes maintain their species-specific response to partial hepatectomy. This finding demonstrates cell autonomous regulation of the timing of hepatocyte progression from G0 to S.
Methods
Animals.
Targeted expression of urokinase-type plasminogen activator (uPA) to mouse hepatocytes is hepatotoxic. uPA-expressing hepatocytes eventually are replaced by progeny of hepatocytes that delete the hepatotoxic transgene, clonally proliferate, and repopulate the liver (8). Transplantation of healthy donor hepatocytes into the spleen of young uPA transgenic mice also can reconstitute a large fraction of the diseased uPA transgenic mouse liver (9). Furthermore, immunodeficient uPA transgenic mice can support extensive parenchymal repopulation (up to 100%) by rat donor hepatocytes despite differences in adult hepatocyte size and ploidy between those hepatocytes (10). We recently described transgenic mice carrying a major urinary protein (MUP)-uPA transgene construct that similarly permits parenchymal repopulation by donor hepatocytes (11), and these mice were used in the studies described below. MUP-uPA transgenic mice are identified by PCR as described (11). Female C57BL/6 transgenic mice were mated with homozygous athymic (nude) Swiss male mice (nu/nu; Taconic Farms) to generate heterozygous nu/+ transgenic mice. The nu/+ transgenic females subsequently were mated with nu/nu nontransgenic males to generate transgenic nu/nu homozygotes (identifiable by lack of hair). These immunocompromised MUP-uPA transgenic mice were used as recipients of hepatocytes isolated from Fisher 344 (F344) rats or from mice carrying a human placental alkaline phosphatase (hPAP) marker transgene (12). F344 rats were obtained from Taconic Farms, and were bred in our colony. The transgenic lines used in these studies have been assigned the following genetic designations: TgN(MUPPlau)1Eps [MUP-uPA line 350–2 (11)] and TgN(R26ALPP)5Eps [R26-hPAP line 808–6 (12)]. Mice and rats were housed and maintained in accordance with the Guide for the Care and Use of Laboratory Animals (44). All experimental procedures were approved by the Animal Care and Use Committee of the School of Veterinary Medicine, University of Wisconsin-Madison.
Liver Cell Isolation and Transplantation and Partial Hepatectomy.
Hepatocytes were isolated from F344 rats or mice carrying a transgene encoding hPAP (12) by using a modified two-step EDTA/collagenase protocol (13). The concentration of viable cells was determined by trypan blue exclusion measurements using a hemacytometer. Recipient mice between 2 and 4 weeks of age were anesthetized with tribromoethanol (Avertin; ICN), the spleen was exteriorized through a small left flank incision, and a Hamilton syringe (no. 81041; Hamilton) with a 26-gauge needle was used to inject 10 μl of the cell suspension into the spleen. The spleen was returned to the abdominal cavity and the incision site was closed with suture and wound clips. The total number of viable hepatocytes transplanted into each recipient mouse was 2–10 × 105. Recipient mice were held for at least 13 weeks to allow time for proliferation and reorganization of donor hepatocytes into normal-appearing parenchyma before partial hepatectomies were performed. Before two-thirds partial hepatectomy, mice were anesthetized with Avertin supplemented with methoxyflurane (Mallinckrodt) as needed. Rats (at least 9 weeks old) were anesthetized with 30 mg/ml ketamine hydrochloride (Fort Dodge Laboratories, Fort Dodge, IA) and 3 mg/ml xylazine (Phoenix Pharmaceuticals, St. Joseph, MO). The left and median lobes of the liver (constituting approximately 70% of total liver mass) were removed by the method of Higgins et al. (14) through a mid-abdominal incision. Experimental animals were allowed to recover on a 37°C warm plate. All surgeries were performed between 10:00 a.m. and 2:00 p.m. to control for diurnal variation
Immunohistochemistry and Histochemistry.
