Abstract
Defining molecular interactions that occur at the interface between “normal” and “abnormal” cell populations represents an important but often underexplored aspect of the pathogenesis of diseases with focal origins. Here, we illustrate an approach for conducting such analyses based on mosaic patterns of Cre recombinase expression in the adult mouse intestinal epithelium. Transgenic mice were generated that express Cre in the stem cell niche of crypts located in specified regions of their intestine. Some of these mice were engineered to allow for doxycycline-inducible Cre expression. Recombination in all pedigrees was mosaic: Cre-expressing crypts that supported recombination in all of their active multipotent stem cells were located adjacent to “control” crypts that did not express Cre at detectable levels. Cre-mediated recombination of a floxed LacZ reporter provided direct evidence that adult small-intestinal crypts contain more than one active multipotent stem cell, and that these cells can be retained in both small-intestinal and colonic crypts for at least 80 d. A method was developed to recover epithelial cells from crypts with or without recombination for subsequent gene expression profiling. Stained sections of intestine were used to create electronic image templates to guide laser capture microdissection (LCM) of adjacent frozen sections. This navigated form of LCM overcomes problems with mRNA degradation encountered when cells are marked directly by immunohistochemical methods. Combining Cre-engineered genetic mosaic mice with navigated-LCM will allow biology and pathobiology to be explored at the junction between normal and perturbed cellular cohorts.
Keywords: mouse intestinal stem cells, genetic mosaic analysis
The ability to manipulate gene function in a subset of cells within a tissue has a number of virtues. Genetic mosaic animal models make it easier to determine whether engineered mutations effect various biological processes through cell autonomous or nonautonomous mechanisms. These models also increase the likelihood of identifying subtle phenotypes produced from mutant alleles, because there is a reference control population of normal cells present in the same microenvironment as the experimental cohort. If a gene is essential, restricting its manipulation to a subpopulation of cells in a selected tissue may permit survival of the host to a later developmental stage or to adulthood.
The adult mouse intestinal epithelium is organized in a manner that makes it well suited for genetic mosaic analyses. Renewal occurs rapidly and continuously throughout life and is sustained by multipotent stem cells located in flask-shaped mucosal structures known as crypts of Lieberkühn (1). All active stem cells in a given crypt are derived from a common ancestor (1–3). These stem cells give rise to daughters that undergo several rounds of division, creating a rapidly cycling transit cell population of oligo- and unipotential lineage precursors (4, 5). In the small intestine, epithelial cells belonging to the enterocytic, goblet, and enteroendocrine cell lineages differentiate as they migrate from a crypt up an adjacent finger-shaped villus. Cells from each crypt move up a villus in coherent vertical columns and are removed once they reach the villus tip. In the colon, epithelial cells migrate from a crypt to a flat surface cuff that surrounds its opening (6).
Typically, genetic mosaic analysis in the gut involves the use of chimeras. For example, adult chimeric mice, produced by introducing genetically manipulated 129/Sv embryonic stem (ES) cells into normal C57BL/6 (B6) blastocysts, will contain monoclonal 129/Sv crypts and monoclonal B6 crypts juxtaposed next to one another. The effects of manipulating a gene's expression in one crypt can be defined by examining the adjacent normal B6 crypt. The impact of a genetic manipulation on the orderliness and rate of epithelial cell migration can also be identified, because some villi are supplied by both 129/Sv ES cell- and B6 blastocyst-derived crypts (7). Instructive epithelial–epithelial and epithelial–mesenchymal interactions important for normal intestinal morphogenesis (8–10) and disease pathogenesis (11) can also be examined in chimeras. However, there are some disadvantages. When embryonic cells from strains with different genetic backgrounds are used to produce chimeras, phenotypic differences observed between cell populations in a given chimeric tissue may be caused by strain effects rather than by an engineered genetic manipulation. Moreover, if ES cells homozygous for a null allele of an essential gene are used to produce chimeras, it may be difficult to obtain viable adults.
In this report, we describe an alternative approach for performing genetic mosaic analyses in the intestine, based on the use of Cre recombinase. This enzyme excises DNA sequences located between 34 bp loxP sites (12). Our approach builds on the work of two groups. Betz et al. (13) were able to bypass the embryonic lethality produced by “conventional” knockout of the DNA polymerase β gene (Polβ) by crossing transgenic mice that expressed Cre in multiple organs to mice that contained an engineered polβ allele with internal loxP sites. Southern blot analysis of tissue DNAs indicated that the resulting viable offspring were mosaic with respect to the null polβ allele, presumably because of mosaic patterns of transgene expression. Margolis and Spradling (14) used a heat-shock-inducible FLP recombinase and a tubulin-LacZ reporter gene with FRT sites to mark and characterize ovarian follicular stem cells in genetic mosaic flies.
The system described in this report utilizes transcriptional regulatory elements that reliably direct region-specific and mosaic patterns of Cre expression in the intestine. The system overcomes two of the major challenges encountered when performing Cre-directed gene knockouts in the gut epithelium. One challenge is to retain a recombined allele for a sustained period in all rapidly renewing epithelial cells that populate a given crypt. This requires that Cre be expressed in crypt stem cells. A second challenge arises from the pronounced variations in epithelial differentiation, microbial ecology, and immune functions along the length of the intestine (15). Because gene disruption may have different effects in different regions, it is important to be able to specify where Cre is produced and to have an internal reference control population of normal crypts in that locale. To complement this system, we have developed a method for retrieving epithelial cells from crypts with and without recombination. This method, based on a generally applicable form of guidance for laser capture microdissection, yields mRNA suitable for profiling gene expression.
