Abstract
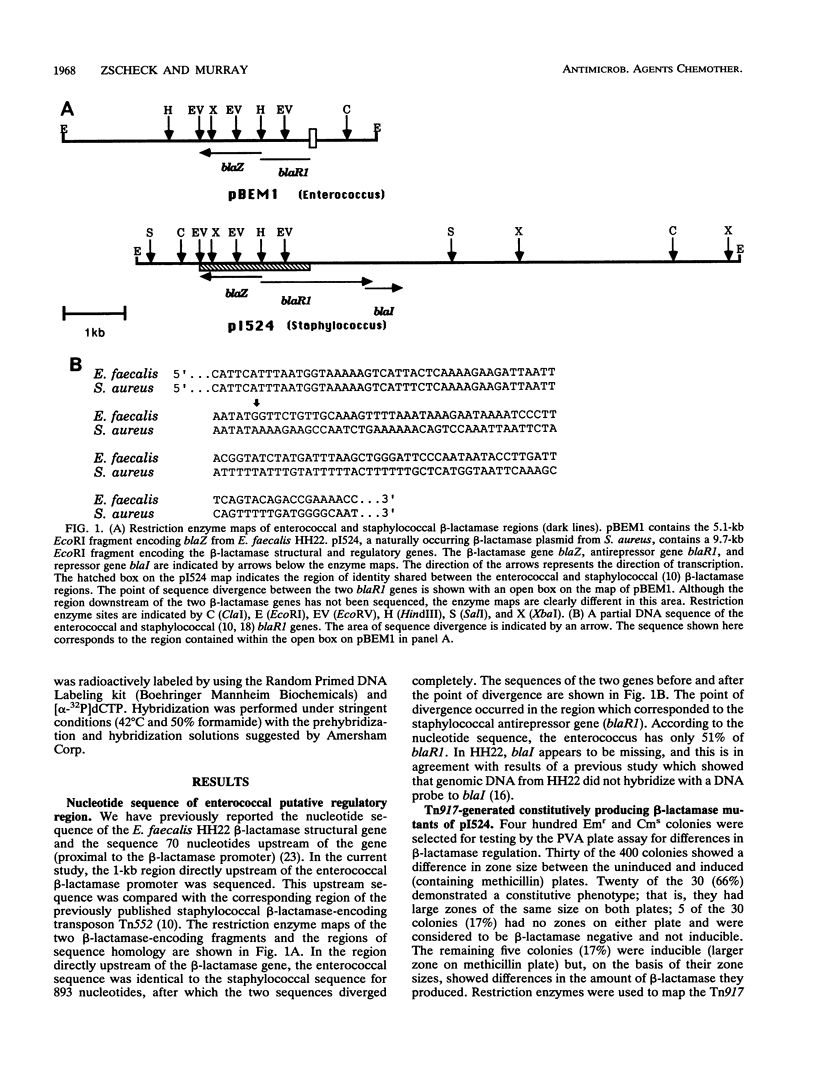
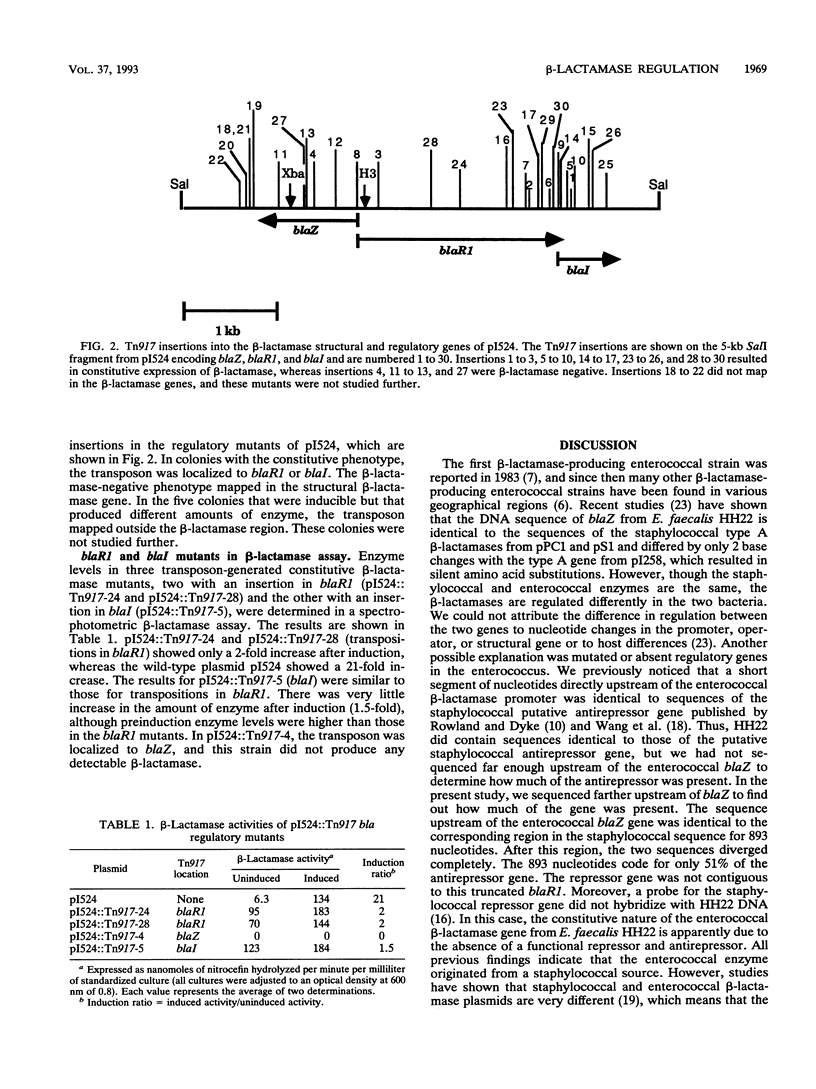
In enterococci, the structural gene for beta-lactamase (blaZ) is identical to blaZ from Staphylococcus aureus. However, in the enterococci studied to date, beta-lactamase is produced constitutively, whereas in staphylococci it is often inducible. Recent reports have revealed the presence of two adjacent genes upstream of the staphylococcal blaZ thought to be the antirepressor (blaR1) and repressor (blaI) genes. In the present study, beta-lactamase expression mutants of the staphylococcal beta-lactamase plasmid pI524 were generated by transposon mutagenesis with the transposon Tn917. Tn917 insertions upstream of blaZ in either blaR1 or blaI resulted in constitutive beta-lactamase production, indicating that the repressor function is lost with insertion of Tn917 into either gene. This finding supports the concept that the staphylococcal beta-lactamase regulatory genes are encoded on a polycistronic mRNA. The corresponding region upstream of the enterococcal blaZ from Enterococcus faecalis HH22 was sequenced and compared with the staphylococcal blaR1 sequence. The two sequences were identical for 893 nucleotides, and then the sequences diverged completely. Therefore, in strain HH22, only 51% of the putative antirepressor gene is present and the repressor gene is also absent. In conclusion, constitutive beta-lactamase production in HH22 appears to be due to a lack of the regulatory genes blaR1 and blaI which regulate expression of blaZ in staphylococci.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Goering R. V., Ruff E. A. Comparative analysis of conjugative plasmids mediating gentamicin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1983 Sep;24(3):450–452. doi: 10.1128/aac.24.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno T., Imanaka T., Aiba S. Nucleotide sequence of the penicillinase repressor gene penI of Bacillus licheniformis and regulation of penP and penI by the repressor. J Bacteriol. 1986 Dec;168(3):1128–1132. doi: 10.1128/jb.168.3.1128-1132.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Himeno T., Aiba S. Cloning and nucleotide sequence of the penicillinase antirepressor gene penJ of Bacillus licheniformis. J Bacteriol. 1987 Sep;169(9):3867–3872. doi: 10.1128/jb.169.9.3867-3872.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Zhu Y. F., Nicholls N. J., Lampen J. O. A second regulatory gene, blaR1, encoding a potential penicillin-binding protein required for induction of beta-lactamase in Bacillus licheniformis. J Bacteriol. 1987 Sep;169(9):3873–3878. doi: 10.1128/jb.169.9.3873-3878.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Novick R. P. Physical mapping of Staphylococcus aureus penicillinase plasmid pI524: characterization of an invertible region. Mol Gen Genet. 1979 Aug;175(1):19–30. doi: 10.1007/BF00267851. [DOI] [PubMed] [Google Scholar]