Animals were administered 200 mg/kg body weight BrdUrd (Sigma) 1 h before sacrifice. Livers were weighed, fixed in formalin overnight or 4% paraformaldehyde for 1–2 h at 4°C, embedded in paraffin, and sectioned at 6 μm. A section of small intestine was collected as a positive control for BrdUrd incorporation. Unstained slides were deparaffinized in xylene and hydrated in graded alcohols. Slides were washed in 0.5% H2O2 in methanol for 20 min to block endogenous peroxidase activity, then boiled in 0.1 M Tris buffer for 10 min in a microwave. To identify rat-derived donor hepatocytes, we used mouse monoclonal antibody 106 (mAb 106, a gift of Ron Faris, Rhode Island Hospital and Brown University, Providence, RI) that is specific for a rat hepatocyte cell adhesion molecule (CAM) present on rat bile canaliculi. This antibody was diluted 1:5 in 0.1% nonfat dry milk in PBS and applied to tissue sections for 12–18 h in a humidified chamber. Slides were washed twice in PBS for 5 min each, incubated with biotinylated goat anti-mouse immunoglobulins (AS1200–16; Innogenex, San Ramon, CA) for 30 min, washed twice in PBS for 5 min each, incubated with peroxidase-conjugated streptavidin (CJ-1005–50; Innogenex) for 30 min, and finally incubated with 3,3′-diaminobenzidine [DAB (D-4293; Sigma)] for 5 min. Next, to identify BrdUrd-labeled nuclei, these same slides were incubated for 12–18 h in a humidified chamber with monoclonal rat anti-BrdUrd (MAS 250; Harlan Sera-lab, Sussex, U.K.) diluted 1:40 in 0.1% nonfat dry milk. Specific stain was detected by using biotinylated goat anti-rat immunoglobulins (AS-2100–16; Innogenex), peroxidase-conjugated streptavidin, and DAB as described above. Tissues were counterstained in hematoxylin for 5 min, then dehydrated and mounted under a coverslip.
To detect connexin 32, a gap junction protein present in hepatocytes, livers were frozen in tissue embedding media (SH75–125D; Fisher Scientific) and sectioned at 11 μm. Slides were fixed in 4°C acetone for 10 min, then washed in 0.5% H2O2 in methanol and boiled as described for BrdUrd immunohistochemistry. Polyclonal rabbit anti-connexin 32 antiserum (90–0500; Zymed) was diluted 1:90 in 0.1% nonfat dry milk in PBS and applied to tissue sections for 12–18 h in a humidified chamber. Slides were rinsed twice in PBS, incubated with anti-rabbit immunoglobulins (AS-1400–16; Innogenex) and peroxidase-conjugated streptavidin (Innogenex) each diluted 1:4 in PBS, then incubated with DAB.
To detect hPAP-marked mouse donor cells, recipient livers were fixed in 4% paraformaldehyde for 1–2 h at 4°C, embedded in paraffin, and sectioned at 6 μm. Slides were heated to 65°C for 30 min in alkaline phosphatase (AP) buffer (pH 9.5) containing 0.1 M Tris⋅HCl, 0.1 M NaCl, and 5 mM MgCl2, then incubated 48 h at 37°C in AP buffer plus 0.17 mg/ml BCIP [5-bromo-4-chloro-3-indolyl phosphate (B-6149; Sigma)]. The hPAP-marked donor hepatocytes displayed a blue reaction product. Slides subsequently were stained immunohistochemically to detect BrdUrd, as described above.
Experimental Design.
Animals in four groups were subjected to two-thirds partial hepatectomy, then killed at 4-h intervals between 20 and 48 h posthepatectomy. Experimental groups were (i) F344 rats, (ii) nu/nu nontransgenic mice or nu/nu MUP-uPA transgenic mice that had not received hepatocyte transplants, (iii) nu/nu MUP-uPA transgenic mice with chimeric livers composed of both endogenous mouse and donor rat hepatocytes, and (iv) nu/nu MUP-uPA transgenic mice with chimeric livers composed of both endogenous mouse and donor hPAP-marked mouse hepatocytes. Several animals from each group also were killed without hepatectomy (0 h). A BrdUrd labeling index was determined by examining approximately 1,000 hepatocyte nuclei for each class of hepatocyte (endogenous and donor) per slide at ×400 magnification, and expressing the number of BrdUrd-labeled nuclei as a percent of all nuclei counted. For each liver, nuclei from all zones of the hepatic lobule were examined.