Materials and Methods
Mice.
To generate Fabp−596 to −21Cre mice, a 1.0-kb SalI/EcoRI fragment encoding Cre recombinase was excised from pML78 (ref. 16; a gift from Gail Martin, University of California, San Francisco) and placed between nucleotides −596 to +21 of rat Fabpl and nucleotides +3 to +2150 of the human growth hormone gene (hGH). Fabpl−596 to +21Cre-hGH was isolated as a 4.0-kb EcoRI fragment and injected into FVB/N oocytes. Fabpl4X at −132Cre mice were generated as described (17). R26R mice (ref. 18; kindly provided by P. Soriano, University of Washington, Seattle) were crossed to both types of FabplCre animals.
Assaying Recombination.
Escherichia coli β-galactosidase (LacZ) produced after recombination of R26R was detected by 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) staining of fixed whole-mount preparations of small intestine, cecum, and colon (7). Five-micrometer-thick serial sections were cut from paraffin-embedded blocks of the X-Gal-stained whole mounts and counterstained with nuclear fast red (NFR). The percentage of crypts that support recombination was defined by scoring 10 sections per mouse, each cut along the cephalocaudal axis of the intestine and separated by 50 μm.
Navigated-LCM.
The lumen of the distal third of the small intestine was perfused with then submerged in OCT compound (Miles) and frozen in Cytocool (Richard Allen Scientific, Kalamazoo, MI). Five-micrometer-thick serial sections were cut parallel to the cephalocaudal axis, placed on numbered Superfrost/Plus slides (Fisher), and put immediately on dry ice. Even-numbered sections were air dried, fixed in 70% EtOH (30 s), incubated in X-Gal solution (10 h, 30°C), washed in dH2O (30 s), and stained with NFR (5 min).
Adjacent odd-numbered sections were prepared for LCM by air drying (30 s), fixation (70% EtOH, 30 s), washing (sterile dH2O, 30 s), NFR staining (60 s), rinsing (dH2O, 30 s), dehydration (graded alcohols, 30 s each), washing (xylene, 3 × 2 min), and air drying before storage (dessicator).
A Zeiss Axioscop with Progress 5.0 camera system was used to capture overlapping digital images from all regions of a stained even-numbered section. Overlapping images were pieced together by using Canvas (Deneba, Miami, FL). Navigated-LCM was performed with a PixCell II apparatus (Arcturus, Mountain View, CA; 7.5-μm-diameter laser spot).
PCR and Reverse Transcription–PCR (RT-PCR) Assays of DNA and mRNA Recovered from Microdissected Ileal Crypt Epithelium.
Genomic DNA was extracted from 2,000 captured cells in a 50-μl reaction [10 mM Tris⋅HCl, pH 8/1 mM EDTA/1% Tween-20/0.04% Proteinase K (Roche Diagnostics, Indianapolis, IN); 10-h incubation at 37°C, then 20 min at 100°C]. PCR was performed using 1 μl of the DNA preparation and primers that distinguish intact from recombined R26R (primer 1 = 5′-AAAGTCGCTCGAGTTGTTAT; primer 2 = 5′-GCGAAGAGTTTGTCCTCAACC; primer 3 = 5′-CGATTAAGTTGGGTAACGCC).
RNA was recovered from 2,000 captured crypt epithelial cells by using the RNA MicroIsolation Kit (Strategene; protocols outlined in http://dir.nichd.nih.gov/lcm/Protocol.htm) and precipitation with Glycoblue (Ambion) plus isopropanol. For RT-PCR, cDNA was synthesized in 20-μl reactions containing 250 ng oligo(dT) and 25 units AMV reverse transcriptase (Roche). PCR primers spanned an intron/exon junction of the mouse E-cadherin gene (5′-GTCAACACCTACAACGCTGCC; 5′-GTTGTGCTCAAGCCTTCGC) or a portion of hGH contained in the FabplCre transcript (5′-AGGTGGCCTTTGACACCTACCAGG; 5′-TCTGTTGTGTTTCCTCCCTGTTGG). Forty cycles of PCR were performed (annealing = 58°C).
SYBR Green-Based Real-Time Quantitative RT-PCR Studies of mRNA Degradation Associated with Immuno-LCM.
Five-micrometer-thick sections were cut from OCT-embedded ileum, thawed (30 s, 23°C), postfixed in 70% EtOH (30 s), and incubated for 1, 10, or 30 min with primary antibodies [rabbit anti-α actin or rat anti-mouse E-cadherin sera (Sigma); diluted 1:100 in sterile PBS with or without pretreatment with RNasin (Roche; 10-unit enzyme/microliter sera)]. Antigen–antibody complexes were visualized as follows: 2-min incubation (23°C) with biotinylated anti-rabbit Ig (ABC kit, Vector Laboratories, diluted 1:100 in PBS); 1-min incubation with avidin and biotin–horseradish peroxidase complexes; 5 min with 3,3′-diaminobenzidine; 60 s with NFR.
