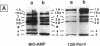Abstract
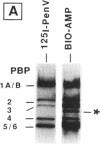
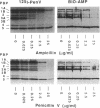
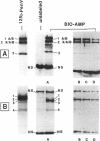
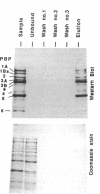
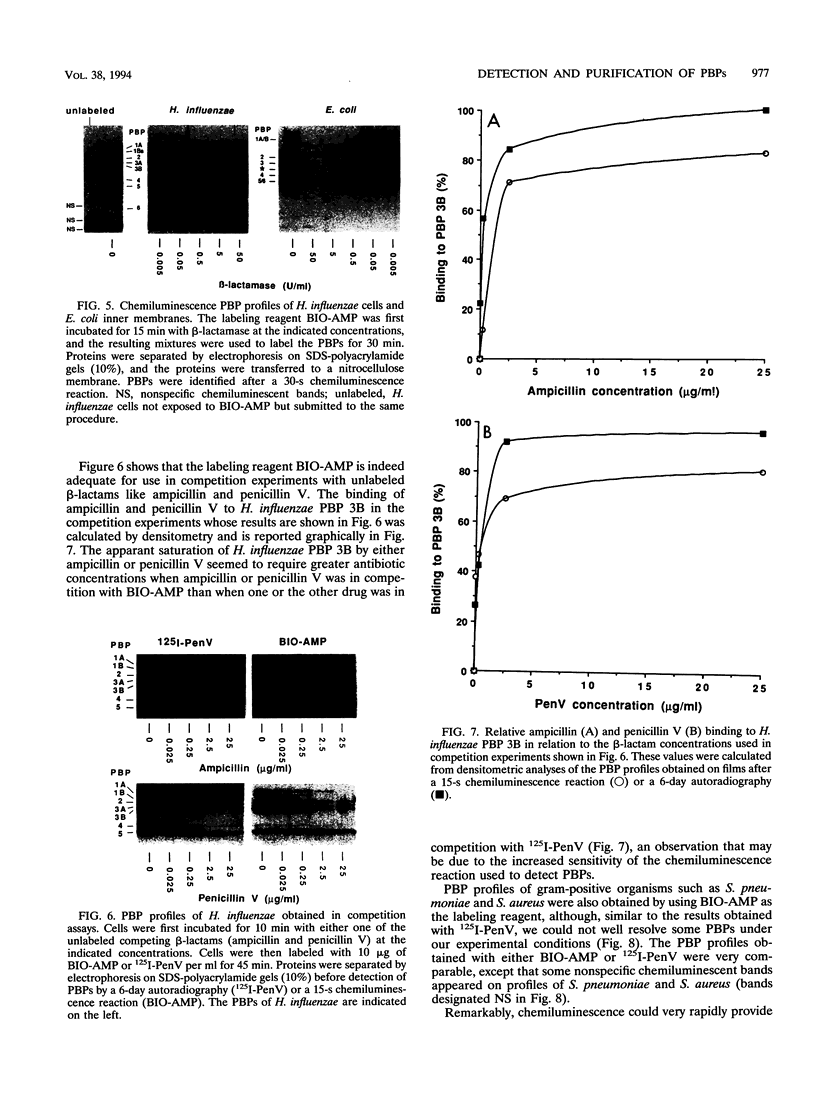
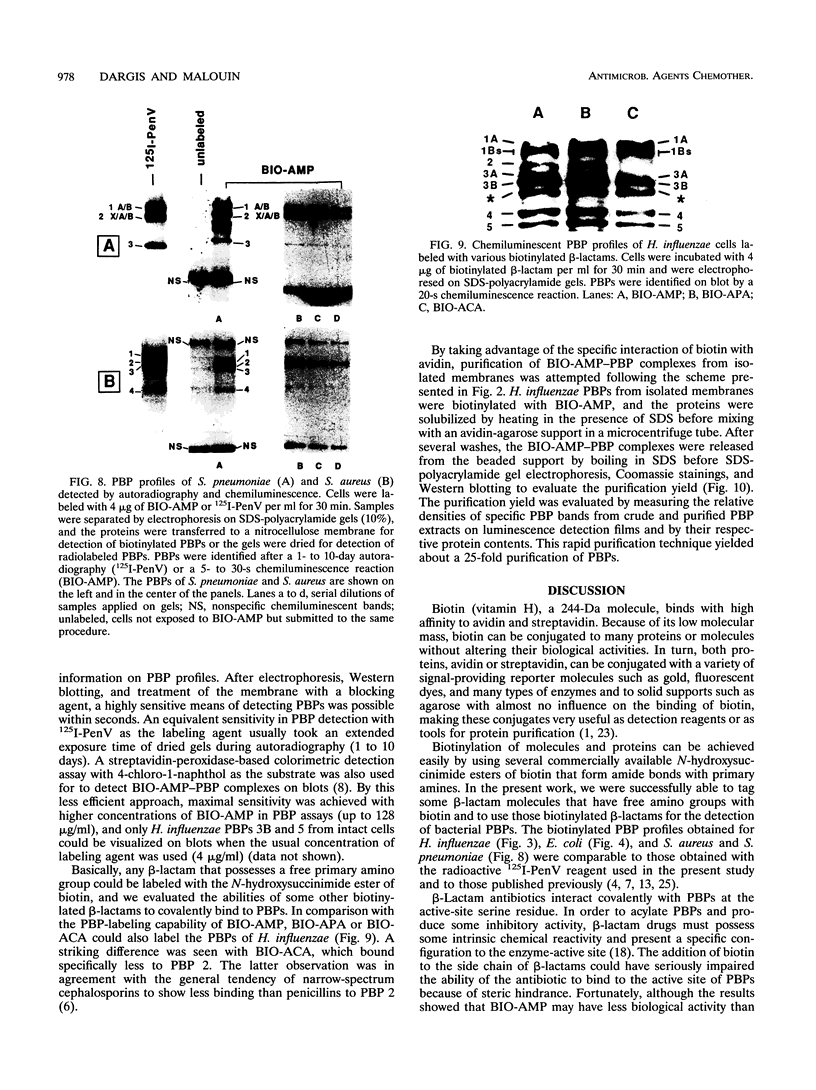
A new reagent for the detection of penicillin-binding proteins (PBPs) was developed. An N-hydroxysuccinimide ester of biotin was used to tag beta-lactam antibiotics with free side chain amino groups such as ampicillin (BIO-AMP), 6-aminopenicillanic acid (BIO-APA), and 7-aminocephalosporanic acid (BIO-ACA). Bacterial PBPs from cells or isolated cytoplasmic membranes of Escherichia coli, Haemophilus influenzae, Staphylococcus aureus, and Streptococcus pneumoniae were labeled with BIO-AMP, subjected to electrophoresis on sodium dodecyl sulfate (SDS)-polyacrylamide gels, and transferred onto nitrocellulose membranes. Electrophoretic PBP profiles were detected on blots, using colorimetric or chemiluminescence systems, on the basis of the interaction of BIO-AMP-PBP complexes and a streptavidin-peroxidase conjugate. The chemiluminescent reaction permitted a high sensitivity of detection, and PBP profiles could be determined within seconds. All PBP profiles were similar to those obtained with a traditional PBP labeling technique with 125I-labeled penicillin V, except that an additional unidentified PBP (approximately 55,000 Da) was labeled with BIO-AMP in E. coli and H. influenzae. Differences in the intensities of labeling for specific PBPs were observed between the chemiluminescent and radioactive labeling agents and were attributed to the differences in their affinities for PBPs. Similarly, BIO-AMP, BIO-APA, and BIO-ACA produced different PBP profiles. We also investigated the use of BIO-AMP in PBP purification. BIO-AMP-PBP complexes from a mixture of H. influenzae proteins were allowed to bind to avidin immobilized on an agarose support in a microcentrifuge tube. After several washes in the presence of salts, PBPs were eluted by boiling and treatment with SDS. The eluted proteins were separated by electrophoresis on SDS-polyacrylamide gels, and biotinylated proteins were identified on blots