Results
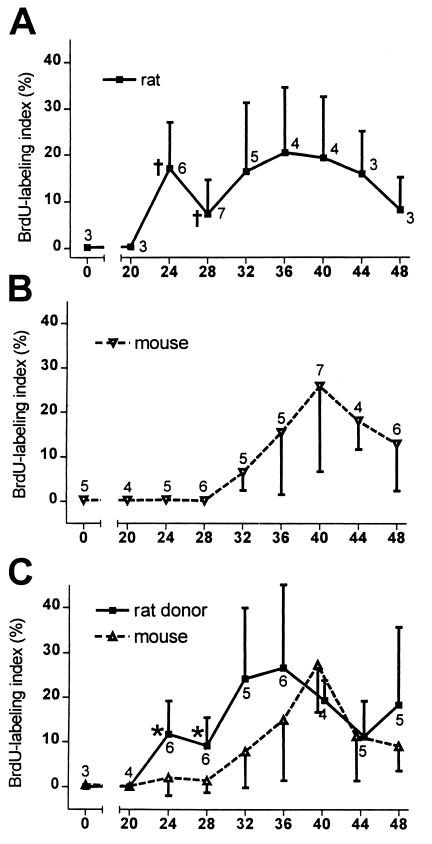
The hepatic responses of rat and mouse livers (groups i and ii) to two-thirds partial hepatectomy were consistent with previous reports (3, 15–17) for each species (Fig. 1 A and B and Fig. 2 A, B, D, and E). Rat hepatocytes displayed an initial peak of BrdUrd incorporation (DNA synthesis) at 24 h posthepatectomy and a second broader peak between 32 and 40 h. In mice, increased labeling became apparent at 32 h and peaked at 40 h. Labeling indices at 24 and 28 h were significantly different between species (P = 0.009 and P = 0.002, respectively; Mann–Whitney U test). The peak percent of labeled cells for each species was less than in some published reports and the second peak in rat cells was larger, as anticipated for animals of the advanced age used in this study (18).
Figure 1.
Kinetics of BrdUrd-labeling in hepatocytes after two-thirds partial hepatectomy (mean ± standard deviation). (A) F344 rat livers, (B) nu/nu mouse livers. (C) MUP-uPA mouse chimeric livers repopulated with donor F344 rat hepatocytes. Number of animals at each time point is indicated on each plot. Statistical differences were identified by using the Mann–Whitney U test. For F344 rat cells versus nu/nu mouse cells, † = P < 0.009. For MUP-uPA endogenous mouse cells versus F344 donor rat hepatocytes in chimeric livers, *, P = 0.015. Data in B represent pooled results from nu/nu nontransgenic mice and nu/nu MUP-uPA transgenic mice that had not received hepatocyte transplants. Values for these groups were not statistically different.