RNA was prepared from ≈20,000 ileal villus epithelial cells harvested by LCM from sections that had no antibody exposure (navigated-LCM control) or had been incubated for 1–30 min with primary antibodies (immuno-LCM). In addition, RNA was extracted from entire ileal sections (each containing ≈1,000 crypt-villus units) that had been recovered with a razor-blade scrape from slides treated with NFR alone or with antibodies.
cDNA was produced from all RNA samples using either oligo(dT) or random hexamer primers (Life Technologies, Grand Island, NY). SYBR green-based real-time RT-PCR (19, 20) was performed (Applied Biosystems 7700 Sequence Detection System) in 25-μl reactions [12.5 μl 2× SYBR Green master mix (Applied Biosystems), 900 nM of each primer, 0.25 units UDP-N-glycosidase (Life Technologies) plus cDNA].
The following primer pairs were employed: for amplification of (i) a portion of the 5′ region of E-cadherin mRNA (5′-GTCAACACCTACAACGCTGCC; 5′-GTTGTGCTCAAGCCTTCGC); (ii) 3′ region of E-cadherin mRNA, 5′-AGGAAATGCACCCCTCCAAT-3′ and 5′-AATCGGCCAGCATTTTCTG-3′; (iii) 5′ region of Adenomatous polyposis coli (Apc) mRNA, 5′-TGACAAGACGGCAGCTGGAG-3′ and 5′-TCTTCGCTGTGCACGCTTC-3′; (iv) middle region of Apc mRNA, 5′-TAGGAAGAGCAGCGCAGACA-3′ and 5′-AGACCCGGAATGGCGTTTAG-3′; (v) Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) mRNA, 5′-TGGCAAAGTGGAGATTGTTGCC-3′ and 5′-AAGATGGTGATGGGCTTCCCG-3′; (vi) 18S rRNA, 5′-CATTCGAACGTCTGCCCTATC-3′ and 5′-CCTGCTGCCTTCCTTGGA-3′. The melting temperatures for amplicons ranged from 79 to 80°C. Control experiments established that the signal for each amplicon was derived from cDNA and not from primer dimers.
The ΔΔCT method (User Bulletin #2, Applied Biosystems) was modified to establish a mRNA integrity index for Apc and E-cadherin mRNAs: because both amplicons were generated from a given mRNA, the CT for the middle (Apc) or 3′ (E-cadherin) amplicon was used as the calibrator.
Results and Discussion
Directing Cre Recombinase to Crypt Stem Cells Located in the Proximal or Distal Intestine.
Pedigrees of FVB/N mice were established that contain Cre under the control of nucleotides −596 to +21 of a rat fatty acid-binding protein gene (Fabpl−596 to +21Cre). RNase protection assays of 8-week-old mice indicated that Cre mRNA levels were highest in the proximal half of their small intestine, fell markedly in the distal half, rose slightly in the cecum, and were barely detectable in the colon. Liver was the only other site of expression. Other pedigrees had Cre under the control of a Fabpl−596 to +21 derivative that contained four extra copies of nucleotides −177 to −132 inserted at position −132 (Fabpl4X at −132). Cre mRNA in these mice is present in the distal small intestine, cecum, colon, and ureter/bladder (17).
The efficacy of Cre-mediated recombination was defined by crossing Fabpl−596 to +21Cre or Fabpl4X at −132Cre transgenics to mice hemizygous for R26R (18). R26R contains a neomycin resistance gene cassette flanked by loxP sites, followed by the lacZ ORF in the ROSA26 locus, which is active in essentially all cells during and after completion of development (7, 18). Bi-transgenic mice were examined at embryonic day (E)13.5, E14.5, E18.5, and 5–8 weeks (n = 10–14 mice/group/time point).
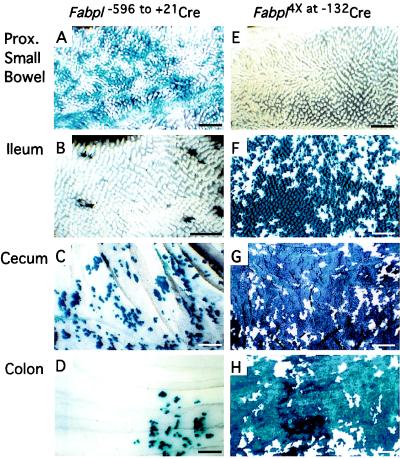
Fabpl−596 to +21Cre produces a patchy pattern of R26R recombination in the intestinal endoderm as early as E13.5 (data not shown). Surveys of X-Gal-stained intestinal whole mounts from 6-week-old bi-transgenic mice (n = 15) revealed a distribution of R26R recombination (Fig. 1 A–D) that paralleled the cephalocaudal distribution of Cre mRNA. The principal site of recombination was the proximal third of the small intestine where 15.2 ± 3.7% of crypts were composed of a wholly LacZ+ population of epithelial cells (n = 3 mice; total of 10,504 crypts scored).
Figure 1.
Regional features of FabplCre-mediated recombination in the intestinal epithelium. Whole mounts, prepared from 6-week-old bi-transgenic mice, were stained with X-Gal to visualize LacZ as blue. (Bars = 1.08 mm.)