by a chemiluminescence reaction. Biotinylation of beta-lactams is rapid, safe, and inexpensive. Our results demonstrate the feasibility of using biotinylated beta-lactams as nonradioactive reagents for the study of PBPs and for the purification of these proteins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayer E. A., Wilchek M. The use of the avidin-biotin complex as a tool in molecular biology. Methods Biochem Anal. 1980;26:1–45. doi: 10.1002/9780470110461.ch1. [DOI] [PubMed] [Google Scholar]
- Chambers H. F. Methicillin-resistant staphylococci. Clin Microbiol Rev. 1988 Apr;1(2):173–186. doi: 10.1128/cmr.1.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clairoux N., Picard M., Brochu A., Rousseau N., Gourde P., Beauchamp D., Parr T. R., Jr, Bergeron M. G., Malouin F. Molecular basis of the non-beta-lactamase-mediated resistance to beta-lactam antibiotics in strains of Haemophilus influenzae isolated in Canada. Antimicrob Agents Chemother. 1992 Jul;36(7):1504–1513. doi: 10.1128/aac.36.7.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis N. A., Orr D., Ross G. W., Boulton M. G. Affinities of penicillins and cephalosporins for the penicillin-binding proteins of Escherichia coli K-12 and their antibacterial activity. Antimicrob Agents Chemother. 1979 Nov;16(5):533–539. doi: 10.1128/aac.16.5.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargis M., Gourde P., Beauchamp D., Foiry B., Jacques M., Malouin F. Modification in penicillin-binding proteins during in vivo development of genetic competence of Haemophilus influenzae is associated with a rapid change in the physiological state of cells. Infect Immun. 1992 Oct;60(10):4024–4031. doi: 10.1128/iai.60.10.4024-4031.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galleni M., Lakaye B., Lepage S., Jamin M., Thamm I., Joris B., Frère J. M. A new, highly sensitive method for the detection and quantification of penicillin-binding proteins. Biochem J. 1993 Apr 1;291(Pt 1):19–21. doi: 10.1042/bj2910019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuysen J. M. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- Hakenbeck R., Briese T., Ellerbrok H. Antibodies against the benzylpenicilloyl moiety as a probe for penicillin-binding proteins. Eur J Biochem. 1986 May 15;157(1):101–106. doi: 10.1111/j.1432-1033.1986.tb09644.x. [DOI] [PubMed] [Google Scholar]