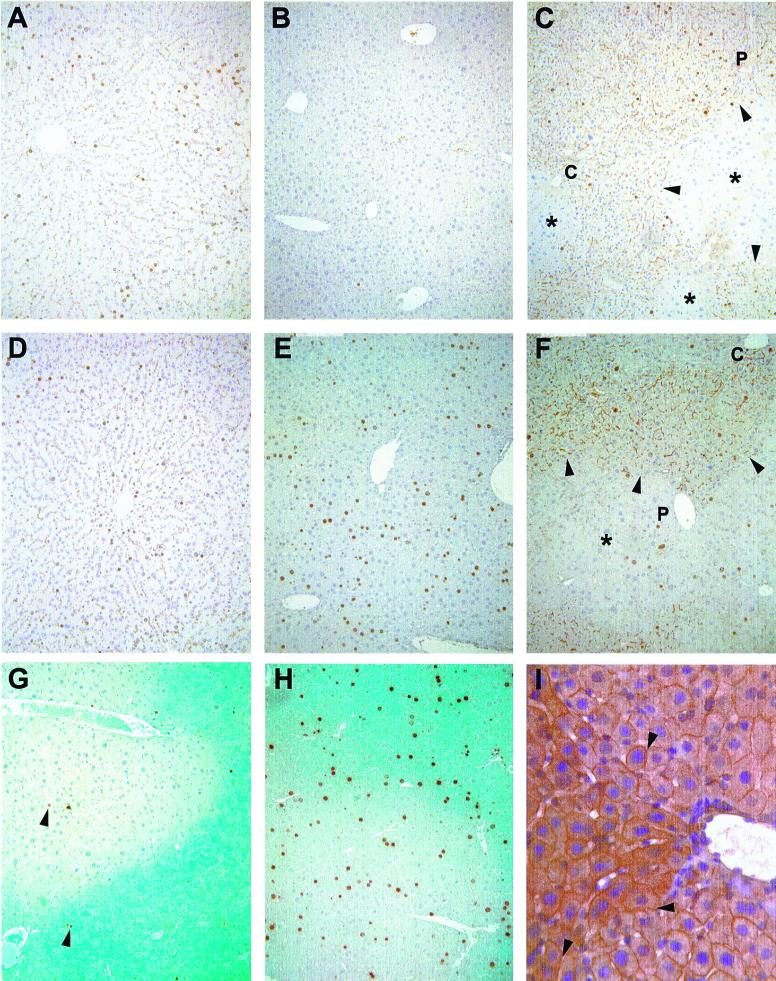
Figure 2.
Immunohistochemistry. Immunohistochemical detection of BrdUrd at 28 (A, B, C, and G) and 44 (D, E, F, and H) h posthepatectomy. (A and D) F344 rat. (B and E) nu/nu mouse. (C and F) MUP-uPA mouse repopulated with donor F344 rat hepatocytes. (G and H) MUP-uPA mouse repopulated with donor hPAP mouse hepatocytes. In C and F, arrowheads point to the edges of donor rat parenchyma, stained with mAb 106 to identify rat bile canilicular antigen (*, focus of endogenous mouse cells; P, portal tract; C, central vein). In G and H, blue staining indicates donor mouse parenchyma expressing the hPAP marker transgene. In G, arrowheads point to BrdUrd-positive hepatocyte nuclei. (A–G original magnification ×100.) (I) Immunohistochemical detection of connexin 32 in chimeric mouse liver. Arrowheads point to cytoplasmic junctions between mouse and rat hepatocytes that show connexin 32 staining (thin brown lines). The triangular patch of cells to the left of the vessel with slightly darker staining is composed of rat hepatocytes. Original magnification ×400.
Hepatocytes in rat/mouse chimeric livers (group iii) displayed BrdUrd labeling indices that were not statistically different from their species of origin at all timepoints (Fig. 1C and Fig. 2 C and F). Rat cells in a mouse host displayed peaks at 24 and 36 h, and mouse cells displayed a peak at 40 h. Rat and mouse hepatocyte labeling indices in chimeric livers were statistically different from one another at 24 and 28 h (P = 0.015 and P = 0.015). To determine whether transplantation altered the response of hepatocytes to partial hepatectomy, we also compared BrdUrd labeling indices between endogenous and donor mouse hepatocytes in MUP-uPA transgenic mice that had received transplanted mouse cells (group iv, Fig. 2 G and H). In 4 mice examined at the 24 and 28 h timepoints, the mean labeling index in donor hepatocytes was 2.1 ± 0.9% and in endogenous hepatocytes was 2.6 ± 1.9%. These values did not differ statistically from each other or from the 24 and 28 h labeling indices of endogenous mouse cells in chimeric livers (P = 0.6; Student's t test). Furthermore, for the 11 mouse/mouse chimeric livers examined between 24 and 44 h, the average ratio of labeling indices in donor versus endogenous hepatocytes was 1.2. Collectively, these data indicate that a history of transplantation does not alter the progression of hepatocytes to S phase after partial hepatectomy.