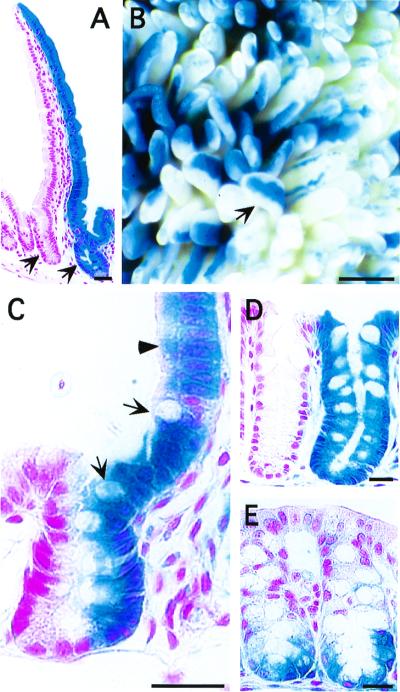
Each small-intestinal villus in the adult mouse is supplied by several crypts. The precise number of active stem cells in these crypts has not been determined (4). The cellular output from juxtaposed monophenotypic LacZ+ and monophenotypic LacZ− crypts gives their associated villus a striped appearance. Each stripe is composed of a coherent column of either wholly LacZ+ or LacZ− cells (Fig. 2 A and B). If a crypt is supplied by more than one active multipotent stem cell, and if Cre-mediated recombination of R26R is not uniform among these lineage progenitors, then that crypt would be composed of a mixture of LacZ+ and LacZ− cells. Such a mixed crypt would supply its associated villus with a cellular cohort having both LacZ+ and LacZ− phenotypes. The homogenous LacZ expression phenotype in crypts and in their upwardly migrating villus epithelial progeny provides evidence for uniform recombination.
Figure 2.
Mosaic patterns of Cre recombination. (A–C) Six-week-old Fabpl−596 to +21Cre/R26R mice. (A) X-Gal- and NFR-stained section from the proximal third of the small intestine. Crypts (e.g., arrows) are monophenotypic: they contain a wholly LacZ+ or LacZ− population of epithelial cells. (B) X-Gal-stained whole mount of proximal small intestine showing striped villi (e.g., arrow). (C) Rare crypts contain segregated cohorts of LacZ+ and LacZ− cells that extend to adjacent villi (arrows and arrowheads point to members of the goblet cell and enterocytic lineages, respectively). We refer to crypts with this distinct sidedness as having a Cheron phenotype, after the protagonists Lokai and Bele in the StarTrek episode “Let that be your last battlefield.” (D and E) X-Gal-stained sections from the proximal colon of 6-week-old Fabpl4X at −132Cre/R26R mice. (D) Monophenotypic crypts. (E) A small fraction of crypts exhibit a distinctive pattern of segregation of LacZ+ cells to their base. (Bar in B = 1.08 mm; elsewhere, 25 μm.)
Only a small percentage (0.33 ± 0.08%) of proximal small-intestinal crypts in Fabpl−596 to +21Cre/R26R mice contained both LacZ+ and LacZ− cells. Fig. 2C shows a representative biphenotypic crypt containing a homogenous population of LacZ+ cells on one side and a homogenous population of LacZ− cells on its other side. Each cohort contains members of each of the intestine's epithelial lineages and extends upward, as an uninterrupted column, to the tip of a villus (Fig. 2C). This finding provides direct evidence that an adult small-intestinal crypt contains more than one active multipotent stem cell. It also reveals that the descendants of each stem cell can maintain a remarkable degree of segregation in a structure where two-thirds of epithelial cells are dividing every 12–19 h (5).
Mice with the other transgene (Fabpl4X at −132Cre) exhibit foci of LacZ+ intestinal endoderm as early as E14.5 (data not shown). The pattern of R26R recombination in 6-week-old bi-transgenic Fabpl4X at −132Cre/R26R animals (n = 23) is shifted to the distal small intestine and colon compared with Fabpl−596 to +21Cre/R26R mice. Wholly LacZ+ crypts represent 2.1 ± 2% of all crypts in the proximal third and 47.2 ± 7.1% in the distal 2 cm of the small intestine, 70.2 ± 1.8% in the cecum, and 67.5 ± 3.5% in the colon (Figs. 1 E–H and 2D) (n = 3 mice; 9,112–25,509 crypts scored/ segment).
Biphenotypic crypts are also rare in these Fabpl4X at −132Cre/ R26R mice (≤0.6%). Two types were encountered in the proximal colon: some had a small scattered population of LacZ+ cells intermingled among LacZ− cells; others contained a homogenous population of LacZ+ cells that filled the lower quarter of the crypt (Fig. 2E). Previous studies have suggested that the stem cell niche in proximal colonic crypts is located above the crypt base (21). The presence of a cohort of LacZ+ cells in the lower quarter of biphenotypic crypts would support this proposal if Cre-mediated R26R recombination in these crypts were limited to a progenitor that gives rise to a downward-migrating lineage. The deep crypt secretory cell progenitor is one candidate (21).
Only one extraintestinal site of R26R recombination was detected in either type of adult bi-transgenic mouse: hepatocytes in the case of Fabpl−596 to +21Cre/R26R, and all layers of the urothelium, from the renal calyces to the ureters and bladder, in the case of Fabpl4X at −132Cre/R26R (n = 15–23 mice/genotype). Access to these sites should be useful when examining genes that participate in enterohepatic functions or for comparing the impact of a genetic manipulation on a rapidly renewing simple epithelium (intestine) and a slowly renewing transitional epithelium (urinary tract).