- Murray B. E., Mederski-Samaroj B. Transferable beta-lactamase. A new mechanism for in vitro penicillin resistance in Streptococcus faecalis. J Clin Invest. 1983 Sep;72(3):1168–1171. doi: 10.1172/JCI111042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E. The life and times of the Enterococcus. Clin Microbiol Rev. 1990 Jan;3(1):46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Murphy E., Gryczan T. J., Baron E., Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979 Jan;2(1):109–129. doi: 10.1016/0147-619x(79)90010-6. [DOI] [PubMed] [Google Scholar]
- Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967 Sep;33(1):155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- Rowland S. J., Dyke K. G. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990 Jun;4(6):961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- Salerno A. J., Lampen J. O. Differential transcription of the bla regulatory region during induction of beta-lactamase in Bacillus licheniformis. FEBS Lett. 1988 Jan 18;227(1):61–65. doi: 10.1016/0014-5793(88)81414-5. [DOI] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt D. J., Collins J. F. Analysis by transformation of the penicillinase system in Bacillus licheniformis. J Gen Microbiol. 1973 May;76(1):217–230. doi: 10.1099/00221287-76-1-217. [DOI] [PubMed] [Google Scholar]
- Smith M. C., Murray B. E. Sequence analysis of the beta-lactamase repressor from Staphylococcus aureus and hybridization studies with two beta-lactamase-producing isolates of Enterococcus faecalis. Antimicrob Agents Chemother. 1992 Oct;36(10):2265–2269. doi: 10.1128/aac.36.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. D., Jr, Archer G. L. Identification and cloning of the conjugative transfer region of Staphylococcus aureus plasmid pGO1. J Bacteriol. 1989 Feb;171(2):684–691. doi: 10.1128/jb.171.2.684-691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Z., Projan S. J., Novick R. P. Nucleotide sequence of beta-lactamase regulatory genes from staphylococcal plasmid pI258. Nucleic Acids Res. 1991 Jul 25;19(14):4000–4000. doi: 10.1093/nar/19.14.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanger A. R., Murray B. E. Comparison of enterococcal and staphylococcal beta-lactamase plasmids. J Infect Dis. 1990 Jan;161(1):54–58. doi: 10.1093/infdis/161.1.54. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zhu Y. F., Curran I. H., Joris B., Ghuysen J. M., Lampen J. O. Identification of BlaR, the signal transducer for beta-lactamase production in Bacillus licheniformis, as a penicillin-binding protein with strong homology to the OXA-2 beta-lactamase (class D) of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):1137–1141. doi: 10.1128/jb.172.2.1137-1141.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zscheck K. K., Murray B. E. Nucleotide sequence of the beta-lactamase gene from Enterococcus faecalis HH22 and its similarity to staphylococcal beta-lactamase genes. Antimicrob Agents Chemother. 1991 Sep;35(9):1736–1740. doi: 10.1128/aac.35.9.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]