We also examined the organization of chimeric liver parenchyma at the time of hepatectomy (t = 0). Donor rat and endogenous mouse hepatocytes were present in large patches, not as small foci, and cells of both types spanned all zones of the hepatic lobule (Fig. 2 C and F). Therefore, differential BrdUrd labeling between rat and mouse hepatocytes in chimeric livers cannot be the result of differential location of cells of each species within the lobule. Staining with anti-connexin 32 antibody demonstrated the continuous and uniform presence of this gap junction protein throughout chimeric parenchyma (Fig. 2I). These findings suggested that, at the time of hepatectomy, chimeric livers displayed normal parenchymal organization.
Discussion
Knowledge of the events that induce cellular proliferation after two-thirds partial hepatectomy remains incomplete, but the process must be initiated by signals extrinsic to the hepatocyte (19, 20). Loss of liver mass and associated changes in metabolism initiate a sequence of events involving TNFα, IL-6, and possibly other circulating molecules that causes hepatocytes to exit G0 (1, 2, 6). In contrast, once DNA synthesis begins, progression through S phase becomes irreversible and therefore not subject to extrinsic control. We find that the timing of progression of rat hepatocytes through G1 to S after partial hepatectomy is unaltered when these cells are present in a mouse, despite the fact that the mouse environment “directs” or “supports” significantly different hepatocyte cell cycle kinetics in which the peak of DNA synthesis is delayed by 12 to 16 h relative to rat. If the timing of cell cycle progression was controlled by circulating factors, then the cell cycle kinetics of rat hepatocytes in chimeric mouse livers should reflect the mouse pattern. Thus, in a permissive environment, cell cycle signaling pathways intrinsic to the hepatocyte must assume regulatory control of this process at an early stage after G0 exit. This defines the process as cell autonomous and provides evidence for a molecular “mitotic clock” in hepatocytes that regulates progression through G1 to S.
If G1 progression is cell autonomous, what influence do growth factors have on this process? There is strong evidence associating several growth factors, notably HGF, TGFα, and EGF, with hepatocyte mitosis (1, 2, 21). First, growth factor signaling through the respective receptors appears necessary for hepatocyte cell cycle progression. Phosphorylation of the c-met receptor for HGF occurs rapidly after two-thirds partial hepatectomy (22). Epidermal growth factor receptor (EGFR)-mediated activation of the MEK/ERK (mitogen-activated protein kinase kinase/extracellular signal-regulated kinase) pathway may be required for passage beyond a late G1 restriction point (23). Second, each factor is present in liver at various times posthepatectomy (24). HGF blood concentration increases shortly after partial hepatectomy (25, 26), hepatic TGFα production peaks at mid to late G1 (27), and, in mouse, circulatory EGF is produced constitutively by salivary glands (17). Based on our data reported above, we suggest that these growth factors have a permissive but not an instructive role during posthepatectomy hepatocyte cell cycle progression (that is, their concentration is not limiting for either rat or mouse hepatocytes during G1 progression in the mouse). In this view, hepatocyte replication is initiated by external events (related to loss of hepatic mass) occurring immediately after hepatectomy. The sequence and timing of subsequent molecular events (constituting G1 progression) is directed by a program intrinsic to the hepatocyte itself as long as the environment is permissive. The precise character of the growth factor/cytokine milieu that constitutes a permissive environment may not be important, provided that ligands for all necessary receptor-mediated signaling pathways are present at a sufficient level during passage through G1 restriction points.