Inducible Cre-Mediated Recombination in Crypt Stem Cells.
The Fabpl/Cre mice described above do not offer the option of regulated expression of Cre. One way of gaining temporal control of recombination is to generate mice that express Cre under the control of the reverse tetracycline-regulated transactivator (rtTA) (17, 22). Members of three pedigrees of Fabpl4X at −132/rtTA transgenics were each crossed to two other types of mice: one containing seven copies of tetO linked to a minimal promoter followed by the Cre ORF (17) and the other R26R. Tri-transgenic mice (4–6 weeks old; n = 29) were given water containing doxycycline (2 mg/ml) and sucrose (5%) for 6 d and then killed. X-Gal-stained whole mounts of their intestines revealed recombination in 19/29 animals (66%). The failure to induce recombination in some of these mice could not be related to differences in their measured consumption of doxycycline, age, or gender. Moreover, recombination was not improved by i.p. injection of drug (n = 10 mice). No recombination was observed in controls (n = 7) that received 5% sucrose alone, as defined by X-Gal staining of intestinal whole mounts.
In tri-transgenic mice generated from two of the Fabpl4X at −132rtTA pedigrees, recombination was restricted to the cecum and colon (5–20% and <5% of crypts, respectively). Only a small subset of these crypts contained a monophenotypic population of LacZ+ cells: most were biphenotypic.
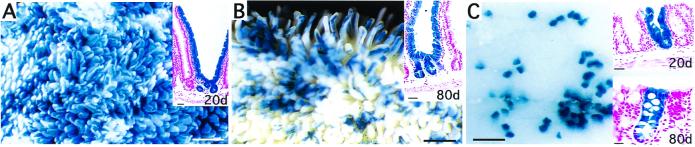
Mice produced from the third Fabpl4X at −132rtTA pedigree exhibited recombination in the proximal, middle, and distal thirds of their small intestine (14 ± 0.9, 6 ± 0.8, and 1.2 ± 0.6% of total crypts, respectively), cecum (3.5 ± 1%), and colon (14 ± 4%) (n = 2 mice; 2,683–6,482 crypts scored/segment). Most importantly, >99% of these crypts were wholly LacZ+. Moreover, when doxycycline was given for 6 d and animals killed 20 d or 80 d later (time for 4 or 16 cycles of epithelial renewal, respectively, in the small intestine and proximal colon), monophenotypic LacZ+ crypts were retained in each case (n = 4/6 mice at the 80 d time point) (Fig. 3). These wholly LacZ+ crypts provide direct evidence that doxycycline-induced Cre-mediated recombination of R26R can occur in all of a crypt's active multipotent stem cells, and that these “marked” stem cells can be retained for at least 80 d.
Figure 3.
Doxycycline-inducible Cre recombination. (A and B) Proximal small intestines from Fabpl4X at −132rtTA/tetO-Cre/R26R mice analyzed 20 d and 80 d after cessation of doxycycline treatment. Insets show that monophenotypic LacZ+ crypts are present in each case. (C) Ceca from mice killed 20 d and 80 d after treatment. (Bars in A and B = 1.08 mm; C = 0.55 mm; Insets = 25 μm.)
An inducible system for Cre-mediated recombination in crypts has the advantage of allowing gene function to be manipulated/audited at selected times, under specified physiologic or pathologic conditions, in all cells of the crypt including its stem cells. This system provides a means for studying stem cell turnover and for conducting simulations of the types of “stochastic” changes in gene expression that occur in subsets of crypts during initiation or progression of a number of focally derived diseases of the intestinal epithelium (e.g., cancer).
Recovering Cells from Crypts by Using Navigated-LCM.
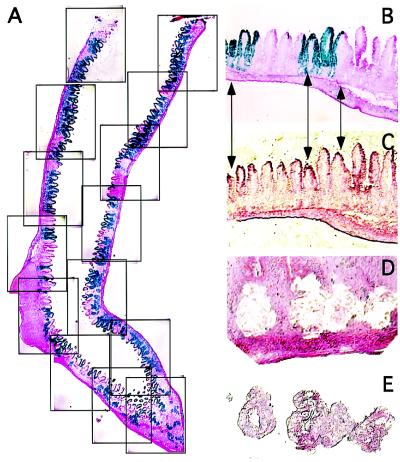
LCM (23, 24) represents a way to recover epithelial cells from selected crypts and to profile the effect of a knockout on gene expression. Cells in fresh-frozen tissue sections can be marked for LCM by direct histo- or immunohistochemical staining (25, 26). Unfortunately, staining protocols that require more than a few minutes to complete compromise the ability to recover intact mRNA (ref. 27 and below). An alternative is to stain an adjacent section (28) and use it as a guide for the microdissection. Fig. 4 illustrates this latter method, which we call navigated-LCM. Serial 5-μm-thick frozen sections are prepared from the ileum. The section preceding the one targeted for LCM is stained with X-Gal and NFR (or antibodies) to identify crypts with and without recombination. Electronic images are captured from the entire stained section and used to assemble a panoramic view (Fig. 4A). The composite image is projected on a computer screen. Regions containing clusters of monophenotypic LacZ+ and LacZ− crypts are magnified and distinctive topologic features referenced to a juxtaposed “live” video image of the adjacent section targeted for LCM (Fig. 4 B and C). The electronic image template is used as a guide to navigate the microdissection (Fig. 4 D and E). The process can be repeated through a group of serial sections.