- Klugman K. P. Pneumococcal resistance to antibiotics. Clin Microbiol Rev. 1990 Apr;3(2):171–196. doi: 10.1128/cmr.3.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G., Spratt B. G., Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991 Aug;5(8):1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- Malouin F., Bryan L. E. Modification of penicillin-binding proteins as mechanisms of beta-lactam resistance. Antimicrob Agents Chemother. 1986 Jul;30(1):1–5. doi: 10.1128/aac.30.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M., Malouin F. Molecular basis of the efficacy of cefaclor against Haemophilus influenzae. Antimicrob Agents Chemother. 1992 Nov;36(11):2569–2572. doi: 10.1128/aac.36.11.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston D. A., Wu C. Y., Blaszczak L. C., Seitz D. E., Halligan N. G. Biological characterization of a new radioactive labeling reagent for bacterial penicillin-binding proteins. Antimicrob Agents Chemother. 1990 May;34(5):718–721. doi: 10.1128/aac.34.5.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schryvers A. B. Identification of the transferrin- and lactoferrin-binding proteins in Haemophilus influenzae. J Med Microbiol. 1989 Jun;29(2):121–130. doi: 10.1099/00222615-29-2-121. [DOI] [PubMed] [Google Scholar]
- Spratt B. G., Cromie K. D. Penicillin-binding proteins of gram-negative bacteria. Rev Infect Dis. 1988 Jul-Aug;10(4):699–711. doi: 10.1093/clinids/10.4.699. [DOI] [PubMed] [Google Scholar]
- Spratt B. G. Properties of the penicillin-binding proteins of Escherichia coli K12,. Eur J Biochem. 1977 Jan;72(2):341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- Tipper D. J., Strominger J. L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Nakagami S., Nitta A., Yamane A., Kawakami S., Sugiura M., Konno M. Rapid detection of the mecA gene in methicillin-resistant staphylococci by enzymatic detection of polymerase chain reaction products. J Clin Microbiol. 1992 Jul;30(7):1728–1733. doi: 10.1128/jcm.30.7.1728-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]