This model is compatible with existing data regarding the effects of growth factor perturbations on posthepatectomy hepatocyte replication. First, transgenic mice constitutively overexpressing either TGFα or HGF in hepatocytes display a steady-state increase in hepatocyte turnover and 1.3- to 2-fold increased hepatic mass (28–30), yet they do not display significant alterations in the timing of posthepatectomy cell cycle progression (29, 30). Thus, elevation in concentration of either growth factor alone does not provide an instructive signal that alters posthepatectomy G1 progression. Second, genetically engineered TGFα-null mice have a normal response (timing and magnitude) to partial hepatectomy (31), indicating that this factor is dispensable for hepatocyte G1 progression. In these mice, circulating EGF may preserve the permissive milieu and thereby compensate for the lack of TGFα in this species (17). [A similar analysis of HGF function cannot be conducted because HGF-null mice die in utero (32, 33).] Third, infusion of these growth factors into rodents, alone or in combination, produces only slight elevations in hepatocyte DNA synthesis 24 to 48 h after the start of infusion. The magnitude of the response to growth factor infusion can be increased 3- to 10-fold if these rodent livers first have been “primed” by stimuli such as one-third partial hepatectomy (34, 35) or a brief introduction of collagenase into the portal vein (36). Nevertheless, the timing of this response remains unaltered relative to the smaller response of primed liver lacking growth factor (34, 35). Thus, in vivo priming may increase the sensitivity of hepatocytes to exogenous growth factors both in vivo and in vitro (37), but does not influence the timing of cell cycle events that require growth factor presence.
This model does not require that G1 progression be resistant to influence by all external stimuli. Several experimental manipulations delay hepatocyte initiation of DNA synthesis after partial hepatectomy, including infusion of TGFβ during mid to late G1 (38, 39), sialoadenectomy in mice (which reduces circulating EGF) (17, 40), and reestablishment of normal liver mass by surgical addition of heterotopic liver during mid G1 (41). These manipulations may render the environment nonpermissive for hepatocyte G1 progression by greatly elevating the concentration of a growth-inhibitory molecule (TGFβ) or by reducing below a critical threshold the concentration of growth-permissive molecules (on a systemic or per hepatocyte basis). A TGFβ gradient in the local hepatic microenvironment has been proposed to orchestrate a periportal to pericentral wave of hepatocyte DNA synthesis after two-thirds partial hepatectomy (42). However, this hypothesis would not be consistent with our data unless a TGFβ gradient was established differentially in rat versus mouse parenchyma in chimeric livers (recall that parenchyma of both species' origin spanned all zones of the hepatic lobule). The observation that heterotopic liver transplantation inhibits hepatocyte replication after two-thirds hepatectomy suggests that continuous reduction of liver metabolic capacity below a certain threshold also is a necessary condition for hepatocyte G1 progression in an otherwise permissive host.
Brinster and colleagues (43) have explored regulatory autonomy during the process of spermatogenesis in vivo, by using spermatogonial transplantation to reconstitute mouse seminiferous tubules with rat spermatogonial stem cells. As in our study, Franca et al. (43) determined that spermatogonial cycle length (different in rats and mice) retained its species-specific character in chimeric seminiferous tubules even though the donor rat spermatogonial cells established a close association with mouse Sertoli cells, demonstrating that regulation of this process is intrinsic to the stem cell genotype. Spermatogenesis involves multiple cell doublings but also produces a series of successively more highly specialized cells. There is a gradual reduction of gene transcription as chromosomes become condensed. In contrast, hepatocyte progression through G1 after hepatectomy involves only one or two cell cycles, produces daughter cells that are identical to the parent cell, and occurs while hepatocyte differentiated functions are retained. Despite these differences, each process displays a characteristic species-specific timing that cannot be altered by placing the relevant cells in a different species context, underscoring the importance of cell autonomous control in diverse biological contexts.
Acknowledgments
This study was supported by grants from The National Institutes of Health (RO1-ES07671) and GlaxoWellcome (to E.P.S.), and by Grant 2 T32 ES7015–21 from the National Institute of Environmental Health Sciences (to T.C.W.).