Figure 4.
Navigated-LCM. (A) Montage of captured electronic images of an X-Gal- and NFR-stained section of terminal ileum from a Fabpl4X at −132Cre/R26R mouse. (B–E) LCM of a cluster of neighboring monophenotypic LacZ+ and LacZ− crypts: (B) electronic image template; (C) adjacent NFR-stained section targeted for LCM; (D) removal of crypts; (E) captured crypts.
PCR and RT-PCR Analysis of R26R Recombination and Cre Expression in LacZ+ and LacZ− Crypts Recovered by Navigated-LCM.
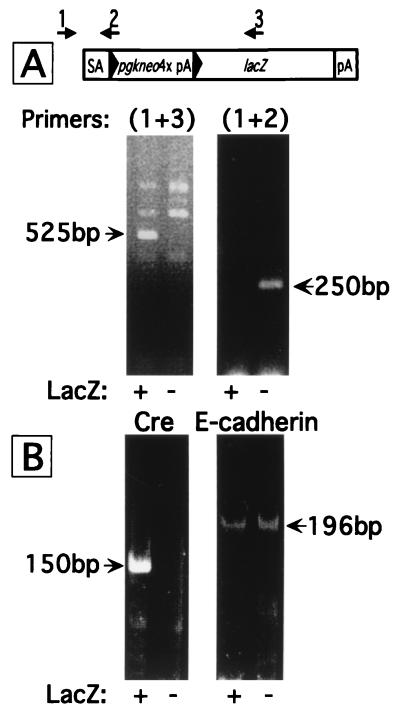
DNA was isolated from epithelial cells that had been dissected from 100 monophenotypic LacZ+ and 100 LacZ− crypts (≈2,000 epithelial cells/phenotype). PCR assays confirmed that R26R locus genotype correlates with LacZ phenotype (Fig. 5A). RNA was purified from epithelial cells populating 50 monophenotypic LacZ+ crypts and 50 LacZ− crypts. RT-PCR established that Cre expression correlates with R26R genotype and LacZ phenotype: Cre mRNA is present in LacZ+ crypts and undetectable, even after 40 cycles of PCR, in LacZ− crypts (Fig. 5B). Control RT-PCR demonstrated that the mRNA encoding the principal intestinal epithelial cadherin (E-cadherin; involved in cell–cell adhesion) was present at equal levels in LacZ+ and LacZ− crypts (Fig. 5B).
Figure 5.
PCR genotyping and RT-PCR profiling of gene expression in juxtaposed LacZ+ and LacZ− crypt epithelial cells recovered by navigated-LCM. (A) R26R genotyping. Primers 1,2 were used to detect the intact locus, primers 1,3, the recombined locus. DNA from monophenotypic LacZ+ crypts yield a 525-bp PCR product from the recombined R26R locus (Left) and no detectable 250-bp product from the intact locus (Right). (B) PCR of cDNA showing that the FabplCre mRNA transcript is detectable only in LacZ+ crypts, whereas E-cadherin mRNA is present in both LacZ+ and LacZ− crypts.
Together, these analyses support our conclusion that recombination produced by FabplCre affects all active stem cells and their progeny in LacZ+ crypts, and that a wholly LacZ− phenotype reflects a failure of these populations to support Cre expression (at least to levels detectable by our RT-PCR assay and necessary for recombination of R26R). In the absence of known molecular or morphologic markers of multipotent crypt stem cells, it is not possible to directly assess Cre and LacZ in this cellular population. Thus, we cannot directly distinguish between the possibility that a wholly LacZ+ crypt phenotype is produced because of a single recombination event occurring in a progenitor that gives rise to a limited set of active multipotential descendants, vs. multiple independent “uniform” recombination events occurring in these descendants.
Comparison of the Integrity of Ileal RNA Prepared After Navigated- LCM and After Immuno-LCM.
To establish the efficacy of navigated- LCM in recovering intact RNA from microdissected cells, frozen sections of ileum were incubated with antibodies directed against α-actin. This abundant epithelial protein can be detected by using fixation conditions compatible with LCM and subsequent RNA isolation. A 10-min incubation with α-actin antibodies was barely sufficient to visualize the protein. A 30-min incubation produced a good signal (data not shown).
Some slides containing ileal sections were stained only for 60 s with NFR before microdissection to recapitulate conditions used for navigated-LCM. Other sets of slides were incubated for 1, 10, or 30 min with the primary antibody. Each of these slides was then subjected to a multistep procedure for visualizing antibody–antigen complexes that requires 8 min to complete, a 60-s counterstain with NFR, and then LCM.
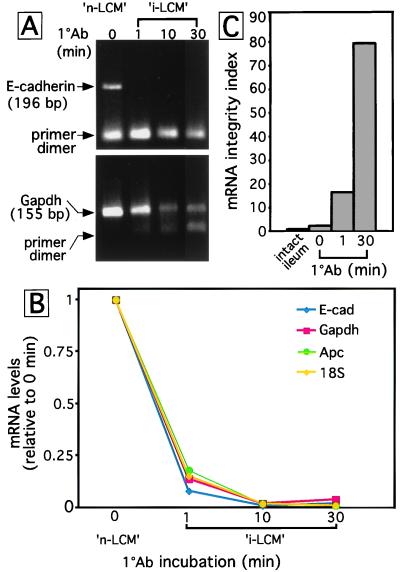
RNA was prepared from ≈20,000 cells dissected from sections subjected to each of these protocols, and E-cadherin mRNA levels were defined by SYBR Green-based quantitative real-time RT-PCR using oligo(dT)-primed cDNA and PCR primers directed to a region derived from the 5′ end of this mRNA species. The results (Fig. 6A) show that E-cadherin mRNA was detectable in the “navigated”-LCM control but in none of the samples prepared after direct immuno-LCM.
Figure 6.
Comparison of the integrity of mRNA isolated after navigated-LCM and immuno-LCM. (A) Real-time RT-PCR. Each cDNA was prepared by using oligo(dT) and RNA from ≈20,000 LCM ileal villus epithelial cells. After 40 cycles of PCR with primers that recognize sequences from the 5′ end of E-cadherin, reaction products were resolved by agarose gel electrophoresis. (B) Effects of navigated (n)- and immuno (i)-LCM protocols on levels of various RNA species. Oligo(dT)-primed cDNA was generated from equivalent number of ileal cells that had no antibody exposure (zero time point; n-LCM conditions) or had been subjected to i-LCM conditions. Real-time RT-PCR used aliquots of cDNA representing ≈200 crypt-villus units and primers directed to regions derived from the 5′ ends of all RNAs surveyed. (C) Real-time RT-PCR assays of Apc mRNA integrity in RNA prepared from intact ileum and from cells scraped from ileal sections subjected to n-LCM or i-LCM conditions. See text for definition of the integrity index.
To increase the number of cells used for quantitative real-time RT-PCR and to extend the analysis to other intestinal epithelial mRNA species, cells subjected to the same staining protocols as above were recovered from entire sections by scraping rather than by LCM. A 1-min incubation with the primary antibody (followed by the 8-min visualization step) produces a dramatic reduction in the level of 18S rRNA and equally pronounced reductions in the levels of mRNAs encoding proteins with a variety of cellular activities: E-cadherin; the glycolytic enzyme glyceraldehyde-3 phosphate dehydrogenase; and adenomatous polyposis coli (Apc), a 2,843-aa residue intracellular protein that participates in several functions including β-catenin signaling (ref. 29; Fig. 6 A and B). Pretreatment of the sera with a ribonuclease inhibitor did not affect the results. Moreover, degradation was not a unique property of the α-actin antibody preparation: incubation with E-cadherin antibodies produced similar results (data not shown).
A real-time quantitative RT-PCR study was performed of the integrity of the 10,381-nt Apc mRNA in cells isolated from ileal sections that had, or had not, been exposed to antibodies. mRNA degradation in mammalian cells can occur 3′ to 5′ or 5′ to 3′ (30). Therefore, the analysis was based on a comparison of the ratios of amplicons generated from the 5′ and middle regions of this mRNA. Random hexanucleotide primers were used to synthesize cDNA so that there would be unbiased representation of intact as well as partially degraded mRNA species. cDNAs prepared from RNAs isolated from (i) snap-frozen ileum and (ii) ileal epithelial cells scraped from sections that had no antibody exposure and just a 60-s NFR stain were used as reference controls. Using the cDNA from snap-frozen ileum as a template, we could demonstrate that amplification of both regions of Apc was linear as a function of increasing cDNA concentration (data not shown).
A mRNA integrity index was derived by comparing the ratio of amplicons, derived from the 5′ and middle regions of Apc mRNA, using a modified ΔΔCT analysis (Materials and Methods). The ratio of the 5′ to middle region amplicons from Apc mRNA in RNA prepared from snap-frozen intact ileum was 1:1 (integrity index = 1 in Fig. 6C). Navigated-LCM conditions produce only modest mRNA degradation (integrity index = 3), whereas the immuno-LCM steps result in pronounced loss of integrity (indices = 17 and 80 at the 1- and 30-min time points, respectively; n = 6 independent determinations/time point, each in triplicate; Fig. 6C). These findings were not unique to Apc mRNA: analysis of the same RNA samples revealed time-dependent degradation of E-cadherin mRNA under immuno-LCM conditions (data not shown).
Prospectus.
Our studies establish that Cre can be used to create a system for auditing gene function in an epithelium well suited for genetic mosaic analysis. Unlike chimeric mice generated by traditional means, all cells in the Cre-based system are otherwise isogenic. Navigated-LCM can be used to recover DNA and intact mRNA from specified crypt epithelial cell populations with and without a recombined allele. Real-time quantitative RT-PCR and/or DNA microarray profiling of gene expression (31) should allow biology and pathobiology to be defined at the interface between normal and manipulated (abnormal) cellular cohorts. These capabilities will enhance our ability to explore the molecular pathogenesis of (intestinal) diseases having focal origins.