Abbreviations
- MUP
major urinary protein
- EGF
epidermal growth factor
- HGF
hepatocyte growth factor
- TGFα
transforming growth factor α
- TGFβ
transforming growth factor β
- uPA
urokinase type plasminogen activator
- hPAP
human placental alkaline phosphatase
- F344
Fisher 344
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220430497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220430497
References
- 1.Michalopoulos G K, DeFrances M C. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 2.Fausto N. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 3.Yamada Y, Kirillova I, Peschon J J, Fausto N. Proc Natl Acad Sci USA. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada Y, Webber E M, Kirillova I, Peschon J J, Fausto N. Hepatology. 1998;28:959–970. doi: 10.1002/hep.510280410. [DOI] [PubMed] [Google Scholar]
- 5.Cressman D E, Greenbaum L E, DeAngelis R A, Ciliberto G, Furth E E, Poli V, Taub R. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 6.Taub R, Greenbaum L E, Peng Y. Semin Liver Dis. 1999;19:117–127. doi: 10.1055/s-2007-1007104. [DOI] [PubMed] [Google Scholar]
- 7.Michalopoulos G K. FASEB J. 1990;4:176–187. [PubMed] [Google Scholar]
- 8.Sandgren E P, Palmiter R D, Heckel J L, Daugherty C C, Brinster R L, Degen J L. Cell. 1991;66:245–256. doi: 10.1016/0092-8674(91)90615-6. [DOI] [PubMed] [Google Scholar]
- 9.Rhim J A, Sandgren E P, Degen J L, Palmiter R D, Brinster R L. Science. 1994;263:1149–1152. doi: 10.1126/science.8108734. [DOI] [PubMed] [Google Scholar]
- 10.Rhim J A, Sandgren E P, Palmiter R D, Brinster R L. Proc Natl Acad Sci USA. 1995;92:4942–4946. doi: 10.1073/pnas.92.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weglarz, T., Degen, J. & Sandgren, E. (2000) Am. J. Pathol. 157, in press. [DOI] [PMC free article] [PubMed]
- 12.Kisseberth W C, Brettingen N T, Lohse J K, Sandgren E P. Dev Biol. 1999;214:128–138. doi: 10.1006/dbio.1999.9417. [DOI] [PubMed] [Google Scholar]
- 13.Klaunig J E, Goldblatt P J, Hinton D E, Lipsky M M, Chacko J, Trump B F. In Vitro. 1981;17:913–925. doi: 10.1007/BF02618288. [DOI] [PubMed] [Google Scholar]
- 14.Higgins G, Anderson R. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 15.Grisham J. Cancer Res. 1962;22:842–849. [PubMed] [Google Scholar]
- 16.Cornell R P, Liljequist B L, Bartizal K F. Hepatology. 1990;11:916–922. doi: 10.1002/hep.1840110603. [DOI] [PubMed] [Google Scholar]
- 17.Noguchi S, Ohba Y, Oka T. J Endocrinol. 1991;128:425–431. doi: 10.1677/joe.0.1280425. [DOI] [PubMed] [Google Scholar]
- 18.Bucher N L R, Swaffield M N, DiTroia J F. Cancer Res. 1964;24:509–512. [PubMed] [Google Scholar]
- 19.Moolten F L, Bucher N L. Science. 1967;158:272–274. doi: 10.1126/science.158.3798.272. [DOI] [PubMed] [Google Scholar]
- 20.Fisher B, Szuch P, Levine M, Fisher E R. Science. 1971;171:575–577. doi: 10.1126/science.171.3971.575. [DOI] [PubMed] [Google Scholar]
- 21.Fausto N, Laird A, Webber E. FASEB J. 1995;9:1527–1536. doi: 10.1096/fasebj.9.15.8529831. [DOI] [PubMed] [Google Scholar]
- 22.Stolz D, Mars W, Petersen B, Kim T, Michalopoulos G. Cancer Res. 1999;59:3954–3960. [PubMed] [Google Scholar]