Characterization of intestinal stem cells has been a “holy grail” to workers in the field. A dividend of this system is that Cre is expressed in multipotent crypt stem cells. We have used Cre to provide direct evidence that small-intestinal crypts contain several active stem cells, and that small-intestinal and colonic crypts can retain these cells for as long as 80 d. The doxycycline-inducible Cre-based recombination system can be used in future studies to directly manipulate expression of specific genes in gut stem cells and thus define the molecular basis of the functions of these cells.
Acknowledgments
We thank Sabrina Wagoner, David O'Donnell, and Maria Karlsson for invaluable technical assistance. This work was supported by National Institutes of Health Grants DK30292 and DK09724.
Abbreviations
- Apc
adenomatous polyposis coli
- LacZ
E. coli β-galactosidase
- LCM
laser capture microdissection
- NFR
nuclear fast red
- R26R
ROSA26 reporter
- rtTA
reverse tetracycline-regulated transactivator
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- RT-PCR
reverse transcription–PCR
- En
embryonic day n
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230237997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230237997
References
- 1.Booth C, Potten C S. J Clin Invest. 2000;105:1493–1499. doi: 10.1172/JCI10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winton D J, Blount M A, Ponder B A J. Nature (London) 1988;333:463–466. doi: 10.1038/333463a0. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt G H, Winton D J, Ponder B A J. Development (Cambridge, UK) 1988;103:785–790. doi: 10.1242/dev.103.4.785. [DOI] [PubMed] [Google Scholar]
- 4.Bjerknes M, Cheng H. Gastroenterology. 1999;116:7–14. doi: 10.1016/s0016-5085(99)70222-2. [DOI] [PubMed] [Google Scholar]
- 5.Potten C S, Loeffler M. Development (Cambridge, UK) 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt G H, Wilkinson M M, Ponder B A J. Cell. 1985;40:425–429. doi: 10.1016/0092-8674(85)90156-4. [DOI] [PubMed] [Google Scholar]
- 7.Wong M H, Rubinfeld B, Gordon J I. J Cell Biol. 1998;141:765–777. doi: 10.1083/jcb.141.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kedinger M, Lefebvre O, Duluc I, Freund J N, Simon-Assmann P. Philos Trans R Soc London Biol Sci. 1998;353:847–856. doi: 10.1098/rstb.1998.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaestner K H, Silberg D G, Traber P G, Schutz G. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- 10.Pabst O, Zweigerdt R, Arnold H H. Development (Cambridge, UK) 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 11.Kinzler K W, Vogelstein B. Science. 1998;280:1036–1037. doi: 10.1126/science.280.5366.1036. [DOI] [PubMed] [Google Scholar]
- 12.Abremski K, Hoess R. J Biol Chem. 1984;259:1509–1514. [PubMed] [Google Scholar]
- 13.Betz U A K, Vosshenrich C A J, Rajewsky K, Muller W. Curr Biol. 1996;6:1307–1316. doi: 10.1016/s0960-9822(02)70717-3. [DOI] [PubMed] [Google Scholar]
- 14.Margolis J, Spradling A. Development (Cambridge, UK) 1995;121:3797–3807. doi: 10.1242/dev.121.11.3797. [DOI] [PubMed] [Google Scholar]
- 15.Falk P G, Hooper L V, Midtvedt T, Gordon J I. Micro Mol Biol Rev. 1998;62:1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewandoski M, Meyer E N, Martin G R. Cold Spring Harbor Symp Quant Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- 17.Saam J R, Gordon J I. J Biol Chem. 1999;274:38071–38082. doi: 10.1074/jbc.274.53.38071. [DOI] [PubMed] [Google Scholar]
- 18.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 19.Heid C A, Stevens J, Livak K J, Williams P M. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 20.Steuerwald N, Cohen J, Herrera R J, Brenner C A. Mol Hum Reprod. 1999;5:1034–1039. doi: 10.1093/molehr/5.11.1034. [DOI] [PubMed] [Google Scholar]
- 21.Altmann G G. Am J Anat. 1983;167:95–117. doi: 10.1002/aja.1001670109. [DOI] [PubMed] [Google Scholar]
- 22.Utomo A R H, Nikitin A Y, Lee W-H. Nat Biotechnol. 1999;17:1091–1096. doi: 10.1038/15073. [DOI] [PubMed] [Google Scholar]
- 23.Emmert-Buck M R, Bonner R F, Smith P D, Chuaqui R F, Zhuang A, Goldstein S R, Weiss R A, Liotta L A. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 24.Simone N L, Bonner R F, Gillespie J W, Emmert-Buck M R, Liotta L A. Trends Genet. 1998;14:272–276. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- 25.Goldsworthy S M, Stockton P S, Trempus C S, Foley J F, Maronpot R R. Mol Carcinog. 1999;25:86–91. [PubMed] [Google Scholar]
- 26.Fend F, Emmert-Buck M R, Chuagui R, Cole K, Lee J, Liotta L A, Raffeld M. Am J Pathol. 1999;154:61–66. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin L, Thompson C A, Qian X, Kuecker S J, Kulig E, Lloyd R V. Lab Invest. 1999;79:511–512. [PubMed] [Google Scholar]
- 28.Darling T N, Yee C, Bauer J W, Hintner H, Yancey K B. J Clin Invest. 1999;103:1371–1377. doi: 10.1172/JCI4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peifer M, Polakis P. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell P, Tollervey D. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 31.Wang E, Miller L D, Ohnmacht G A, Liu E T, Marincola F M. Nat Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]