- 23.Talarmin H, Rescan C, Cariou S, Glaise D, Zanninelli G, Bilodeau M, Loyer P, Guguen-Guillouzo C, Baffet G. Mol Cell Biol. 1999;19:6003–6011. doi: 10.1128/mcb.19.9.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomiya T, Ogata I, Fujiwara K. Am J Pathol. 1998;153:955–961. doi: 10.1016/s0002-9440(10)65637-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinoshita T, Hirao S, Matsumoto K, Nakamura T. Biochem Biophys Res Commun. 1991;177:330–335. doi: 10.1016/0006-291x(91)91987-n. [DOI] [PubMed] [Google Scholar]
- 26.Zarnegar R, DeFrances M C, Kost D P, Lindroos P, Michalopoulos G K. Biochem Biophys Res Commun. 1991;177:559–565. doi: 10.1016/0006-291x(91)92020-k. [DOI] [PubMed] [Google Scholar]
- 27.Webber E M, FitzGerald M J, Brown P I, Bartlett M H, Fausto N. Hepatology. 1993;18:1422–1431. [PubMed] [Google Scholar]
- 28.Sandgren E P, Luetteke N C, Palmiter R D, Brinster R L, Lee D C. Cell. 1990;61:1121–1135. doi: 10.1016/0092-8674(90)90075-p. [DOI] [PubMed] [Google Scholar]
- 29.Webber E M, Wu J C, Wang L, Merlino G, Fausto N. Am J Pathol. 1994;145:398–408. [PMC free article] [PubMed] [Google Scholar]
- 30.Sakata H, Takayama H, Sharp R, Rubin J S, Merlino G, LaRochelle W J. Cell Growth Differ. 1996;7:1513–1523. [PubMed] [Google Scholar]
- 31.Russell W E, Kaufmann W K, Sitaric S, Luetteke N C, Lee D C. Mol Carcinog. 1996;15:183–189. doi: 10.1002/(SICI)1098-2744(199603)15:3<183::AID-MC4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Nature (London) 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- 33.Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, Kitamura N. Nature (London) 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- 34.Ishiki Y, Ohnishi H, Muto Y, Matsumoto K, Nakamura T. Hepatology. 1992;16:1227–1235. [PubMed] [Google Scholar]
- 35.Webber E M, Godowski P J, Fausto N. Hepatology. 1994;19:489–497. [PubMed] [Google Scholar]
- 36.Liu M L, Mars W M, Zarnegar R, Michalopoulos G K. Hepatology. 1994;19:1521–1527. [PubMed] [Google Scholar]
- 37.Mead J, Braun L, Martin D, Fausto N. Cancer Res. 1990;50:7023–7030. [PubMed] [Google Scholar]
- 38.Thoresen G, Refsnes M, Christoffersen T. Cancer Res. 1992;52:3598–3603. [PubMed] [Google Scholar]
- 39.Russell W E, Coffey R J, Jr, Ouellette A J, Moses H L. Proc Natl Acad Sci USA. 1988;85:5126–5130. doi: 10.1073/pnas.85.14.5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambotte L, Saliez A, Triest S, Maiter D, Baranski A, Barker A, Li B. Hepatology. 1997;25:607–612. doi: 10.1002/hep.510250319. [DOI] [PubMed] [Google Scholar]
- 41.Lambotte L, Saliez A, Triest S, Tagliaferri E M, Barker A P, Baranski A G. Am J Physiol. 1997;273:G905–G912. doi: 10.1152/ajpgi.1997.273.4.G905. [DOI] [PubMed] [Google Scholar]
- 42.Jirtle R L, Carr B I, Scott C D. J Biol Chem. 1991;266:22444–22450. [PubMed] [Google Scholar]
- 43.Franca L R, Ogawa T, Avarbock M R, Brinster R L, Russell L D. Biol Reprod. 1998;59:1371–1377. doi: 10.1095/biolreprod59.6.1371. [DOI] [PubMed] [Google Scholar]
- 44.Committee on Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals. Bethesda: Natl. Inst. Health; 1985. , DHHS Publ. No. (NIH) 85–23. [Google Scholar]